Overview of Methylation and Demethylation Mechanisms and Influencing Factors of Mercury in Water
Abstract
:1. Introduction
2. Methylation of Mercury in the Aquatic Environment
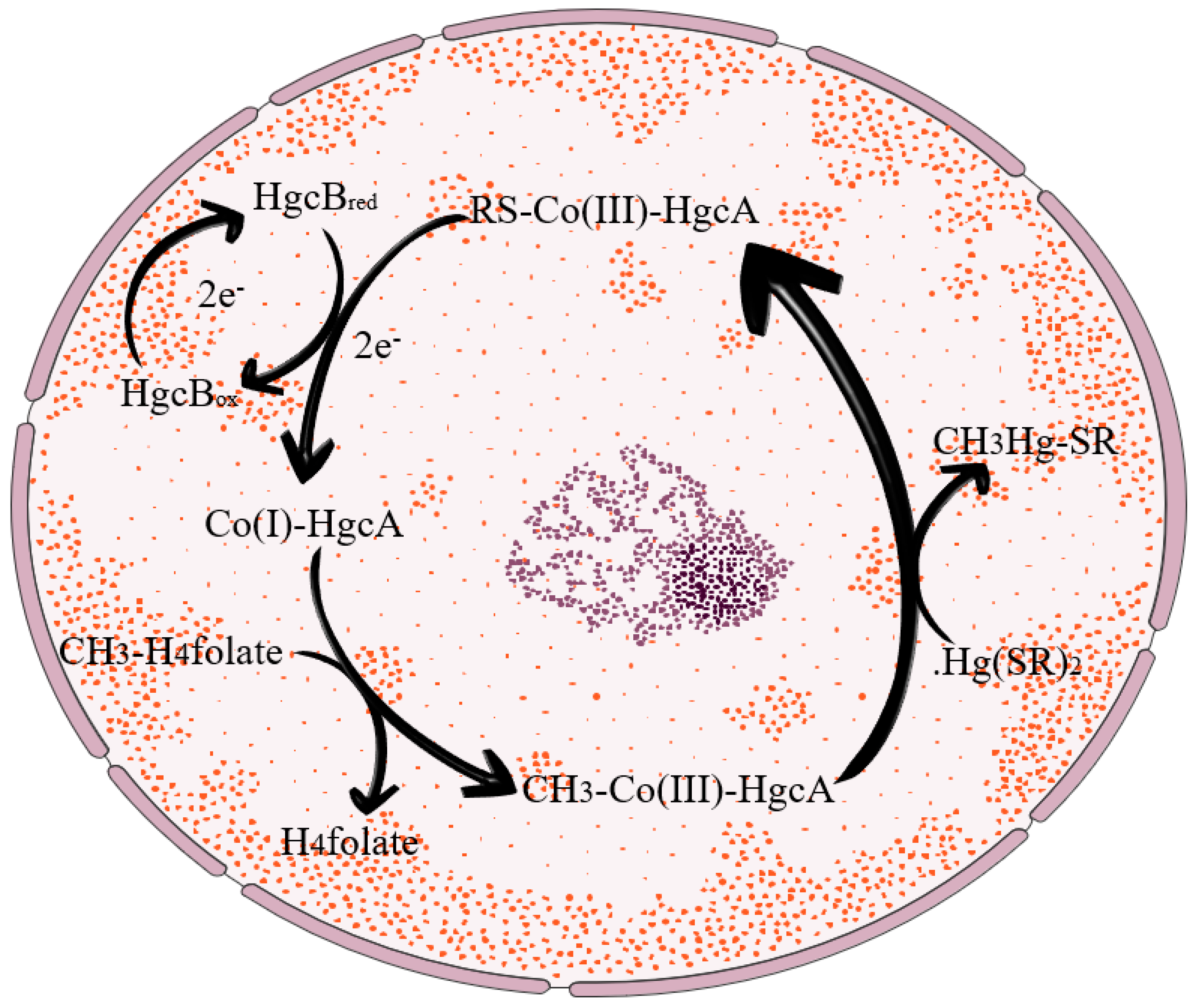
2.1. Microbial Pathway Methylation
2.2. Abiotic Pathway Methylation
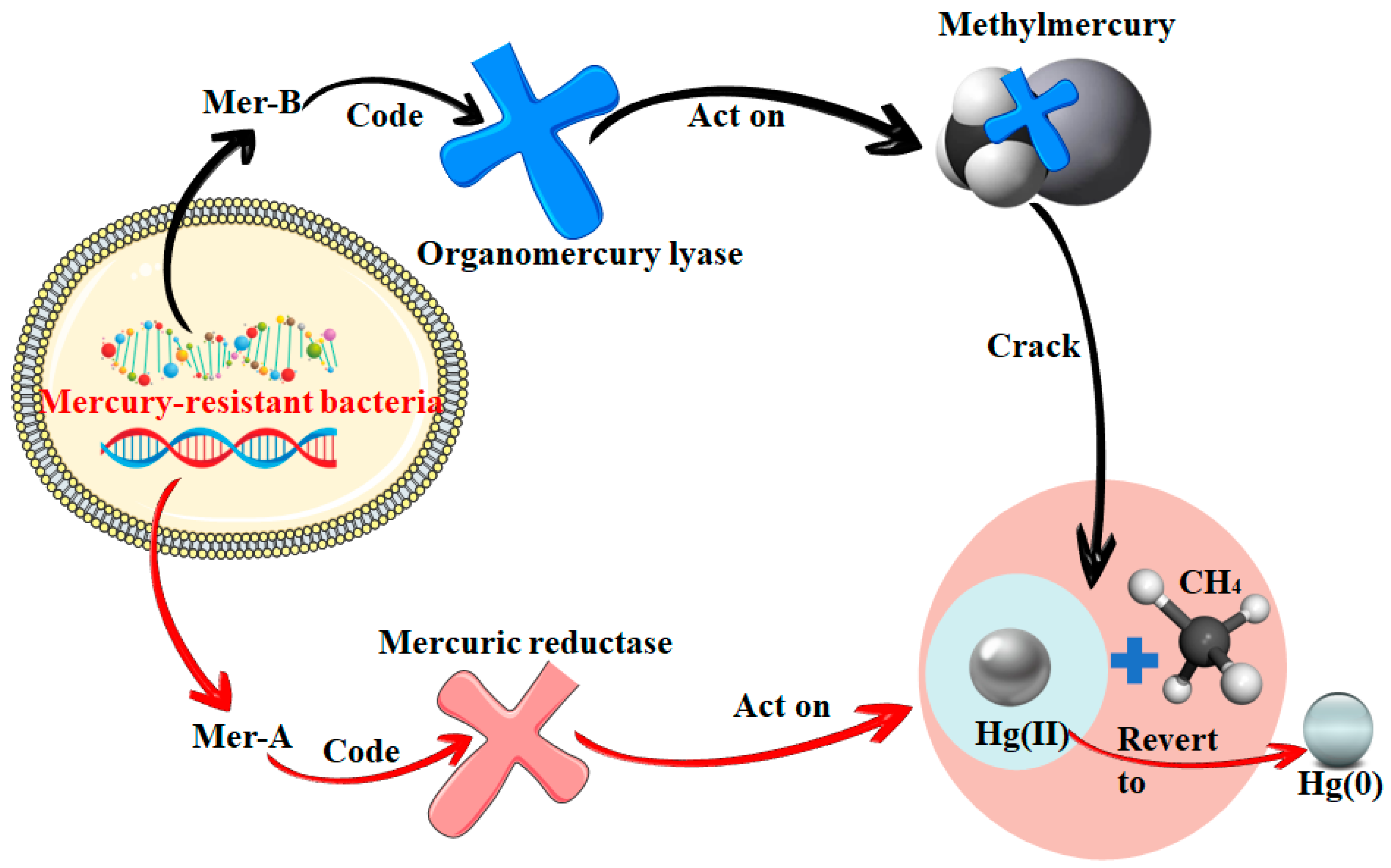
3. Demethylation of Mercury in Aquatic Environments
4. Factors Influencing Mercury Methylation and Demethylation
4.1. Redox Conditions
4.2. Organic Substances
4.3. Sulphide
4.4. Temperature
4.5. pH
4.6. Iron and Manganese Oxides
5. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bjørklund, G.; Dadar, M.; Mutter, J.; Aaseth, J. The toxicology of mercury: Current research and emerging trends. Environ. Res. 2017, 159, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Clarkson, T.W. The toxicology of mercury. Crit. Rev. Clin. Lab. Sci. 1997, 34, 369–403. [Google Scholar] [CrossRef] [PubMed]
- Mahaffey, K.R.; Clickner, R.P.; Bodurow, C.C. Blood organic mercury and dietary mercury intake: National Health and Nutrition Examination Survey, 1999 and 2000. Environ. Health Perspect. 2004, 112, 562–570. [Google Scholar] [CrossRef]
- Steffan, R.J.; Korthals, E.T.; Winfrey, M.R. Effects of acidification on mercury methylation, demethylation, and volatilization in sediments from an acid-susceptible lake. Appl. Environ. Microbiol. 1988, 54, 2003–2009. [Google Scholar] [CrossRef] [PubMed]
- Hudelson, K.E.; Drevnick, P.E.; Wang, F.Y.; Armstrong, D.; Fisk, A.T. Mercury methylation and demethylation potentials in Arctic lake sediments. Chemosphere 2020, 248, 126001. [Google Scholar] [CrossRef] [PubMed]
- Siciliano, S.D.; O’Driscoll, N.J.; Tordon, R.; Hill, J.; Beauchamp, S.; Lean, D.R.S. Abiotic production of methylmercury by solar radiation. Environ. Sci. Technol. 2005, 39, 1071–1077. [Google Scholar] [CrossRef]
- Tang, W.L.; Liu, Y.R.; Guan, W.Y.; Zhong, H.; Qu, X.M.; Zhang, T. Understanding mercury methylation in the changing environment: Recent advances in assessing microbial methylators and mercury bioavailability. Sci. Total Environ. 2020, 714, 136827. [Google Scholar] [CrossRef]
- Yin, X.X.; Wang, L.J.; Zhang, L.J.; Chen, H.M.; Liang, X.J.; Lu, X.; DiSpirito, A.A.; Semrau, J.D.; Gu, B.H. Synergistic Effects of a Chalkophore, Methanobactin, on Microbial Methylation of Mercury. Appl. Environ. Microbiol. 2020, 86, e00122-20. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.Y.; Yang, Y.; Yang, S.; Li, L.; Song, L.Y. Recent advance of microbial mercury methylation in the environment. Appl. Microbiol. Biotechnol. 2024, 108, 235. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Guan, W.Y.; Ji, Y.Y.; He, X.; Chen, W.; Alvarez, P.J.J.; Zhang, T. Microbial methylation potential of mercury sulfide particles dictated by surface structure. Nat. Geosci. 2021, 14, 409–416. [Google Scholar] [CrossRef]
- Gionfriddo, C.M.; Tate, M.T.; Wick, R.R.; Schultz, M.B.; Zemla, A.; Thelen, M.P.; Schofield, R.; Krabbenhoft, D.P.; Holt, K.E.; Moreau, J.W. Microbial mercury methylation in Antarctic sea ice. Nat. Microbiol. 2016, 1, 16127. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.X.; Li, Y.L.; Zhao, D.Y.; Zhuang, L.; Yang, G.Q.; Gong, Y.Y. Immobilization of mercury by iron sulfide nanoparticles alters mercury speciation and microbial methylation in contaminated groundwater. Chem. Eng. J. 2019, 381, 122664. [Google Scholar] [CrossRef]
- Goñi-Urriza, M.; Klopp, C.; Ranchou-Peyruse, M.; Ranchou-Peyruse, A.; Monperrus, M.; Khalfaoui-Hassani, B.; Guyoneaud, R. Genome insights of mercury methylation among Desulfovibrio and Pseudodesulfovibrio strains. Res. Microbiol. 2019, 171, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Munson, K.M.; Lamborg, C.H.; Boiteau, R.M.; Saito, M.A. Dynamic mercury methylation and demethylation in oligotrophic marine water. Biogeosciences 2018, 15, 6451–6460. [Google Scholar] [CrossRef]
