Arecoline-Induced Hepatotoxicity in Rats: Screening of Abnormal Metabolic Markers and Potential Mechanisms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Animals
2.1.1. Instrument
2.1.2. Reference Standards and Reagents
2.1.3. Animal Treatment
2.2. Manifestation of Hepatotoxicity Induced by Arecoline
2.2.1. Animal Status (Behavioral) Analysis
2.2.2. Serum Biochemical Indexes Detection
2.2.3. Histopathological Evaluation
2.3. Metabolomics Study on Hepatotoxicity Induced by Arecoline
2.3.1. Sample Preparation
Serum Sample Preparation
QC Sample Preparation
2.3.2. Analysis Conditions
Chromatographic Conditions
Mass Spectrometry Conditions
2.3.3. Methodological Investigation
Instrument Precision Test
Method Reproducibility Test
Sample Stability Test
2.3.4. Data Processing
2.4. Network Toxicology Study on Arecoline-Induced Hepatotoxicity
2.4.1. Acquisition of Arecoline Targets
2.4.2. Acquisition of Hepatotoxicity Targets
2.4.3. Construction of Common Targets
2.4.4. Protein–Protein Interaction (PPI) Network Construction
2.4.5. GO Bioprocess Analysis and KEGG Enrichment Analysis
3. Results
3.1. Manifestations of Hepatotoxicity Induced by Arecoline
3.1.1. Animal Status (Behavioral) Analysis
3.1.2. Analysis of Serum Biochemical Indexes
3.1.3. Histopathological Evaluation
3.2. Metabolomics Study on Arecoline-Induced Hepatotoxicity
3.2.1. Methodological Investigation
3.2.2. Metabolic Profiling Analysis
3.2.3. Identification of Differential Metabolites
3.2.4. Metabolic Pathway Analysis
3.3. Network Toxicology Study on Arecoline-Induced Hepatotoxicity
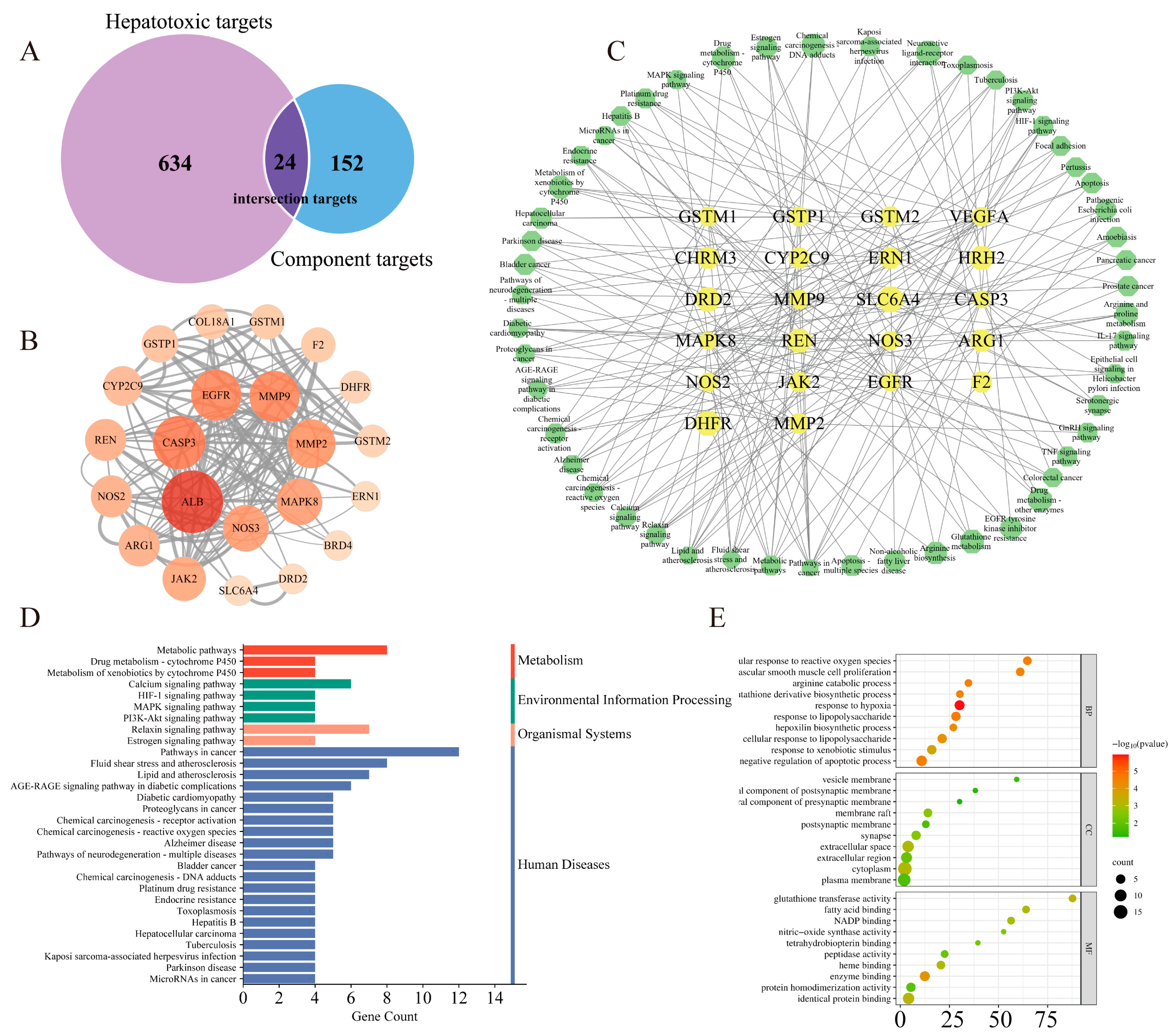
3.3.1. Potential Targets Prediction of Arecoline
3.3.2. Protein–Protein Interaction (PPI) Network Construction
3.3.3. GO Bioprocess Analysis and KEGG Enrichment Analysis
4. Discussion
4.1. The Manifestation of Arecoline-Induced Hepatotoxicity in Rats
4.2. Network Toxicology Study on Arecoline-Induced Hepatotoxicity
4.3. Limitations and Prospects of the Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peng, W.; Liu, Y.J.; Wu, N.; Sun, T.; He, X.Y.; Gao, Y.X.; Wu, C.J. Areca catechu L. (Arecaceae): A review of its traditional uses, botany, phytochemistry, pharmacology and toxicology. J. Ethnopharmacol. 2015, 164, 340–356. [Google Scholar] [CrossRef]
- Gupta, A.K.; Tulsyan, S.; Thakur, N.; Sharma, V.; Sinha, D.N.; Mehrotra, R. Chemistry, metabolism and pharmacology of carcinogenic alkaloids present in areca nut and factors affecting their concentration. Regul. Toxicol. Pharmacol. 2020, 110, 104548. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Peng, W.; Hu, M.B.; Xu, M.; Wu, C.J. The pharmacology, toxicology and potential applications of arecoline: A review. Pharm. Biol. 2016, 54, 2753–2760. [Google Scholar] [CrossRef] [PubMed]
- Myers, A.L. Metabolism of the areca alkaloids - toxic and psychoactive constituents of the areca (betel) nut. Drug Metab. Rev. 2022, 54, 343–360. [Google Scholar] [CrossRef] [PubMed]
