Triphenyltin Influenced Carotenoid-Based Coloration in Coral Reef Fish, Amphiprion ocellaris, by Disrupting Carotenoid Metabolism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Fish Experimental Treatment
2.3. Samples
2.4. Carotenoid Measurement
2.5. Histological Detection
2.6. Antioxidant Capacity Analysis
2.7. Bioinformatics Analysis
2.8. Quantitative Real-Time PCR Analysis
2.9. Statistical Analysis
3. Results
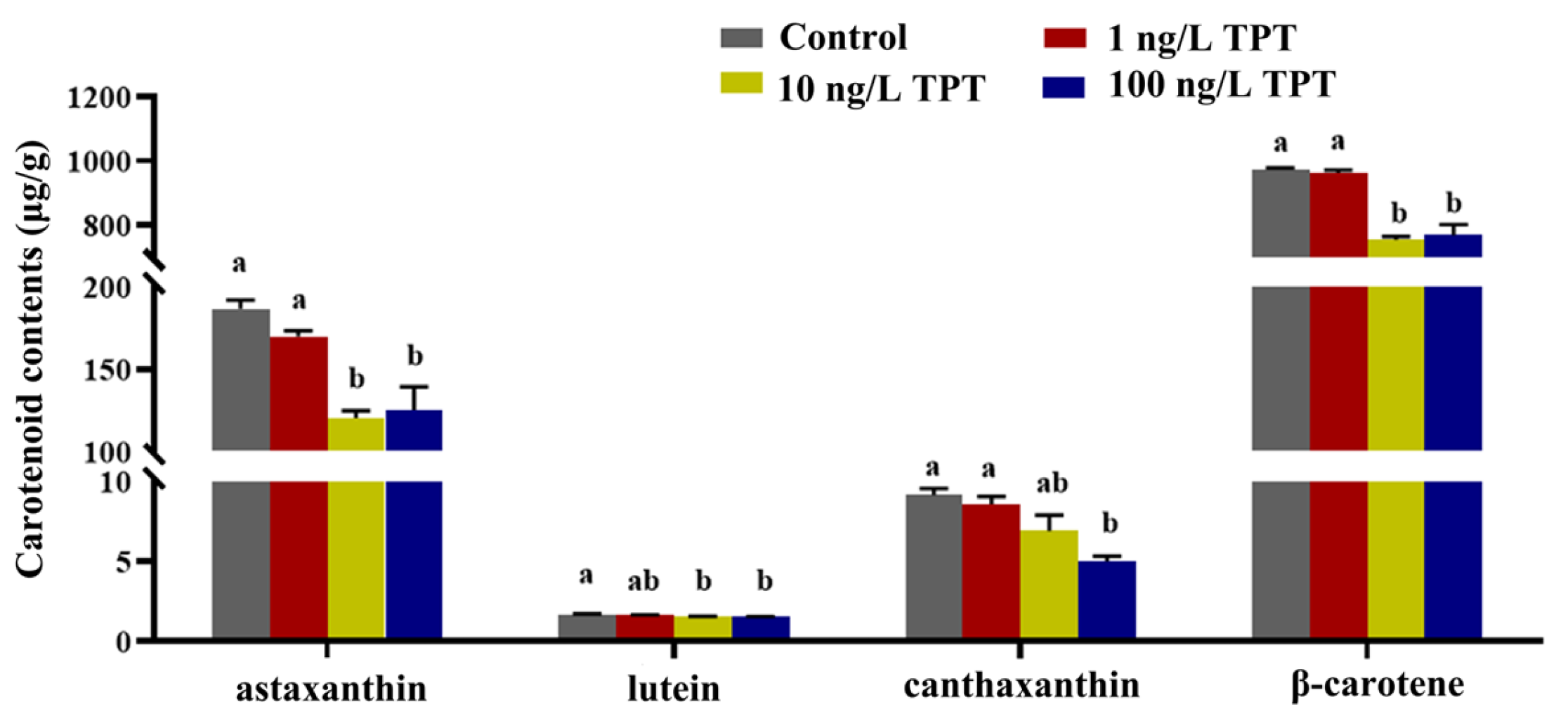
3.1. Carotenoid Contents
3.2. Liver Histology
3.3. Transcriptome Analysis
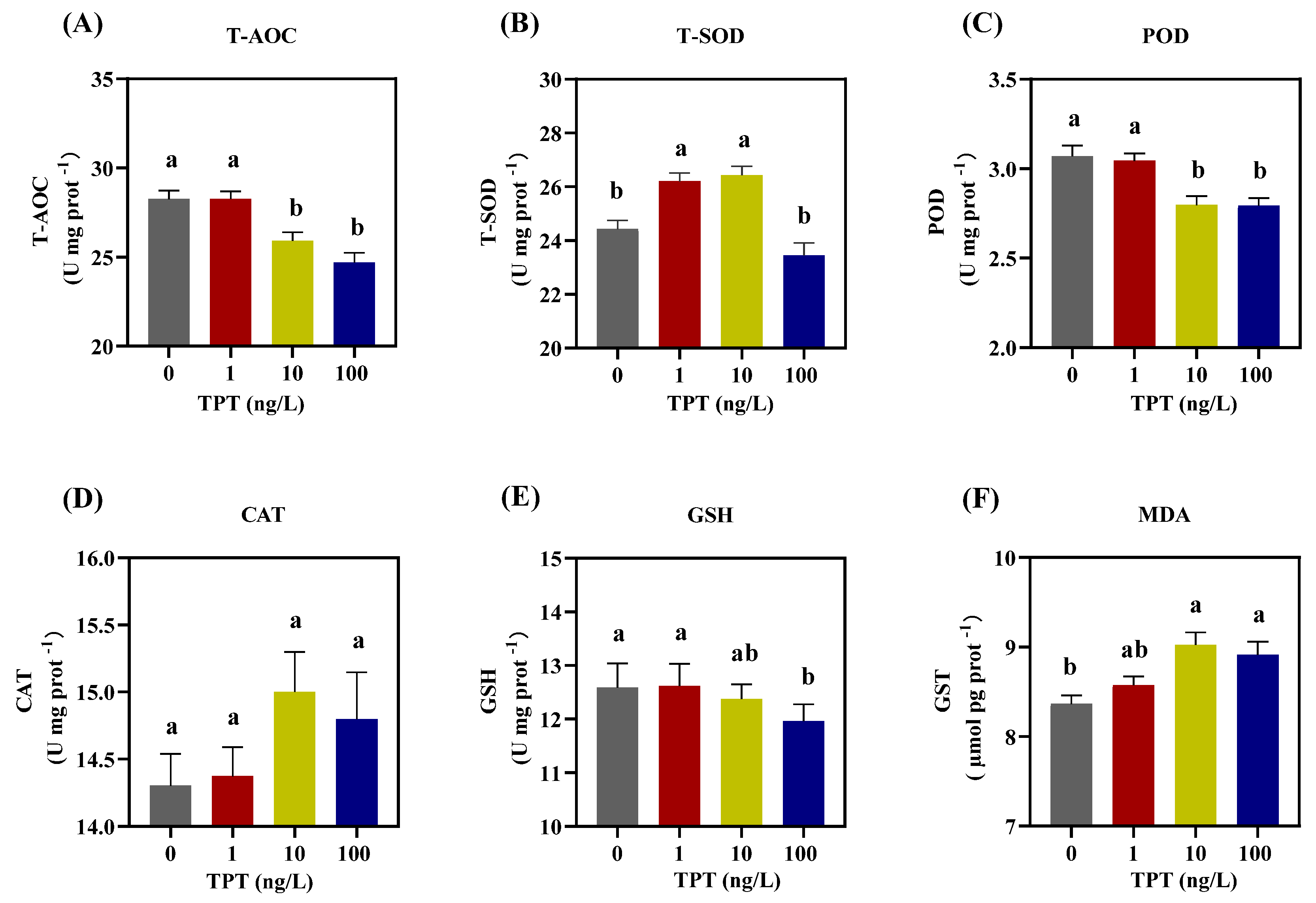
3.4. Hepatic Oxidative Damage
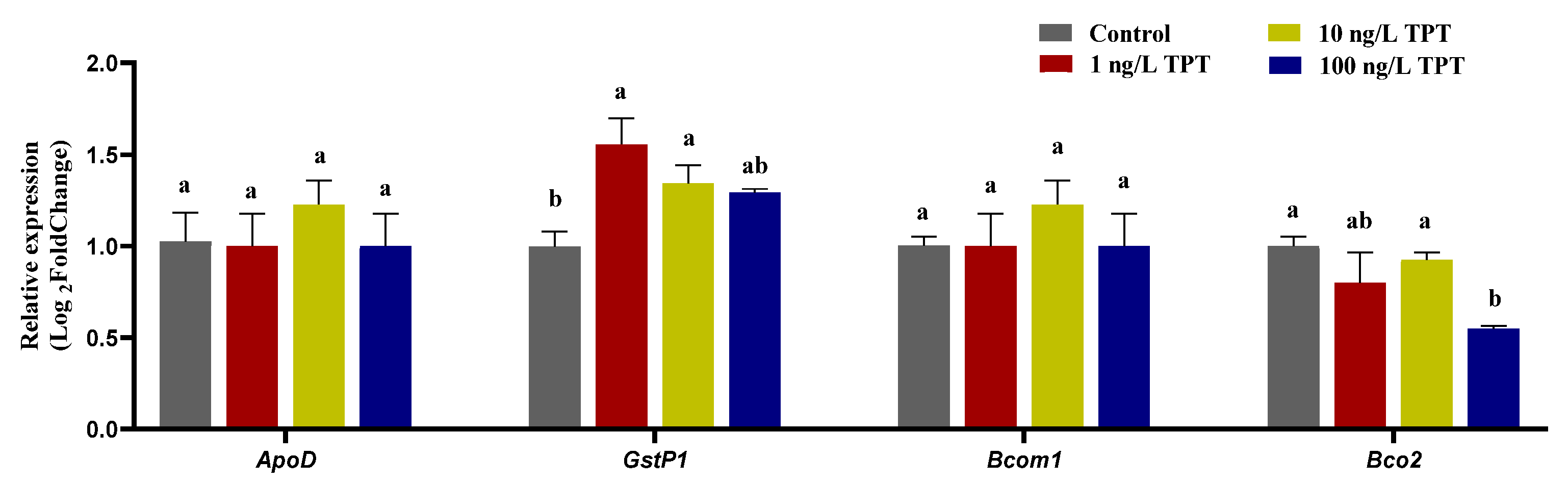
3.5. Genes Related to Carotenoid Absorption and Transport
3.6. Genes Related to Carotenoid Deposition and Decomposition
4. Discussion
4.1. Carotenoid Contents
4.2. Liver Histology
4.3. Transcriptome Analysis
4.4. Hepatic Oxidative Damage
4.5. Genes Related to Carotenoid Metabolism
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Fent, K. Ecotoxicology of organotin compounds. Crit. Rev. Toxicol. 1996, 26, 3–117. [Google Scholar] [CrossRef]
- Ren, X.; Zhang, X.; Ma, X.; Liu, H.; Wang, L. Triphenyltin (TPT) exposure causes SD rat liver injury via lipid metabolism disorder and ER stress revealed by transcriptome analysis. Toxicol. Lett. 2023, 381, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Antes, F.G.; Krupp, E.; Flores, E.M.; Dressler, V.L.; Feldmann, J. Speciation and degradation of triphenyltin in typical paddy fields and its uptake into rice plants. Environ. Sci. Technol. 2011, 45, 10524–10530. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Jiang, J.; Hu, W.; Zhao, Y.; Hu, J. Toxicity of triphenyltin on the development of retinal axons in zebrafish at low dose. Aquat. Toxicol. 2017, 189, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhen, H.; Wan, Y.; Gao, J.; An, W.; An, L.; Jin, F.; Jin, X. Trophic magnification of triphenyltin in a marine food web of Bohai Bay, North China: Comparison to tributyltin. Environ. Sci. Technol. 2006, 40, 3142–3147. [Google Scholar] [CrossRef]
