Metabolomics Provides Novel Insights into the Potential Toxicity Associated with Heated Tobacco Products, Electronic Cigarettes, and Tobacco Cigarettes on Human Bronchial Epithelial BEAS-2B Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Cell Culture and Experimental Design for Cell Exposure
2.3. Cytotoxicity Evaluation
2.4. Sample Preparation
2.5. LC–MS Conditions
2.6. Extraction of Raw Data and Pre-Processing
2.7. Data Processing and Statistical Analysis
2.8. Feature Annotation and Pathway Analysis
3. Results
3.1. Evaluation of Cytotoxicity
3.2. Evaluation of Metabolomics Data Pre-Treatment
3.3. Impact of the Type of Emission
3.4. Impact of the Exposure Dose
3.5. Feature Identification
3.6. Exogenous Compounds
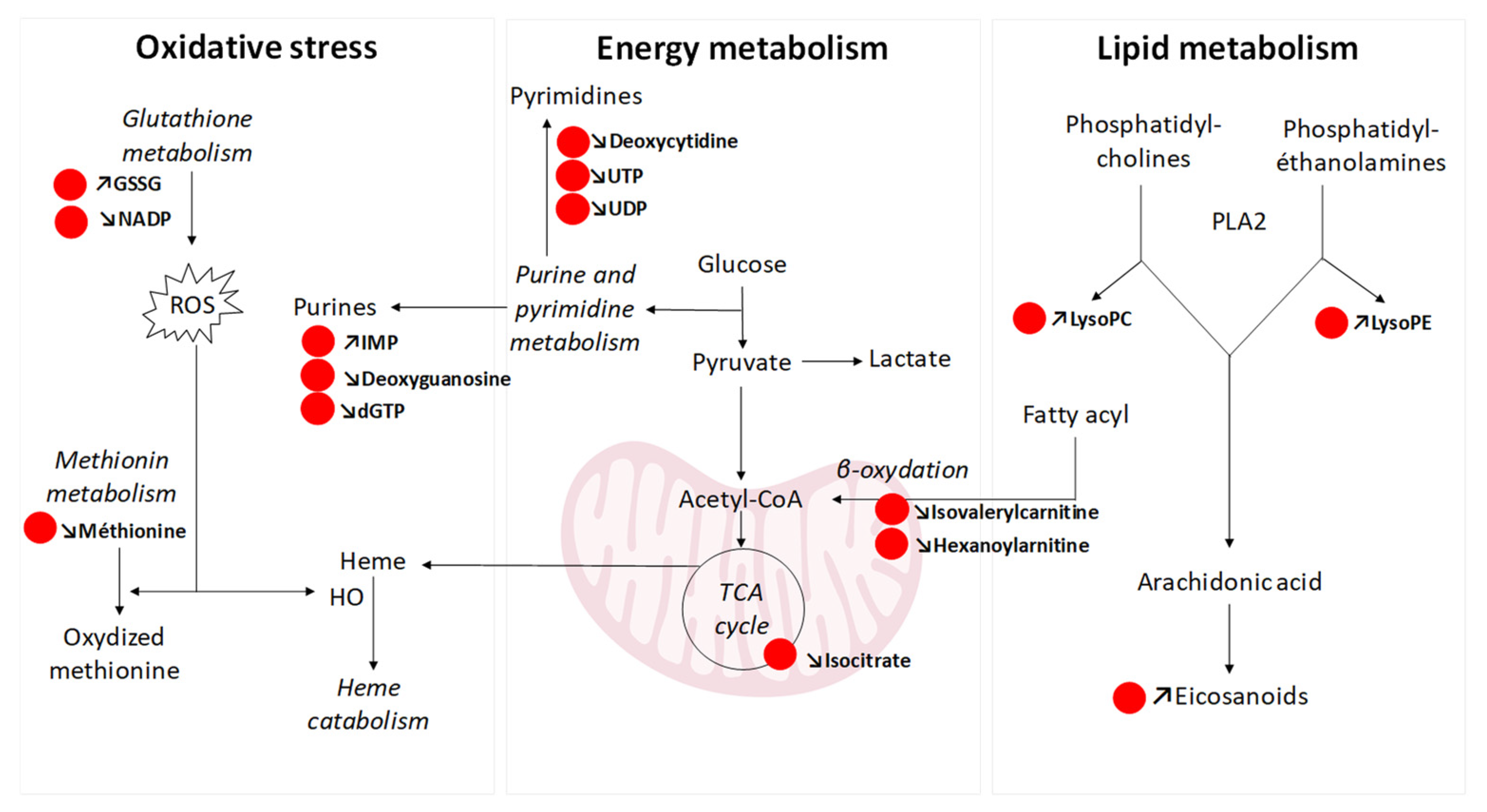
3.7. Endogenous Compounds: Pathway Analysis and Biological Interpretation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALI | Air–liquid interface |
| CCS | Collision cross-section |
| e-cigs | Electronic cigarettes |
| ESI | Electrospray ionization |
| FDR | False discovery rate |
| GSSG | Oxidized glutathione |
| HMDB | Human metabolome database |
| HO | Heme oxygenase |
| HRMS | High-resolution mass spectrometry |
| HTPs | Heated tobacco products |
| IMS | Ion mobility spectrometry |
| LC | Liquid chromatography |
| LDH | Lactate dehydrogenase |
| Log2(FCD1) | Log2(FC) D0 vs. D1 |
| Log2(FCD2) | Log2(FC) D0 vs. D2 |
| LysoPC | Lysophosphatidylcholine |
| LysoPE | Lysophosphatidylethanolamine |
| m/z | Mass-to-charge ratio |
| Mb-18W | Modbox e-cig model set at 18 W |
| Mb-30W | Modbox e-cig model set at 30 W |
| MS | Mass spectrometry |
| Nrf2 | Nuclear factor erythroid 2-related factor 2 |
| PBS | Phosphate buffer solution |
| PLA2 | Phospholipase A2 |
| QC | Quality control |
| QToF | Quadrupole time-of-flight |
| ROS | Reactive oxygen species |
| Rt | Retention time |
| TCA cycle | Tricarboxylic acid cycle |
| UPLC | Ultra high-performance liquid chromatography |
| VIP | Variable importance in the projection |
| PLS-DA | Partial least-squares discriminant analysis |
References
- Reitsma, M.B.; Kendrick, P.J.; Ababneh, E.; Abbafati, C.; Abbasi-Kangevari, M.; Abdoli, A.; Abedi, A.; Abhilash, E.S.; Abila, D.B.; Aboyans, V.; et al. Spatial, Temporal, and Demographic Patterns in Prevalence of Smoking Tobacco Use and Attributable Disease Burden in 204 Countries and Territories, 1990–2019: A Systematic Analysis from the Global Burden of Disease Study 2019. Lancet 2021, 397, 2337–2360. [Google Scholar] [CrossRef]
- National Center for Chronic Disease Prevention and Health Promotion (US) Office on Smoking and Health. The Health Consequences of Smoking—50 Years of Progress: A Report of the Surgeon General; Reports of the Surgeon General; Centers for Disease Control and Prevention (US): Atlanta, GA, USA, 2014. [Google Scholar]