- Al-Tayar, B.A.; Ahmad, A.; Yusoff, M.E.; Abdullah, S.F.; Mohamad, N.K.; Md Hashim, S.N.; Kishida, S.; Kishida, M.; Nakamura, N.; Kibe, T.; et al. Cytotoxic Effects of Betel Quid and Areca Nut Aqueous Extracts on Mouse Fibroblast, Human Mouth-Ordinary-Epithelium 1 and Human Oral Squamous Cell Carcinoma Cell Lines. Asian Pac. J. Cancer Prev. 2020, 21, 1005–1009. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Yang, D.; Liang, Y.; Gao, W.; Ren, Z.; Zeng, W.; Wang, B.; Han, J.; Guo, D. Alkaloids from areca (betel) nuts and their effects on human sperm motility in vitro. J. Food Sci. 2012, 77, T70–T78. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, Z.Y.; Huang, L.; Yu, T.L.; Wan, S.Q.; Song, J.; Zhang, B.L.; Hu, M. Do betel quid and areca nut chewing deteriorate prognosis of oral cancer? A systematic review, meta-analysis, and research agenda. Oral. Dis. 2021, 27, 1366–1375. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, N.G.; Ramos, D.L.; Dinis-Oliveira, R.J. Genetic toxicology and toxicokinetics of arecoline and related areca nut compounds: An updated review. Arch. Toxicol. 2021, 95, 375–393. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Wang, C.; Jia, Z.; Xiong, H.; Xue, X.; Liu, M.; Xu, X.; Qu, W.; Li, X. UPLC-HDMS-based on serum metabolomics reveals the toxicity of arecae semen. J. Ethnopharmacol. 2020, 247, 112223. [Google Scholar] [CrossRef]
- Choudhury, Y.; Sharan, R.N. Ultrastructural alterations in liver of mice exposed chronically and transgenerationally to aqueous extract of betel nut: Implications in betel nut-induced carcinogenesis. Microsc. Res. Tech. 2010, 73, 530–539. [Google Scholar] [CrossRef]
- Wu, G.H.; Boucher, B.J.; Chiu, Y.H.; Liao, C.S.; Chen, T.H. Impact of chewing betel-nut (Areca catechu) on liver cirrhosis and hepatocellular carcinoma: A population-based study from an area with a high prevalence of hepatitis B and C infections. Public. Health Nutr. 2009, 12, 129–135. [Google Scholar] [CrossRef]
- Chou, W.W.; Guh, J.Y.; Tsai, J.F.; Hwang, C.C.; Chen, H.C.; Huang, J.S.; Yang, Y.L.; Hung, W.C.; Chuang, L.Y. Arecoline-induced growth arrest and p21WAF1 expression are dependent on p53 in rat hepatocytes. Toxicology 2008, 243, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chacko, S.; Haseeb, Y.B.; Haseeb, S. Metabolomics Work Flow and Analytics in Systems Biology. Curr. Mol. Med. 2022, 22, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Yan, G.L.; Han, Y.; Sun, H.; Zhang, A.H.; Li, X.N.; Wang, X.J. UPLC-Q-TOF/MS-based metabolomic studies on the toxicity mechanisms of traditional Chinese medicine Chuanwu and the detoxification mechanisms of Gancao, Baishao, and Ganjiang. Chin. J. Nat. Med. 2015, 13, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Xie, C.; Wang, K.; Takahashi, S.; Krausz, K.W.; Lu, D.; Wang, Q.; Luo, Y.; Gong, X.; Mu, X.; et al. Comprehensive analysis of transcriptomics and metabolomics to understand triptolide-induced liver injury in mice. Toxicol. Lett. 2020, 333, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, T.; Strigun, A.; Verlohner, A.; Huener, H.A.; Peter, E.; Herold, M.; Bordag, N.; Mellert, W.; Walk, T.; Spitzer, M.; et al. Prediction of liver toxicity and mode of action using metabolomics in vitro in HepG2 cells. Arch. Toxicol. 