- Yao, F.; Li, Y.; Ru, H.; Wu, L.; Xiao, Z.; Ni, Z.; Chen, D.; Zhong, L. Thyroid disruption and developmental toxicity caused by triphenyltin (TPT) in zebrafish embryos/larvae. Toxicol. Appl. Pharmacol. 2020, 394, 114957. [Google Scholar] [CrossRef]
- Ho, K.K.; Zhou, G.J.; Xu, E.G.; Wang, X.; Leung, K.M. Long-Term Spatio-Temporal Trends of Organotin Contaminations in the Marine Environment of Hong Kong. PLoS ONE 2016, 11, e0155632. [Google Scholar] [CrossRef]
- Gao, J.M.; Wu, L.; Chen, Y.P.; Zhou, B.; Guo, J.S.; Zhang, K.; Ouyang, W.J. Spatiotemporal distribution and risk assessment of organotins in the surface water of the Three Gorges Reservoir Region, China. Chemosphere 2017, 171, 405–414. [Google Scholar] [CrossRef]
- Wen, J.; Cui, X.; Gibson, M.; Li, Z. Water quality criteria derivation and ecological risk assessment for triphenyltin in China. Ecotoxicol. Environ. Saf. 2018, 161, 397–401. [Google Scholar] [CrossRef]
- Chung, S.W.C.; Lau, J.S.Y.; Lau, J.P.K. Occurrence of organotin compounds in seafood from Hong Kong market. Mar. Pollut. Bull. 2020, 154, 111116. [Google Scholar] [CrossRef]
- He, S.; Yu, D.; Li, P.; Zhang, M.; Xing, S.; Sun, C.; Li, Z.H. Triphenyltin exposure causes changes in health-associated gut microbiome and metabolites in marine medaka. Environ. Pollut. 2021, 288, 117751. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Yu, D.; Li, P.; Zhang, M.; Xing, S.; Liu, B.; Sun, C.; Liu, L.; Li, Z.H. A new perspective on endocrine disrupting effects of triphenyltin on marine medaka: From brain transcriptome, gut content metabolome and behavior. Chemosphere 2022, 307, 136190. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, Z.H. Toxicity evaluation of triphenyltin in zebrafish larvae by embryonic malformation, retinal development, and GH/IGF axis. Fish Physiol. Biochem. 2020, 46, 2101–2107. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hu, J.; Zhen, H.; Wu, X.; Huang, C. Reproductive inhibition and transgenerational toxicity of triphenyltin on medaka (Oiyzias latipes) at environmentally relevant levels. Environ. Sci. Technol. 2008, 42, 8133–8139. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.J. Chemical contamination of a coral reef by the grounding of a cruise ship in Bermuda. Mar. Pollut. Bull. 2007, 54, 905–911. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, L.J.; Jin, Y.H.; Guo, R.Y.; Yang, L.; Li, E.C.; Zhang, J.L. Triphenyltin exposure induced abnormal morphological colouration in adult male guppies (Poecilia reticulata). Ecotoxicol. Environ. Saf. 2022, 242, 113912. [Google Scholar] [CrossRef]
- Selkoe, K.A.; Gaggiotti, O.E.; Treml, E.A.; Wren, J.L.; Donovan, M.K.; Toonen, R.J. The DNA of coral reef biodiversity: Predicting and protecting genetic diversity of reef assemblages. Proc. Biol. Sci. 2016, 283, 20160354. [Google Scholar] [CrossRef]
- Roberts, C.M.; McClean, C.J.; Veron, J.E.; Hawkins, J.P.; Allen, G.R.; McAllister, D.E.; Mittermeier, C.G.; Schueler, F.W.; Spalding, M.; Wells, F.; et al. Marine biodiversity hotspots and conservation priorities for tropical reefs. Science 2002, 295, 1280–1284. [Google Scholar] [CrossRef]
- Carpenter, K.E.; Abrar, M.; Aeby, G.; Aronson, R.B.; Banks, S.; Bruckner, A.; Chiriboga, A.; Cortés, J.; Delbeek, J.C.; Devantier, L.; et al. One-third of reef-building corals face elevated extinction risk from climate change and local impacts. Science 2008, 321, 560–563. [Google Scholar] [CrossRef]
- Cortesi, F.; Mitchell, L.J.; Tettamanti, V.; Fogg, L.G.; de Busserolles, F.; Cheney, K.L.; Marshall, N.J. Visual system diversity in coral reef fishes. Semin. Cell Dev. Biol. 2020, 106, 31–42. [Google Scholar] [CrossRef]
- Brandl, S.J.; Johansen, J.L. Extreme environmental conditions reduce coral reef fish biodiversity and productivity. Nat. Commun. 2020, 11, 3832. [Google Scholar] [CrossRef] [PubMed]
- Manning, J.C. Movement, Space Use, and the Responses of Coral Reef Fish to Climate Change. Integr. Comp. Biol. 2022, 62, 1725–1733. [Google Scholar] [CrossRef] [PubMed]
- Roux, N.; Salis, P.; Lambert, A.; Logeux, V.; Soulat, O.; Romans, P.; Frédérich, B.; Lecchini, D.; Laudet, V. Staging and normal table of postembryonic development of the clownfish (Amphiprion ocellaris). Dev. Dyn. 2019, 248, 545–568. [Google Scholar] [CrossRef] [PubMed]