- Lázaro, W.L.; Díez, S.; da Silva, C.J.; Ignácio, Á.R.A.; Guimarães, J.R.D. Waterscape determinants of net mercury methylation in a tropical wetland. Environ. Res. 2016, 150, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, G.; Cai, Y. Possible pathways for mercury methylation in oxic marine waters. Crit. Rev. Environ. Sci. Technol. 2021, 52, 3997–4015. [Google Scholar] [CrossRef]
- Akagi, H.; Takabatake, E.; Fujita, Y. Photochemical methylation of inorganic mercury in the presence of solid sulfur. Chem. Lett. 1974, 3, 761–764. [Google Scholar] [CrossRef]
- Zhang, T.; Kim, B.; Levard, C.; Reinsch, B.C.; Lowry, G.V.; Deshusses, M.A.; Hsu-Kim, H. Methylation of mercury by bacteria exposed to dissolved, nanoparticulate, and microparticulate mercuric sulfides. Environ. Sci. Technol. 2012, 46, 6950–6958. [Google Scholar] [CrossRef]
- Ramlal, P.S.; Rudd, J.W.; Hecky, R.E. Methods for measuring specific rates of mercury methylation and degradation and their use in determining factors controlling net rates of mercury methylation. Appl. Environ. Microbiol. 1986, 51, 110–114. [Google Scholar] [CrossRef]
- Tulasi, D.; Fajon, V.; Kotnik, J.; Shlyapnikov, Y.S.; Adotey, D.K.; Serfor-Armah, Y.; Horvat, M. Mercury methylation in cyanide influenced river sediments: A comparative study in Southwestern Ghana. Environ. Monit. Assess. 2021, 193, 180. [Google Scholar] [CrossRef]
- Gilmour, C.C.; Henry, E.A. Mercury methylation in aquatic systems affected by acid deposition. Environ. Pollut. 1991, 71, 131–169. [Google Scholar] [CrossRef] [PubMed]
- Demissie, T.B.; Garabato, B.D.; Ruud, K.; Kozlowski, P.M. Mercury Methylation by Cobalt Corrinoids: Relativistic Effects Dictate the Reaction Mechanism. Angew. Chem. Int. Ed. 2016, 55, 11503–11506. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, Y.; Zhang, L.; Zheng, J.Q.; Pierce, E.M.; Gu, B.H. Mercury Adsorption on Minerals and Its Effect on Microbial Methylation. ACS Earth Space Chem. 2019, 3, 1338–1345. [Google Scholar] [CrossRef]
- Gallorini, A.; Loizeau, J.L. Mercury methylation in oxic aquatic macro-environments: A review. J. Limnology. 2021, 10, 4081. [Google Scholar] [CrossRef]
- Schaefer, J.K.; Morel, F.M.M. High methylation rates of mercury bound to cysteine by Geobacter sulfurreducens. Nat. Geosci. 2009, 2, 123–126. [Google Scholar] [CrossRef]
- Cesário, R.; Hintelmann, H.; Mendes, R.; Eckey, K.; Dimock, B.; Araújo, B.; Mota, A.M.; Canário, J. Evaluation of mercury methylation and methylmercury demethylation rates in vegetated and non-vegetated saltmarsh sediments from two Portuguese estuaries. Environ. Pollut. 2017, 226, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Guo, P.; Xu, X. Effect of selenium on mercury methylation in facultative lake sediments. Toxicol. Environ. Chem. 1999, 69, 255–261. [Google Scholar] [CrossRef]
- Song, W.; Xiong, H.; Qi, R.; Wang, S.Z.; Yang, Y.Y. Effect of salinity and algae biomass on mercury cycling genes and bacterial communities in sediments under mercury contamination: Implications of the mercury cycle in arid regions. Environ. Pollut. 2020, 269, 116141. [Google Scholar] [CrossRef]
- Li, Y.; Li, D.; Song, B.B.; Li, Y.B. The potential of mercury methylation and demethylation by 15 species of marine microalgae. Water Res. 2022, 215, 118266. [Google Scholar] [CrossRef]
- Tohyama, C. Comment on “Rethinking the Minamata Tragedy: What Mercury Species Was Really Responsible?”. Environ. Sci. Technol. 2020, 54, 8486–8487. [Google Scholar] [CrossRef]
- Yang, W.L. Minamata disease in Japan. Environmental 2006, 3, 96–97. [Google Scholar]
- National Centre for Food Safety Risk Assessment (China). Risk Assessment of Methylmercury in Dietary Animal Fish and Fishery Products for Chinese Residents (Abstract) Exposure Risk Assessment. [EB/OL]. Chin. Med. 2023, 18, 73. [Google Scholar]
- Xu, X.; Han, J.L.; Pang, J.; Wang, X.; Lin, Y.; Wang, Y.J.; Qiu, G.L. Methylmercury and inorganic mercury in Chinese commercial rice: Implications for overestimated human exposure and health risk. Environ. Pollut. 2020, 258, 113706. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, Y.; Omura, Y.; Okazaki, E. Total mercury and methylmercury levels in commercially important fishes in Japan. Fish. Sci. 2005, 71, 1029–1035. [Google Scholar] [CrossRef]