2018, 92, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.Q.; Chen, H.; Chen, L.; Tang, D.D.; Miao, H.; Zhao, Y.Y. Metabolomic application in toxicity evaluation and toxicological biomarker identification of natural product. Chem. Biol. Interact. 2016, 252, 114–130. [Google Scholar] [CrossRef]
- Bauermeister, A.; Mannochio-Russo, H.; Costa-Lotufo, L.V.; Jarmusch, A.K.; Dorrestein, P.C. Mass spectrometry-based metabolomics in microbiome investigations. Nat. Rev. Microbiol. 2022, 20, 143–160. [Google Scholar] [CrossRef]
- Kim, S.J.; Song, H.E.; Lee, H.Y.; Yoo, H.J. Mass Spectrometry-based Metabolomics in Translational Research. Adv. Exp. Med. Biol. 2021, 1310, 509–531. [Google Scholar] [CrossRef]
- Miao, Y.J.; Shi, Y.Y.; Li, F.Q.; Shan, C.X.; Chen, Y.; Chen, J.W.; Li, X. Metabolomics study on the toxicity of Annona squamosa by ultraperformance liquid-chromatography high-definition mass spectrometry coupled with pattern recognition approach and metabolic pathways analysis. J. Ethnopharmacol. 2016, 184, 187–195. [Google Scholar] [CrossRef]
- Fang, Z.Z.; Gonzalez, F.J. LC-MS-based metabolomics: An update. Arch. Toxicol. 2014, 88, 1491–1502. [Google Scholar] [CrossRef]
- Zhao, Z.; Du, J.F.; Wang, Q.L.; Qiu, F.N.; Chen, X.Y.; Liu, F.J.; Li, P.; Jiang, Y.; Li, H.J. An integrated strategy combining network toxicology and feature-based molecular networking for exploring hepatotoxic constituents and mechanism of Epimedii Folium-induced hepatotoxicity in vitro. Food Chem. Toxicol. 2023, 176, 113785. [Google Scholar] [CrossRef]
- Xiao, F.; Qiu, J.; Zhao, Y. Exploring the Potential Toxicological Mechanisms of Vine Tea on the Liver Based on Network Toxicology and Transcriptomics. Front. Pharmacol. 2022, 13, 855926. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, K.; Jiao, M.; Jiao, J.; Chen, D.; Yin, Y.; Zhang, J.; Li, F. Study on the Mechanism of Mesaconitine-Induced Hepatotoxicity in Rats Based on Metabonomics and Toxicology Network. Toxins 2022, 14, 486. [Google Scholar] [CrossRef]
- Selvan, R.S.; Selvakumaran, M.; Rao, A.R. Influence of arecoline on immune system: II. Suppression of thymus-dependent immune responses and parameter of non-specific resistance after short-term exposure. Immunopharmacol. Immunotoxicol. 1991, 13, 281–309. [Google Scholar] [CrossRef]
- Abduh, M.; Saghir, S.; Al-Gabri, N.; Ahmeda, A.; Abdelkarim, M.; Aldaqal, S.; Alshawsh, M. Interleukin-35 and Thymoquinone nanoparticle-based intervention for liver protection against paracetamol-induced liver injury in rats. Saudi J. Biol. Sci. 2023, 30, 103806. [Google Scholar] [CrossRef]