- Duarte, R.C.; Flores, A.A.V.; Stevens, M. Camouflage through colour change: Mechanisms, adaptive value and ecological significance. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160342. [Google Scholar] [CrossRef] [PubMed]
- Endler, J.A.; Mappes, J. The current and future state of animal coloration research. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160352. [Google Scholar] [CrossRef] [PubMed]
- Merilaita, S.; Scott-Samuel, N.E.; Cuthill, I.C. How camouflage works. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160341. [Google Scholar] [CrossRef] [PubMed]
- Shawkey, M.D.; D’Alba, L. Interactions between colour-producing mechanisms and their effects on the integumentary colour palette. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160536. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, X.; Zhao, B.; Ren, X.; Chen, Y.; Si, J.; Zhou, R.; Gan, L.; Zhang, H. Early embryonic exposure of ionizing radiations disrupts zebrafish pigmentation. J. Cell. Physiol. 2018, 234, 940–949. [Google Scholar] [CrossRef]
- Richterová, Z.; Máchová, J.; Stará, A.; Tumová, J.; Velíšek, J.; Sevčíková, M.; Svobodová, Z. Effects of cyhalothrin-based pesticide on early life stages of common carp (Cyprinus carpio L.). Biomed. Res. Int. 2014, 2014, 107373. [Google Scholar] [CrossRef]
- Andrade, P.; Pinho, C.; Pérez, I.d.L.G.; Afonso, S. Regulatory changes in pterin and carotenoid genes underlie balanced color polymorphisms in the wall lizard. Proc. Natl. Acad. Sci. USA 2019, 116, 5633–5642. [Google Scholar] [CrossRef]
- Plonka, P.M.; Passeron, T.; Brenner, M.; Tobin, D.J.; Shibahara, S.; Thomas, A.; Slominski, A.; Kadekaro, A.L.; Hershkovitz, D.; Peters, E.; et al. What are melanocytes really doing all day long...? Exp. Dermatol. 2009, 18, 799–819. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
- Baatrup, E.; Junge, M. Antiandrogenic pesticides disrupt sexual characteristics in the adult male guppy Poecilia reticulata. Environ. Health Perspect. 2001, 109, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Arellano-Aguilar, O.; Macías Garcia, C. Exposure to pesticides impairs the expression of fish ornaments reducing the availability of attractive males. Proc. Biol. Sci. 2008, 275, 1343–1350. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.; Ahmed, I. Effect of Supplementation of Vitamin A on Growth, Haemato-Biochemical Composition, and Antioxidant Ability in Cyprinus carpio var. communis. Aquac. Nutr. 2022, 2022, 8446092. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liang, J.; Dai, H.; Wan, X.; Wang, Z. Effects of vitamin A supplementation in the diet of breeding geese on offspring intestinal tissue morphology and immune performance. Asian Australas. J. Anim. Sci. 2020, 33, 1463–1469. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.R.; Kim, U.J.; Lee, I.S.; Choi, M.; Oh, J.E. Assessment of organotin and tin-free antifouling paints contamination in the Korean coastal area. Mar. Pollut. Bull. 2015, 99, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Wiegertjes, G. Properties of Carotenoids in Fish Fitness: A Review. Mar. Drugs 2020, 18, 568. [Google Scholar] [CrossRef]
- Sefc, K.M.; Brown, A.C.; Clotfelter, E.D. Carotenoid-based coloration in cichlid fishes. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2014, 173, 42–51. [Google Scholar] [CrossRef]
- Yanong, R.P. Nutrition of ornamental fish. Vet. Clin. N. Am. Exot. Anim. Pract. 1999, 2, 19–42. [Google Scholar] [CrossRef]
- Alonso-Alvarez, C.; Pérez-Rodríguez, L.; Mateo, R.; Chastel, O.; Viñuela, J. The oxidation handicap hypothesis and the carotenoid allocation trade-off. J. Evol. Biol. 2008, 21, 1789–1797. [Google Scholar] [CrossRef] [PubMed]
- Sham, R.C.; Ho, K.K.Y.; Hui, T.T.Y.; Zhou, G.J.; Chan, J.K.Y.; Leung, K.M.Y. Tissue distribution of triphenyltin compounds in marine teleost fishes. J. Hazard. Mater. 2021, 401, 123426. [Google Scholar] [CrossRef] [PubMed]
- Trefts, E.; Gannon, M.; Wasserman, D.H. The liver. Curr. Biol. 2017, 27, R1147–R1151. [Google Scholar] [CrossRef] [PubMed]
- Blahova, J.; Dobsikova, R.; Enevova, V.; Modra, H.; Plhalova, L.; Hostovsky, M.; Marsalek, P.; Mares, J.; Skoric, M.; Vecerek, V.; et al. Comprehensive fitness evaluation of common carp (Cyprinus carpio L.) after twelve weeks of atrazine exposure. Sci. Total Environ. 2020, 718, 135059. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Finelli, R. Total antioxidant capacity-Relevance, methods and clinical implications. Andrologia 2021, 53, e13624. [Google Scholar] [CrossRef] [PubMed]