- Kružíková, K.; Blahová, J.; Kenšová, R.; Jurčíková, J.; Hypr, D.; Svobodová, Z. Mercury and methylmercury content in muscle of chub and in sediment from the Svitava and Svratka rivers. Toxicol. Lett. 2009, 189, S190. [Google Scholar] [CrossRef]
- Mbanga, O.; Ncube, S.; Tutu, H.; Chimuka, L.; Cukrowska, E. Mercury accumulation and biotransportation in wetland biota affected by gold mining. Environ. Monit. Assess. 2019, 191, 186. [Google Scholar] [CrossRef]
- Maršálek, P.; Svobodová, Z.; Randák, T. The content of total mercury and methylmercury in common carp from selected Czech ponds. Aquac. Int. 2007, 15, 299–304. [Google Scholar] [CrossRef]
- Hajeb, P.; Jinap, S.; Ahmad, I. Biomagnifications of mercury and methylmercury in tuna and mackerel. Environ. Monit. Assess. 2009, 171, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Han, J.L.; Abeysinghe, K.S.; Atapattu, A.J.; Silva, P.M.C.S.D.; Xu, Z.D.; Long, S.T.; Qiu, G.L. Dietary exposure assessment of total mercury and methylmercury in commercial rice in Sri Lanka. Chemosphere 2019, 239, 124749. [Google Scholar] [CrossRef]
- Jensen, S.; Jernelöv, A. Biological Methylation of Mercury in Aquatic Organisms. Nature 1969, 223, 753–754. [Google Scholar] [CrossRef]
- Wood, J.M.; Kennedy, F.S.; Rosen, C.G. Synthesis of Methyl-mercury Compounds by Extracts of a Methanogenic Bacterium. Nature 1968, 220, 173–174. [Google Scholar] [CrossRef] [PubMed]
- DeSimone, R.E.; Penley, M.W.; Charbonneau, L.; Smith, S.G.; Wood, J.M.; Hill, H.A.O.; Pratt, J.M.; Ridsdale, S.; Williams, R.J.P. The kinetics and mechanism of cobalamin-dependent methyl and ethyl transfer to mercuric ion. Biochim. Biophys. Acta (BBA) Gen. Subj. 1973, 304, 851–863. [Google Scholar] [CrossRef]
- Ridley, W.P.; Dizikes, L.J.; Wood, J.M. Biomethylation of toxic elements in the environment. Science 1977, 197, 329–332. [Google Scholar] [CrossRef]
- Imura, N.; Sukegawa, E.; Pan, S.K.; Nagao, K.; KIM, J.Y.; Kwan, T.; Ukita, T. Chemical methylation of inorganic mercury with methylcobalamin, a vitamin B12 analog. Science 1971, 172, 1248–1249. [Google Scholar] [CrossRef]
- Parks, J.M.; Johs, A.; Podar, M.; Bridou, R.; Hurt, R.A.; Smith, S.D.; Tomanicek, S.J.; Qian, Y.; Brown, S.D.; Brandt, C.C.; et al. The Genetic Basis for Bacterial Mercury Methylation. Science 2013, 339, 1332–1335. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.W.; Roth, S.; Schaefer, J.K.; Reinfelder, J.R.; Yee, N. Production of methylmercury by methanogens in mercury contaminated estuarine sediments. FEMS Microbiol. Lett. 2020, 367, fnaa196. [Google Scholar] [CrossRef]
- Date, S.S.; Parks, J.M.; Rush, K.W.; Wall, J.D.; Ragsdale, S.W.; Johs, A. Kinetics of Enzymatic Mercury Methylation at Nanomolar Concentrations Catalyzed by HgcAB. Appl. Environ. Microbiol. 2019, 85, e00438-19. [Google Scholar] [CrossRef]
- Lin, H.; Ascher, D.B.; Myung, Y.; Lamborg, C.H.; Hallam, S.J.; Gionfriddo, C.M.; Holt, K.E.; Moreau, J.W. Mercury methylation by metabolically versatile and cosmopolitan marine bacteria. ISME J. 2021, 15, 1810–1825. [Google Scholar] [CrossRef]
- Cassarini, C.; Zhang, Y.; Lens, P.N.L. Pressure Selects Dominant Anaerobic Methanotrophic Phylotype and Sulfate Reducing Bacteria in Coastal Marine Lake Grevelingen Sediment. Front. Environ. Sci. 2019, 06, 00162. [Google Scholar] [CrossRef]
- Shao, D.D.; Kang, Y.; Wu, S.C.; Wong, M.H. Effects of sulfate reducing bacteria and sulfate concentrations on mercury methylation in freshwater sediments. Sci. Total Environ. 2012, 424, 331–336. [Google Scholar] [CrossRef]
- Yin, X.; Wang, L.; Liang, X.; Zhang, L.J.; Zhao, J.T.; Gu, B.H. Contrary effects of phytoplankton Chlorella vulgaris and its exudates on mercury methylation by iron- and sulfate-reducing bacteria. J. Hazard. Mater. 2022, 433, 128835. [Google Scholar] [CrossRef] [PubMed]