- Yang, Y.; Jin, Y.; Zhu, X.; Rao, Q.; Zhao, Z.; Yang, J. Hepatotoxicity evaluation and possible mechanisms of decabrominated diphenyl ethers (BDE-209) in broilers: Oxidative stress, inflammatory, and transcriptomics. Ecotoxicol. Environ. Saf. 2023, 264, 115460. [Google Scholar] [CrossRef]
- Mihaylova, R.; Gevrenova, R.; Stefanova, A.; Zheleva-Dimitrova, D.; Balabanova, V.; Zengin, G.; Simeonova, R.; Momekov, G. Prenanthes purpurea. The Phytochemical Profiling, In Vitro Antioxidant, and Hepatoprotective Activity of L. and Caffeoylquinic Acids in Diclofenac-Induced Hepatotoxicity on HEP-G2 Cells. Int. J. Mol. Sci. 2023, 24, 14148. [Google Scholar] [CrossRef]
- Molavinia, S.; Moosavi, M.; Hejazi, S.; Azadnasab, R.; Mansouri, E.; Khodayar, M. Metformin alleviates sodium arsenite-induced hepatotoxicity and glucose intolerance in mice by suppressing oxidative stress, inflammation, and apoptosis. J. Trace Elem. Med. Biol. Organ Soc. Miner. Trace Elem. (GMS) 2023, 80, 127299. [Google Scholar] [CrossRef]
- Jaeschke, H.; Ramachandran, A. Central mechanisms of acetaminophen hepatotoxicity: Mitochondrial dysfunction by protein adducts and oxidant stress. Drug Metab. Dispos. Biol. Fate Chem. 2023. [Google Scholar] [CrossRef]
- Xu, R.; Xu, P.; Wei, H.; Huang, Y.; Zhu, X.; Lin, C.; Yan, Z.; Xin, L.; Li, L.; Lv, W.; et al. Ticlopidine induces embryonic development toxicity and hepatotoxicity in zebrafish by upregulating the oxidative stress signaling pathway. Ecotoxicol. Environ. Saf. 2023, 262, 115283. [Google Scholar] [CrossRef]
- Demir, M.; Altinoz, E.; Koca, O.; Elbe, H.; Onal, M.; Bicer, Y.; Karayakali, M. Antioxidant and anti-inflammatory potential of crocin on the doxorubicin mediated hepatotoxicity in Wistar rats. Tissue Cell 2023, 84, 102182. [Google Scholar] [CrossRef]
- Indumathi, M.; Swetha, K.; Abhilasha, K.; Siddappa, S.; Kumar, S.; Prasad, G.; Chen, C.; Marathe, G. Selenium Ameliorates Acetaminophen-Induced Oxidative Stress via MAPK and Nrf2 Pathways in Mice. Biol. Trace Elem. Res. 2023. [Google Scholar] [CrossRef]
- El-Mowafy, A.; Katary, M.; Pye, C.; Ibrahim, A.; Elmarakby, A. Novel molecular triggers underlie valproate-induced liver injury and its alleviation by the omega-3 fatty acid DHA: Role of inflammation and apoptosis. Heliyon 2016, 2, e00130. [Google Scholar] [CrossRef]
- Fei, W.; Zhang, J.; Yu, S.; Yue, N.; Ye, D.; Zhu, Y.; Tao, R.; Chen, Y.; Chen, Y.; Li, A.; et al. Antioxidative and Energy Metabolism-Improving Effects of Maca Polysaccharide on Cyclophosphamide-Induced Hepatotoxicity Mice via Metabolomic Analysis and Keap1-Nrf2 Pathway. Nutrients 2022, 14, 4264. [Google Scholar] [CrossRef]
- Xu, F.; Liu, Y.; Zhao, H.; Yu, K.; Song, M.; Zhu, Y.; Li, Y. Aluminum chloride caused liver dysfunction and mitochondrial energy metabolism disorder in rat. J. Inorg. Biochem. 2017, 174, 55–62. [Google Scholar] [CrossRef]
- Zhang, R.; Lu, W.; Yao, Y.; Tu, L.; Yu, T.; Luan, T.; Chen, B. Metabolomics analysis of the 3D L-02 cell cultures revealing the key role of metabolism of amino acids in ameliorating hepatotoxicity of perfluorooctanoic acid. Sci. Total Environ. 2022, 806, 150438. [Google Scholar] [CrossRef]