- Munteanu, I.G.; Apetrei, C. Analytical Methods Used in Determining Antioxidant Activity: A Review. Int. J. Mol. Sci. 2021, 22, 3380. [Google Scholar] [CrossRef]
- De Oliveira, F.K.; Santos, L.O.; Buffon, J.G. Mechanism of action, sources, and application of peroxidases. Food Res. Int. 2021, 143, 110266. [Google Scholar] [CrossRef] [PubMed]
- Landis, G.N.; Tower, J. Superoxide dismutase evolution and life span regulation. Mech. Ageing Dev. 2005, 126, 365–379. [Google Scholar] [CrossRef]
- Zhao, H.; Zhang, R.; Yan, X.; Fan, K. Superoxide dismutase nanozymes: An emerging star for anti-oxidation. J. Mater. Chem. B 2021, 9, 6939–6957. [Google Scholar] [CrossRef]
- Rao, A.V.; Rao, L.G. Carotenoids and human health. Pharmacol. Res. 2007, 55, 207–216. [Google Scholar] [CrossRef]
- Fraternale, A.; Paoletti, M.F.; Casabianca, A.; Nencioni, L.; Garaci, E.; Palamara, A.T.; Magnani, M. GSH and analogs in antiviral therapy. Mol. Asp. Med. 2009, 30, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Gaweł, S.; Wardas, M.; Niedworok, E.; Wardas, P. Malondialdehyde (MDA) as a lipid peroxidation marker. Wiad. Lek. 2004, 57, 453–455. [Google Scholar] [PubMed]
- Del Rio, D.; Stewart, A.J.; Pellegrini, N. A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutr. Metab. Cardiovasc. Dis. 2005, 15, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Rajeshkumar, S.; Naik, P. Synthesis and biomedical applications of Cerium oxide nanoparticles—A Review. Biotechnol. Rep. 2018, 17, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Evangelista-Silva, P.H.; Prates, R.P.; Leite, J.S.M.; Moreno, L.G.; Goulart-Silva, F.; Esteves, E.A. Intestinal GLUT5 and FAT/CD36 transporters and blood glucose are reduced by a carotenoid/MUFA-rich oil in high-fat fed mice. Life Sci. 2021, 279, 119672. [Google Scholar] [CrossRef]
- Alquraini, A.; El Khoury, J. Scavenger receptors. Curr. Biol. 2020, 30, R790–R795. [Google Scholar] [CrossRef]