- Compeau, G.C.; Bartha, R. Sulfate-reducing bacteria: Principal methylators of mercury in anoxic estuarine sediment. Appl. Environ. Microbiol. 1985, 50, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.C.; Bartha, R. Cobalamin-mediated mercury methylation by Desulfovibrio desulfuricans LS. Appl. Environ. Microbiol. 1993, 59, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Fleming, E.J.; Mack, E.E.; Green, P.G.; Nelson, D.C. Mercury Methylation from Unexpected Sources: Molybdate-Inhibited Freshwater Sediments and an Iron-Reducing Bacterium. Appl. Environ. Microbiol. 2006, 72, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Richard, A.; Hurt, R.A.; Johs, A.; Parks, J.N.; Morrell-Falvey, J.L.; Liang, L.Y.; Elias, D.A.; Gu, B.H. Unexpected Effects of Gene Deletion on Interactions of Mercury with the Methylation-Deficient Mutant Δ hgcAB. Environ. Sci. Technol. Lett. 2014, 1, 271–276. [Google Scholar] [CrossRef]
- Christensen, G.A.; Somenahally, A.C.; Moberly, J.G.; Miller, C.M.; King, A.J.; Gilmour, C.C.; Brown, S.D.; Podar, M.; Brandt, C.C.; Brooks, S.C.; et al. Carbon Amendments Alter Microbial Community Structure and Net Mercury Methylation Potential in Sediments. Appl. Environ. Microbiol. 2017, 84, e01049-17. [Google Scholar] [CrossRef]
- Celo, V.; Lean, D.R.S.; Scott, S.L. Abiotic methylation of mercury in the aquatic environment. Sci. Total Environ. 2006, 368, 126–137. [Google Scholar] [CrossRef]
- Jiménez-Moreno, M.; Perrot, V.; Epov, V.N.; Monperrus, M.; Amouroux, D. Chemical kinetic isotope fractionation of mercury during abiotic methylation of Hg (II) by methylcobalamin in aqueous chloride media. Chem. Geol. 2013, 336, 26–36. [Google Scholar] [CrossRef]
- Rouleau, C.; Pelletier, É.; Tjälve, H. Short-term bioconcentration and distribution of methylmercury, tributyltin and corresponding inorganic species in the starfish leptasterias polaris. Appl. Organomet. Chem. 2004, 9, 327–334. [Google Scholar] [CrossRef]
- Yin, Y.; Chen, B.; Mao, Y.; Wang, T.; Liu, J.F.; Cai, Y.; Jiang, G.B. Possible alkylation of inorganic Hg (II) by photochemical processes in the environment. Chemosphere 2012, 88, 8–16. [Google Scholar] [CrossRef]
- Hayashi, K.; Kawai, S.; Ohno, T.; MAKI, Y. Photoalkylation of inorganic mercury in the presence of amino acids. II. Photomethylation of inorganic mercury by aliphatic alpha-amino acids (author’s transl). Yakugaku Zasshi 1979, 99, 1250–1253. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.B.; Tuovinen, O.H. Mechanisms of Microbial Resistance and Detoxification of Mercury and Organomercury Compounds: Physiological, Biochemical, and Genetic Analyses. Microbiol. Rev. 1984, 48, 95–124. [Google Scholar] [CrossRef] [PubMed]
- Marvin-DiPasquale, M.; Agee, J.; McGowan, C.; Oremland, R.S.; Thomas, M.; Krabbenhoft, D.; Gilmour, C.C. Methyl-mercury degradation pathways: A comparison among three mercury-impacted ecosystems. Environ. Sci. Technol. 2000, 34, 4908–4916. [Google Scholar] [CrossRef]
- Oremland, R.S.; Culbertson, C.W.; Winfrey, M.R. Methylmercury decomposition in sediments and bacterial cultures: Involvement of methanogens and sulfate reducers in oxidative demethylation. Appl. Environ. Microbiol. 1991, 57, 130–137. [Google Scholar] [CrossRef]
- Baldi, F.; Pepi, M.; Filippelli, M. Methylmercury Resistance in Desulfovibrio desulfuricans Strains in Relation to Methylmercury Degradation. Appl. Environ. Microbiol. 1993, 59, 8–2479. [Google Scholar] [CrossRef]
- Hamelin, S.; Amyot, M.; Barkay, T.; Wang, Y.P.; Planas, D. Methanogens: Principal methylators of mercury in lake periphyton. Environ. Sci. Technol. 2011, 45, 7693–7700. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Gu, W.Y.; Zhao, L.D.; Haque, M.F.U.; DiSpirito, A.A.; Semrau, J.D.; Gu, B.H. Methylmercury uptake and degradation by methanotrophs. Sci. Adv. 2017, 3, e1700041. [Google Scholar] [CrossRef]
- Kritee, K.; Motta, L.C.; Blum, J.D.; Tsui, M.T.K.; Reinfelder, J.R. Photomicrobial Visible Light-Induced Magnetic Mass Independent Fractionation of Mercury in a Marine Microalga. ACS Earth Space Chem. 2017, 2, 432–440. [Google Scholar] [CrossRef]