- Hu, C.; Li, H.; Wu, L.; Ke, J.; Yu, X.; Xiong, Y.; Tang, X. Metabolic profiling of 19 amino acids in triptolide-induced liver injured rats by gas chromatography-triple quadrupole mass spectrometry. Hum. Exp. Toxicol. 2021, 40, 1685–1697. [Google Scholar] [CrossRef]
- Ji, Y.; Gao, Y.; Chen, H.; Yin, Y.; Zhang, W. Indole-3-Acetic Acid Alleviates Nonalcoholic Fatty Liver Disease in Mice via Attenuation of Hepatic Lipogenesis, and Oxidative and Inflammatory Stress. Nutrients 2019, 11, 2062. [Google Scholar] [CrossRef]
- Abulikemu, A.; Zhao, X.; Xu, H.; Li, Y.; Ma, R.; Yao, Q.; Wang, J.; Sun, Z.; Li, Y.; Guo, C. Silica nanoparticles aggravated the metabolic associated fatty liver disease through disturbed amino acid and lipid metabolisms-mediated oxidative stress. Redox Biol. 2023, 59, 102569. [Google Scholar] [CrossRef]
- Teplova, V.V.; Kruglov, A.G.; Kovalyov, L.I.; Nikiforova, A.B.; Fedotcheva, N.I.; Lemasters, J.J. Glutamate contributes to alcohol hepatotoxicity by enhancing oxidative stress in mitochondria. J. Bioenerg. Biomembr. 2017, 49, 253–264. [Google Scholar] [CrossRef]
- Katyare, S.S.; Satav, J.G. Impaired mitochondrial oxidative energy metabolism following paracetamol-induced hepatotoxicity in the rat. Br. J. Pharmacol. 1989, 96, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Yiew, N.; Vazquez, J.; Martino, M.; Kennon-McGill, S.; Price, J.; Allard, F.; Yee, E.; Layman, A.; James, L.; McCommis, K.; et al. Hepatic pyruvate and alanine metabolism are critical and complementary for maintenance of antioxidant capacity and resistance to oxidative insult. Mol. Metab. 2023, 77, 101808. [Google Scholar] [CrossRef] [PubMed]
- Leo, M.A.; Lieber, C.S. Alcohol, vitamin A, and beta-carotene: Adverse interactions, including hepatotoxicity and carcinogenicity. Am. J. Clin. Nutr. 1999, 69, 1071–1085. [Google Scholar] [CrossRef] [PubMed]
- Bray, B.J.; Rosengren, R.J. Retinol potentiates acetaminophen-induced hepatotoxicity in the mouse: Mechanistic studies. Toxicol. Appl. Pharmacol. 2001, 173, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yi, S.; Yu, H.; Zhang, T.; Dong, F.; Zhu, L. Underlying Mechanisms for the Sex-and Chemical-Specific Hepatotoxicity of Perfluoroalkyl Phosphinic Acids in Common Carp (Cyprinus carpio). Environ. Sci. Technol. 2023, 57, 14515–14525. [Google Scholar] [CrossRef] [PubMed]
- Haroun, A.; El-Sayed, W.; Hassan, R. Quercetin and L-Arginine Ameliorated the Deleterious Effects of Copper Oxide Nanoparticles on the Liver of Mice Through Anti-inflammatory and Anti-apoptotic Pathways. Biol. Trace Elem. Res. 2023. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Li, H.; Wang, H.; Wang, R.; Song, W.; Li, D.; Wei, C.; Guo, Y.; He, X.; Deng, Y. Nickel Nanoparticles Induced Hepatotoxicity in Mice via Lipid-Metabolism-Dysfunction-Regulated Inflammatory Injury. Molecules 2023, 28, 5757. [Google Scholar] [CrossRef]