- Alpy, F.; Tomasetto, C. Give lipids a START: The StAR-related lipid transfer (START) domain in mammals. J. Cell Sci. 2005, 118, 2791–2801. [Google Scholar] [CrossRef]
- Pajaud, J.; Ribault, C.; Ben Mosbah, I.; Rauch, C.; Henderson, C.; Bellaud, P.; Aninat, C.; Loyer, P.; Morel, F.; Corlu, A. Glutathione transferases P1/P2 regulate the timing of signaling pathway activations and cell cycle progression during mouse liver regeneration. Cell Death Dis. 2015, 6, e1598. [Google Scholar] [CrossRef]
- Zhang, Y.; Xue, W.; Zhang, W. Histone methyltransferase G9a protects against acute liver injury through GSTP1. Cell Death Differ. 2020, 27, 1243–1258. [Google Scholar] [CrossRef]
- Raghuvanshi, S.; Reed, V.; Blaner, W.S.; Harrison, E.H. Cellular localization of β-carotene 15,15′ oxygenase-1 (BCO1) and β-carotene 9′,10′ oxygenase-2 (BCO2) in rat liver and intestine. Arch. Biochem. Biophys. 2015, 572, 19–27. [Google Scholar] [CrossRef]
- Rassart, E.; Desmarais, F.; Najyb, O.; Bergeron, K.F.; Mounier, C. Apolipoprotein D. Gene 2020, 756, 144874. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Cai, X.; Hou, Y.; Chen, W.; Zhang, J. Triphenyltin Influenced Carotenoid-Based Coloration in Coral Reef Fish, Amphiprion ocellaris, by Disrupting Carotenoid Metabolism. Toxics 2024, 12, 13. https://doi.org/10.3390/toxics12010013
Zhang Y, Cai X, Hou Y, Chen W, Zhang J. Triphenyltin Influenced Carotenoid-Based Coloration in Coral Reef Fish, Amphiprion ocellaris, by Disrupting Carotenoid Metabolism. Toxics. 2024; 12(1):13. https://doi.org/10.3390/toxics12010013
Chicago/Turabian StyleZhang, Yan, Xingwei Cai, Yu Hou, Wenming Chen, and Jiliang Zhang. 2024. "Triphenyltin Influenced Carotenoid-Based Coloration in Coral Reef Fish, Amphiprion ocellaris, by Disrupting Carotenoid Metabolism" Toxics 12, no. 1: 13. https://doi.org/10.3390/toxics12010013
APA StyleZhang, Y., Cai, X., Hou, Y., Chen, W., & Zhang, J. (2024). Triphenyltin Influenced Carotenoid-Based Coloration in Coral Reef Fish, Amphiprion ocellaris, by Disrupting Carotenoid Metabolism. Toxics, 12(1), 13. https://doi.org/10.3390/toxics12010013