- Liang, X.J.; Zhong, H.; Johs, A.; Lei, P.; Zhang, J.; Taş, N.; Zhang, L.J.; Zhao, L.D.; Zhu, N.; Yin, X.X.; et al. Light-independent phytoplankton degradation and detoxification of methylmercury in water. Nat. Water 2023, 1, 705–715. [Google Scholar] [CrossRef]
- Klapstein, S.J.; Ziegler, S.E.; O’Driscoll, N.J. Methylmercury photodemethylation is inhibited in lakes with high dissolved organic matter. Environ. Pollut. 2017, 232, 392–401. [Google Scholar] [CrossRef]
- Benoit, J.M.; Gilmour, C.C.; Mason, R.P. The Influence of Sulfide on Solid-Phase Mercury Bioavailability for Methylation by Pure Cultures of Desulfobulbus propionicus (1pr3). Environ. Sci. Technol. 2001, 35, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.K.; Wang, F. Chemical Demethylation of Methylmercury by Selenoamino Acids. Chem. Res. Toxicol. 2010, 23, 1202–1206. [Google Scholar] [CrossRef]
- Asaduzzaman AMd Schreckenbach, G. Degradation Mechanism of Methyl Mercury Selenoamino Acid Complexes: A Computational Study. Inorg. Chem. 2011, 50, 2366–2372. [Google Scholar] [CrossRef]
- Desrosiers, M.; Planas, D.; Mucci, A. Mercury methylation in the epilithon of boreal shield aquatic ecosystems. Environ. Sci. Technol. 2006, 40, 1540–1546. [Google Scholar] [CrossRef] [PubMed]
- Seller, P.; Kelly, C.A.; Rudd, J.W.M.; MacHutchon, A.R. Photodegradation of methylmercury in lakes. Nature 1996, 380, 694–697. [Google Scholar] [CrossRef]
- Lehnherr, I.; St Louis, V.L. Importance of ultraviolet radiation in the photodemethylation of methylmercury in freshwater ecosystems. Environ. Sci. Technol. 2009, 43, 5692–5698. [Google Scholar] [CrossRef] [PubMed]
- Eckley, C.S.; Luxton, T.P.; Knightes, C.D.; Shah, V. Methylmercury Production and Degradation under Light and Dark Conditions in the Water Column of the Hells Canyon Reservoirs, USA. Environ. Toxicol. Chem. 2021, 40, 1827–1837. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Gómez, C.; Drott, A.; Björn, E.; Díez, S.; Bayona, J.M.; Tesfalidet, S.; Lindfors, A.; Skyllberg, U. Towards universal wavelength-specific photodegradation rate constants for methyl mercury in humic waters, exemplified by a Boreal lake-wetland gradient. Environ. Sci. Technol. 2013, 47, 6279–6287. [Google Scholar] [CrossRef]
- Black, F.J.; Poulin, B.A.; Flegal, A.R. Factors controlling the abiotic photo-degradation of monomethylmercury in surface waters. Geochim. Cosmochim. Acta 2012, 84, 492–507. [Google Scholar] [CrossRef]
- Rose, C.H.; Ghosh, S.; Blum, J.D.; Bergquist, B.A. Effects of ultraviolet radiation on mercury isotope fractionation during photo-reduction for inorganic and organic mercury species. Chem. Geol. 2015, 405, 102–111. [Google Scholar] [CrossRef]
- Bergquist, B.A.; Blum, J.D. Mass-Dependent and -Independent Fractionation of Hg Isotopes by Photoreduction in Aquatic Systems. Science 2007, 318, 417–420. [Google Scholar] [CrossRef] [PubMed]
- Donovan, P.M.; Blum, J.D.; Singer, M.B.; Marvin-DiPasquale, M.; Tsui, M.T.K. Methylmercury degradation and exposure pathways in streams and wetlands impacted by historical mining. Sci. Total Environ. 2016, 568, 1192–1203. [Google Scholar] [CrossRef]
- Blum, J.D.; Drazen, J.C.; Johnson, M.W.; Popp, B.N.; Motta, L.C.; Jamieson, A.J. Mercury isotopes identify near-surface marine mercury in deep-sea trench biota [Earth, Atmospheric, and Planetary Sciences. Proc. Natl. Acad. Sci. USA 2020, 117, 29292–29298. [Google Scholar] [CrossRef] [PubMed]
- Bonsignore, M.; Manta, D.S.; Barsanti, M.; Conte, F.; Delbono, I.; Horvat, M.; Quinci, E.M.; Schirone, A.; Shlyapnikov, Y.; Sprovieri, M. Mercury isotope signatures in sediments and marine organisms as tracers of historical industrial pollution. Chemosphere 2020, 258, 127435. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Yin, Y.; Li, Y.; Cai, Y.; Liu, J. Critical role of natural organic matter in photodegradation of methylmercury in water: Molecular weight and interactive effects with other environmental factors. Sci. Total Environ. 2017, 578, 535–541. [Google Scholar] [CrossRef]