- Hong, Y.; Kim, G.; Park, Y.; Jo, H.; Nam, M.; Kim, D.; Cho, H.; Shim, H.; Jin, J.; Rho, H.; et al. Suaeda glauca Attenuates Liver Fibrosis in Mice by Inhibiting TGFβ1-Smad2/3 Signaling in Hepatic Stellate Cells. Nutrients 2023, 15, 3740. [Google Scholar] [CrossRef]
- Alqahtani, L.; Abd-Elhakim, Y.; Mohamed, A.; Khalifa, N.; Khamis, T.; Alotaibi, B.; Alosaimi, M.; El-Kholy, S.; Abuzahrah, S.; ElAshmouny, N.; et al. Curcumin-loaded chitosan nanoparticles alleviate fenpropathrin-induced hepatotoxicity by regulating lipogenesis and pyroptosis in rats. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2023, 180, 114036. [Google Scholar] [CrossRef]
- Zhang, Y.; Cen, J.; Jia, Z.; Hsiao, C.D.; Xia, Q.; Wang, X.; Chen, X.; Wang, R.; Jiang, Z.; Zhang, L.; et al. Hepatotoxicity Induced by Isoniazid-Lipopolysaccharide through Endoplasmic Reticulum Stress, Autophagy, and Apoptosis Pathways in Zebrafish. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef]
- Sui, Y.; Lu, Y.; Zuo, S.; Wang, H.; Bian, X.; Chen, G.; Huang, S.; Dai, H.; Liu, F.; Dong, H. Aflatoxin B(1) Exposure in Sheep: Insights into Hepatotoxicity Based on Oxidative Stress, Inflammatory Injury, Apoptosis, and Gut Microbiota Analysis. Toxins 2022, 14, 840. [Google Scholar] [CrossRef]
- Temel, Y.; Kucukler, S.; Yıldırım, S.; Caglayan, C.; Kandemir, F.M. Protective effect of chrysin on cyclophosphamide-induced hepatotoxicity and nephrotoxicity via the inhibition of oxidative stress, inflammation, and apoptosis. Naunyn Schmiedebergs Arch. Pharmacol. 2020, 393, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Liu, Q.; Liang, Q.; Gao, S.; Zhuang, K.; Zhang, Y.; Zhang, H.; Liu, K.; She, G.; Xia, Q. Toosendanin triggered hepatotoxicity in zebrafish via inflammation, autophagy, and apoptosis pathways. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2021, 250, 109171. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Chen, S.; Seo, J.E.; Guo, X.; Li, D.; Ning, B.; Guo, L. Mitochondrial dysfunction and apoptosis underlie the hepatotoxicity of perhexiline. Toxicol. Vitr. 2020, 69, 104987. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Gao, M.; Yang, N.; Ai, L.; Guo, L.; Xue, X.; Sheng, Z. Trimethyltin chloride induces apoptosis and DNA damage via ROS/NF-κB in grass carp liver cells causing immune dysfunction. Fish. Shellfish Immunol. 2023, 142, 109082. [Google Scholar] [CrossRef]



| Grouping | Number | Drug | Dose | Administration Mode | Administration Time |
|---|---|---|---|---|---|
| NS | 10 | 0.9% NaCl | 8.0 mL/kg/d | p.o., continuous administration | 7 Days |
| Arecoline hydrobromide—low | 10 | Arecoline hydrobromide | 500 mg/kg/d | p.o., continuous administration | 7 Days |
| Arecoline hydrobromide—high | 10 | Arecoline hydrobromide | 1000 mg/kg/d | p.o., continuous administration | 7 Days |
| NO. | RT/min | Metabolites | Obsed m/z | Calcd m/z | Error ppm | Formula | HMDB | Content Variance | MS/MS |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.705 | D-Lysine | 147.11280 | 147.11299 | 1.29 | C6H14N2O2 | HMDB0003405 | ↑** | 84.08101, 130.08664, 157.62578, 86.99315 |
| 2 | 0.735 | N4-Acetylaminobutanal | 130.08625 | 130.08662 | 2.84 | C6H11NO2 | HMDB0004226 | ↑** | 84.08099, 130.08662, 56.04992, 72.93741, 100.02428, 112.07578 |
| 3 | 0.819 | L-Arginine | 175.11895 | 175.11945 | 2.86 | C6H14N4O2 | HMDB0000517 | ↓** | 70.06542, 60.05606, 116.07096, 175.11945 |
| 4 | 0.847 | D-Valine | 118.08626 | 118.08651 | 2.12 | C5H11NO2 | HMDB0250806 | ↑** | 118.08651, 72.08102, 59.07337, 55.05469, 109.05436, 91.05466 |
| 5 | 0.859 | L-HypoglycinA | 142.08626 | 142.08662 | 2.53 | C7H11NO2 | HMDB0029427 | ↑** | 142.08662, 69.04483, 53.03899, 81.03365, 99.04432 |
| 6 | 0.876 | Prolinebetaine | 144.10191 | 144.10190 | −0.07 | C7H13NO2 | HMDB0004827 | ↑** | 60.93843, 59.93072, 98.06031, 99.04432, 116.97234 |
| 7 | 0.932 | Acetyl-D-carnitine | 204.12303 | 204.12318 | 0.73 | C9H17NO4 | HMDB0240771 | ↓* | 85.02861, 145.04994, 204.12344, 60.08119 |
| 8 | 0.988 | N-Methylnicotinamide | 137.07094 | 137.07112 | 1.31 | C7H8N2O | HMDB0003152 | ↑** | 137.07137, 108.04454, 81.07011, 80.04967, 116.96632 |
| 9 | 1.742 | Propionylcarnitine | 218.13868 | 218.13911 | 1.97 | C10H19NO4 | HMDB0000824 | ↑** | 85.02863, 218.13911, 159.06560, 177.87805 |
| 10 | 1.954 | Indoleacetaldehyde | 160.07569 | 160.07610 | 2.56 | C10H9NO | HMDB0001190 | ↓** | 118.94285, 113.96400, 160.07616, 132.08118 |
| 11 | 2.321 | Pantothenicacid | 220.11795 | 220.11812 | 0.77 | C9H17NO5 | HMDB0000210 | ↓* | 90.05518, 132.10226, 202.10800, 116.03451, 184.09737, 160.09740 |
| 12 | 2.968 | Isobutyryl-L-carnitine | 232.15433 | 232.15439 | 0.26 | C11H21NO4 | HMDB0000736 | ↑* | 152.03506, 231.99182, 108.04536, 187.81517, 79.95724, 87.92511 |
| 13 | 3.183 | (±)-Tryptophan | 227.07910 | 227.07864 | −2.03 | C11H12N2O2 | HMDB0030396 | ↓* | 146.06044, 142.06558, 118.06540, 159.09207, 188.07112, 205.09775 |
| 14 | 3.210 | Indole-3-carboxaldehyde | 146.06004 | 146.06047 | 2.94 | C9H7NO | HMDB0029737 | ↓* | 65.32993, 91.05457, 118.06541, 99.04428, 126.09157, 146.06047 |
| 15 | 3.229 | 6-Chloro-N-(1-methylethyl)-1,3,5-triazine-2,4-diamine | 188.06975 | 188.07066 | 4.84 | C6H10ClN5 | HMDB0033249 | ↓* | 146.06049, 188.07111, 118.06544, 170.06049, 181.06914 |
| 16 | 3.638 | Nopalinicacid | 263.12376 | 263.12411 | 1.33 | C10H18N2O6 | HMDB0029437 | ↑** | 70.06539, 172.07944, 263.12411, 132.10220 |
| 17 | 4.003 | 2-Methylbutyroylcarnitine | 246.16998 | 246.17030 | 1.30 | C12H23NO4 | HMDB0000378 | ↑** | 85.02861, 187.09685, 229.28825, 246.17030 |
| 18 | 4.156 | 5-Hydroxy-6-methoxyindoleglucuronide | 340.10269 | 340.10281 | 0.35 | C15H17NO8 | HMDB0010363 | ↓** | 164.07094, 113.02357, 230.08168, 85.02859, 267.29990, 340.10281 |
| 19 | 5.467 | Vanillylamine | 154.08626 | 154.08661 | 2.27 | C8H11NO2 | HMDB0012309 | ↑** | 131.97456, 113.96394, 122.06030, 53.03911, 94.06535 |
| 20 | 9.444 | CavipetinC | 389.26863 | 389.26941 | 2.00 | C24H36O4 | HMDB0030366 | ↓** | 353.24783, 335.23724, 317.22671, 371.25845, 389.26941, 227.14314, |