- Suda, I.; Suda, M.; Hirayama, K. Degradation of methyl and ethyl mercury by singlet oxygen generated from sea water exposed to sunlight or ultraviolet light. Arch. Toxicol. 1993, 67, 365–368. [Google Scholar] [CrossRef]
- Tai, C.; Zhang, S.; Wang, J.; Yin, Y.; Shi, J.; Wu, H.; Mao, Y. Solar-induced generation of singlet oxygen and hydroxyl radical in sewage. Environ. Chem. 2017, 15, 515–523. [Google Scholar] [CrossRef]
- Qian, Y.; Yin, X.P.; Lin, H.; Rao, B.; Brooks, S.C.; Liang, L.Y.; Gu, B.H. Why dissolved organic matter enhances photodegradation of methylmercury. Environ. Sci. Technol. 2014, 1, 426–431. [Google Scholar] [CrossRef]
- Sonke, J.E.; Heimbürger, L.E.; Dommergue, A. Mercury biogeochemistry: Paradigm shifts, outstanding issues and research needs. Comptes Rendus Geosci. 2013, 345, 213–224. [Google Scholar] [CrossRef]
- Monperrus, M.; Tessier, E.; Amouroux, D.; Leynaert, A.; Huonnic, P.; Donard, O.F.X. Mercury methylation, demethylation and reduction rates in coastal and marine surface waters of the Mediterranean Sea. Mar. Chem. 2007, 107, 49–63. [Google Scholar] [CrossRef]
- Lehnherr, I.; St Louis, V.L.; Hintelmann, H.; Kirk, J.L. Methylation of inorganic mercury in polar marine waters. Nat. Geosci. 2011, 4, 298–302. [Google Scholar] [CrossRef]
- Heimbürger, L.E.; Cossa, D.; Marty, J.C.; Migon, C.; Averty, B.; Dufour, A.; Ras, J. Methyl mercury distributions in relation to the presence of nano- and picophytoplankton in an oceanic water column (Ligurian Sea, North-western Mediterranean). Geochim. Cosmochim. Acta 2010, 74, 5549–5559. [Google Scholar] [CrossRef]
- Liu, B.; Schaider, L.A.; Mason, R.P.; Shine, J.P.; Rabalais, N.N.; Senn, D.B. Controls on methylmercury accumulation in northern Gulf of Mexico sediments. Estuar. Coast. Shelf Sci. 2015, 159, 50–59. [Google Scholar] [CrossRef]
- Correia, R.R.S.; Miranda, M.R.; Guimarães, J.R.D. Mercury methylation and the microbial consortium in periphyton of tropical macrophytes: Effect of different inhibitors. Environ. Res. 2011, 112, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Korthals, E.T.; Winfrey, M.R. Seasonal and spatial variations in mercury methylation and demethylation in an oligotrophic lake. Appl. Environ. Microbiol. 1987, 53, 2397–2404. [Google Scholar] [CrossRef]
- Eckley, C.S.; Hintelmann, H. Determination of mercury methylation potentials in the water column of lakes across Canada. Sci. Total Environ. 2006, 368, 111–125. [Google Scholar] [CrossRef]
- Ullrich, S.M.; Tanton, T.W.; Abdrashitova, S.A. Mercury in the Aquatic Environment: A Review of Factors Affecting Methylation. Crit. Rev. Environ. Sci. Technol. 2001, 31, 241–293. [Google Scholar] [CrossRef]
- Miskimmin, B.M.; Rudd, J.W.M.; Kelly, C.A. Influence of Dissolved Organic Carbon, pH, and Microbial Respiration Rates on Mercury Methylation and Demethylation in Lake Water. Can. J. Fish. Aquat. Sci. 1992, 49, 17–22. [Google Scholar] [CrossRef]
- Barkay, T.; Gillman, M.; Turner, R.R. Effects of dissolved organic carbon and salinity on bioavailability of mercury. Appl. Environ. Microbiol. 1997, 63, 4267–4271. [Google Scholar] [CrossRef]
- Selvendiran, P.; Driscoll, C.T.; Bushey, J.T.; Montesdeoca, M.R. Wetland influence on mercury fate and transport in a temperate forested watershed. Environ. Pollut. 2008, 154, 46–55. [Google Scholar] [CrossRef]
- Waples, J.S.; Nagy, K.L.; Aiken, G.R.; Ryan, J.N. Dissolution of cinnabar (HgS) in the presence of natural organic matter. Geochim. Cosmochim. Acta 2005, 69, 1575–1588. [Google Scholar] [CrossRef]
- Jones, D.S.; Johnson, N.W.; Mitchell, C.P.J.; Walker, G.M.; Bailey, J.V.; Pastor, J.; Swain, E.B. Diverse Communities of hgcAB+ Microorganisms Methylate Mercury in Freshwater Sediments Subjected to Experimental Sulfate Loading. Environ. Sci. Technol. 2020, 54, 14265–14274. [Google Scholar] [CrossRef] [PubMed]