| 21 | 9.507 | 7-Ketodeoxycholicacid | 407.27920 | 407.27884 | −0.88 | C24H38O5 | HMDB0000391 | ↓** | 81.07018, 353.24780, 307.24228, 243.13841 |
| 22 | 13.364 | 1-Lyso-2-arachidonoyl-phosphatidate | 459.25062 | 459.25089 | 0.59 | C23H39O7P | HMDB0012496 | ↓** | 287.23724, 269.22675, 361.27374, 457.23322, 189.16426, 98.98443 |
| 23 | 14.172 | LysoPC(16:0/0:0) | 496.33976 | 496.34085 | 2.20 | C24H50NO7P | HMDB0010382 | ↓* | 184.07381, 104.10723, 86.09664, 313.27386, 60.08119 |
| 24 | 14.692 | VitaminA2 | 285.22129 | 285.22162 | 1.16 | C20H28O | HMDB0013117 | ↓** | 241.19538, 267.21115, 129.07010, 131.08566 |
| 25 | 18.113 | Docosahexaenoicacid | 329.24750 | 329.24774 | 0.73 | C22H32O2 | HMDB0002183 | ↓** | 67.05453, 81.07032, 133.10173, 311.23730, 163.14865, 184.88966 |
| 26 | 24.445 | Imidazoleacetol-phosphate | 111.01973 | 111.02024 | 4.59 | C6H9N2O5P | HMDB0012236 | ↓** | 88.500332, 97.00977, 100.01137, 115.96432, 133.97482, 126.06840, 138.98064, 111.09196 |
| NO. | Pathway Name | Metabolite | p | −Log(p) | Holm p | FDR | Impact |
|---|---|---|---|---|---|---|---|
| 1 | Arginine and proline metabolism | L-Arginine; N4-Acetylaminobutanal | 0.018907 | 1.7234 | 1.0 | 1.0 | 0.06998 |
| 2 | Arginine biosynthesis | L-Arginine | 0.07861 | 1.1045 | 1.0 | 1.0 | 0.07614 |
| 3 | Retinol metabolism | Vitamin A2 | 0.094722 | 1.0235 | 1.0 | 1.0 | 0 |
| 4 | Pentose and glucuronate interconversions | 3-Methoxy-4-hydroxyphenylglycol glucuronide | 0.10004 | 0.99984 | 1.0 | 1.0 | 0.14062 |
| 5 | Pantothenate and CoA biosynthesis | Pantothenic acid | 0.10532 | 0.97747 | 1.0 | 1.0 | 0.00714 |
| 6 | Biosynthesis of unsaturated fatty acids | Docosahexaenoic acid | 0.19108 | 0.71879 | 1.0 | 1.0 | 0 |
| 7 | Glycerophospholipid metabolism | LysoPC (16:0/0:0) | 0.19108 | 0.71879 | 1.0 | 1.0 | 0.01736 |
| 8 | Tryptophan metabolism | Indole-3-carboxaldehyde | 0.21487 | 0.66783 | 1.0 | 1.0 | 0.0139 |
| 9 | Aminoacyl-tRNA biosynthesis | L-Arginine | 0.24713 | 0.60708 | 1.0 | 1.0 | 0 |
| 10 | Purine metabolism | Indole-3-carboxaldehyde | 0.32062 | 0.494 | 1.0 | 1.0 | 0.00234 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, J.; Zhang, K.; Yin, Y.; Qi, Y.; Li, S.; Sun, H.; Luo, M.; Sun, Y.; Yu, Z.; Yang, J.; et al. Arecoline-Induced Hepatotoxicity in Rats: Screening of Abnormal Metabolic Markers and Potential Mechanisms. Toxics 2023, 11, 984. https://doi.org/10.3390/toxics11120984
Sun J, Zhang K, Yin Y, Qi Y, Li S, Sun H, Luo M, Sun Y, Yu Z, Yang J, et al. Arecoline-Induced Hepatotoxicity in Rats: Screening of Abnormal Metabolic Markers and Potential Mechanisms. Toxics. 2023; 11(12):984. https://doi.org/10.3390/toxics11120984
Chicago/Turabian StyleSun, Jing, Kai Zhang, Yihui Yin, Yunpeng Qi, Siyuan Li, Haonan Sun, Min Luo, Yixuan Sun, Zhiying Yu, Jie Yang, and et al. 2023. "Arecoline-Induced Hepatotoxicity in Rats: Screening of Abnormal Metabolic Markers and Potential Mechanisms" Toxics 11, no. 12: 984. https://doi.org/10.3390/toxics11120984