- King, J.K.; Kostka, J.E.; Frischer, M.E.; Jahnke, R.A. A quantitative relationship that demonstrates mercury methylation rates in marine sediments are based on the community composition and activity of sulfate-reducing bacteria. Environ. Sci. Technol. 2001, 35, 2491–2496. [Google Scholar] [CrossRef] [PubMed]
- Benoit, J.M.; Gilmour, C.C.; Mason, R.P. Sulfide Controls on Mercury Speciation and Bioavailability to Methylating Bacteria in Sediment Pore Waters. Environ. Sci. Technol. 1999, 33, 951–957. [Google Scholar] [CrossRef]
- Jeremiason, J.D.; Engstrom, D.R.; Swain, E.B.; Nater, E.A.; Johnson, B.M.; Almendinger, J.E.; Monson, B.A.; Kolka, R.K. Sulfate addition increases methylmercury production in an experimental wetland. Environ. Sci. Technol. 2006, 40, 3800–3806. [Google Scholar] [CrossRef]
- Coleman Wasik, J.K.; Mitchell, C.P.J.; Engstrom, D.R.; Swain, E.B.; Monson, B.A.; Balogh, S.J.; Jeremiason, J.D.; Branfireun, B.A.; Eggert, S.L.; Kolka, R.K.; et al. Methylmercury declines in a boreal peatland when experimental sulfate deposition decreases. Environ. Sci. Technol. 2012, 46, 6663–6671. [Google Scholar] [CrossRef]
- Mitchell, C.P.J.; Branfireun, B.A.; Kolka, R.K. Assessing sulfate and carbon controls on net methylmercury production in peatlands: An in situ mesocosm approach. Appl. Geochem. 2008, 23, 503–518. [Google Scholar] [CrossRef]
- Jordan, M.P.; Stewart, A.R.; Eagles-Smith, C.A.; Strecker, A.L. Nutrients mediate the effects of temperature on methylmercury concentrations in freshwater zooplankton. Sci. Total Environ. 2019, 667, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Devai, I.; Delaune, R.D.; Patrick, W.H., Jr.; Gambrell, R.P. Changes in methylmercury concentration during storage: Effect of temperature. Org. Geochem. 2001, 32, 755–758. [Google Scholar] [CrossRef]
- Curtis, A.N.; Bourne, K.; Borsuk, M.E.; Buckman, K.L.; Demidenko, E.; Taylor, V.F.; Chen, C.Y. Effects of temperature, salinity, and sediment organic carbon on methylmercury bioaccumulation in an estuarine amphipod. Sci. Total Environ. 2019, 687, 907–916. [Google Scholar] [CrossRef]
- Buckman, K.L.; Seelen, E.A.; Mason, R.P.; Balcom, P.; Taylor, V.F.; Ward, J.E.; Chen, C.Y. Sediment organic carbon and temperature effects on methylmercury concentration: A mesocosm experiment. Sci. Total Environ. 2019, 666, 1316–1326. [Google Scholar] [CrossRef] [PubMed]
- Braaten, H.F.V.; de Wit, H.A.; Fjeld, E.; Rognerud, S.; Lydersen, E.; Larssen, T. Environmental factors influencing mercury speciation in Subarctic and Boreal lakes. Sci. Total Environ. 2014, 476, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Kotnik, J.; Horvat, M.; Jereb, V. Modelling of mercury geochemical cycle in Lake Velenje, Slovenia. Environ. Model. Softw. 2002, 17, 593–611. [Google Scholar] [CrossRef]
- Wang, T.; Driscoll, C.T.; Hwang, K.; Chandler, D.; Montesdeoca, M. Total and methylmercury concentrations in ground and surface waters in natural and restored freshwater wetlands in northern New York. Ecotoxicology 2020, 29, 1602–1613. [Google Scholar] [CrossRef] [PubMed]
- Bravo, A.G.; Bouchet, S.; Guédron, S.; Amouroux, D.; Dominik, J.; Zopfi, J. High methylmercury production under ferruginous conditions in sediments impacted by sewage treatment plant discharges. Water Res. 2015, 80, 245–255. [Google Scholar] [CrossRef]
- Chen, B.; Chen, P.; He, B.; Yin, Y.G.; Fang, L.C.; Wang, X.W.; Liu, H.T.; Yang, L.H.; Luan, T.G. Identification of mercury methylation product by tert-butyl compounds in aqueous solution under light irradiation. Mar. Pollut. Bull. 2015, 98, 40–46. [Google Scholar] [CrossRef]
- Faganeli, J.; Hines, M.E.; Covelli, S.; Emili, A.; Giani, M. Mercury in lagoons: An overview of the importance of the link between geochemistry and biology. Estuar. Coast. Shelf Sci. 2012, 113, 126–132. [Google Scholar] [CrossRef]
| Type | Average Methylmercury Content | Reference |
|---|---|---|
| Freshwater Crab | 0.028 mg/kg | [32] |
| Freshwater Fish | 0.034 mg/kg | [32] |
| Marine Fish | 0.031 mg/kg | [32] |
| Rice | 0.00137 ± 0.00118 mg/kg | [33] |
| Beryx splendens | 0.78 ± 0.56 mg/kg | [34] |
| Atlantic Thunnus thynnus | 0.42 ± 0.06 mg/kg | [34] |
| Thunnus obesus | 0.98 ± 0.34 mg/kg | [34] |
| Tetraptrus audax | 0.51 ± 0.08 mg/kg | [34] |
| Hypophthalmichthys moritrix | 0.18 ± 0.09 mg/kg | [35] |
| Sediment | 0.06 mg/kg to 1.38 mg/kg | [35] |
| Australian Reed | 0.618 mg/kg | [36] |
| Carps | 0.019 mg/kg to 0.063 mg/kg | [37] |
| Long-tailed Tuna | 0.180 mg/kg to 1.460 mg/kg | [38] |
| Sri Lankan Rice | 0.0051 ± 0.37 mg/kg | [39] |
| Type of Demethylation | Mechanism of Action | Characteristics | References |
|---|---|---|---|
| Mer operon demethylation | Genes encode lytic and reductive enzymes that lyse methylmercury to methyl and mercury(II) and reduce mercury(II) to mercury(0) | Widespread in mercury-resistant bacteria | [61,62] |
| Demethylation of methane nutrients | Bacteria produce methanobactin molecules to promote methane oxidation and methylmercury degradation | Uses the methyl group in methylmercury as an auxiliary C1 carbon source for microorganisms | [65] |
| Phytoplankton demethylation | Phytoplankton utilise endogenous reactive oxygen species as the main driver of demethylation | Can degrade methylmercury by dark reaction | [66,67] |
| Selenium amino acid demethylation | Methylmercury and selenoamino acids through the formation of bis(methylmercury) selenide and dimethylmercury as intermediates, with HgSe(s) as the final degradation product | Laboratory stage, not proven in natural environment | [70,71] |
| Photodemethylation pathway 1 | Methylmercury causes C-Hg bond breaking by direct absorption of light energy | Most important demethylation pathway in the aquatic environment, about 56–80% of methylmercury is photodegradable (pathway1, 2, 3) | [85] |
| Photodemethylation pathway 2 | Reactive oxygen species (ROS) and other photoactive substances, produced by organic molecules, ions, suspended solids, etc., attack the C-Hg bond and degrade MeHg when exposed to sunlight | [86,87] | |
| Photodemethylation pathway 3 | When MeHg is complexed with photochemically active dissolved organic matter (DOM) and exposed to light, the excited DOM-MeHg complex may undergo intramolecular electron transfer, leading to C-Hg bond breaking | [88] |
| Influencing Factors | Effects on Methylation |
|---|---|
| Redox conditions | The anoxic environment is the primary environment for methylation, but methylation under anoxic and oxidative conditions both may occur. |
| Organic substances | Several recent studies have demonstrated a clear correlation between certain organic substances and methylmercury concentrations. Organic substances act in a number of ways, by stimulating microbial activity and by providing methyl groups. |
| Sulphide | Increased sulphate may stimulate methylation. |
| Temperature | The rate of methylmercury production is generally positively correlated with temperature. |
| PH | May be negatively correlated, with higher levels of methylmercury prevalent at low pH. |
| Iron and manganese oxides | Methylation levels are highest in areas rich in dissolved iron and organic matter, but it is still not possible to determine their exact effect or impact. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, W.; Gan, R.; Xian, B.; Wu, T.; Wu, G.; Huang, S.; Wang, R.; Liu, Z.; Zhang, Q.; Bai, S.; et al. Overview of Methylation and Demethylation Mechanisms and Influencing Factors of Mercury in Water. Toxics 2024, 12, 715. https://doi.org/10.3390/toxics12100715
Zhao W, Gan R, Xian B, Wu T, Wu G, Huang S, Wang R, Liu Z, Zhang Q, Bai S, et al. Overview of Methylation and Demethylation Mechanisms and Influencing Factors of Mercury in Water. Toxics. 2024; 12(10):715. https://doi.org/10.3390/toxics12100715
Chicago/Turabian StyleZhao, Wenyu, Runjie Gan, Bensen Xian, Tong Wu, Guoping Wu, Shixin Huang, Ronghua Wang, Zixuan Liu, Qin Zhang, Shaoyuan Bai, and et al. 2024. "Overview of Methylation and Demethylation Mechanisms and Influencing Factors of Mercury in Water" Toxics 12, no. 10: 715. https://doi.org/10.3390/toxics12100715