- WHO Report on the Global Tobacco Epidemic. 2023. Available online: https://www.who.int/teams/health-promotion/tobacco-control/global-tobacco-report-2023 (accessed on 18 December 2023).
- Le Foll, B.; Piper, M.E.; Fowler, C.D.; Tonstad, S.; Bierut, L.; Lu, L.; Jha, P.; Hall, W.D. Tobacco and Nicotine Use. Nat. Rev. Dis. Primers 2022, 8, 19. [Google Scholar] [CrossRef]
- Jha, P. The Hazards of Smoking and the Benefits of Cessation: A Critical Summation of the Epidemiological Evidence in High-Income Countries. eLife 2020, 9, e49979. [Google Scholar] [CrossRef] [PubMed]
- United States Public Health Service Office of the Surgeon General; National Center for Chronic Disease Prevention and Health Promotion (US) Office on Smoking and Health. Smoking Cessation: A Report of the Surgeon General; US Department of Health and Human Services: Washington, DC, USA, 2020. [Google Scholar]
- Dusautoir, R.; Zarcone, G.; Verriele, M.; Garçon, G.; Fronval, I.; Beauval, N.; Allorge, D.; Riffault, V.; Locoge, N.; Lo-Guidice, J.-M.; et al. Comparison of the Chemical Composition of Aerosols from Heated Tobacco Products, Electronic Cigarettes and Tobacco Cigarettes and Their Toxic Impacts on the Human Bronchial Epithelial BEAS-2B Cells. J. Hazard. Mater. 2021, 401, 123417. [Google Scholar] [CrossRef] [PubMed]
- Beauval, N.; Antherieu, S.; Soyez, M.; Gengler, N.; Grova, N.; Howsam, M.; Hardy, E.M.; Fischer, M.; Appenzeller, B.M.R.; Goossens, J.-F.; et al. Chemical Evaluation of Electronic Cigarettes: Multicomponent Analysis of Liquid Refills and Their Corresponding Aerosols. J. Anal. Toxicol. 2017, 41, 670–678. [Google Scholar] [CrossRef] [PubMed]
- Zarcone, G.; Lenski, M.; Martinez, T.; Talahari, S.; Simonin, O.; Garçon, G.; Allorge, D.; Nesslany, F.; Lo-Guidice, J.-M.; Platel, A.; et al. Impact of Electronic Cigarettes, Heated Tobacco Products and Conventional Cigarettes on the Generation of Oxidative Stress and Genetic and Epigenetic Lesions in Human Bronchial Epithelial BEAS-2B Cells. Toxics 2023, 11, 847. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. “Metabonomics”: Understanding the Metabolic Responses of Living Systems to Pathophysiological Stimuli via Multivariate Statistical Analysis of Biological NMR Spectroscopic Data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Chaumont, M.; Tagliatti, V.; Channan, E.M.; Colet, J.-M.; Bernard, A.; Morra, S.; Deprez, G.; Van Muylem, A.; Debbas, N.; Schaefer, T.; et al. Short Halt in Vaping Modifies Cardiorespiratory Parameters and Urine Metabolome: A Randomized Trial. Am. J. Physiol. Lung Cell Mol. Physiol. 2020, 318, L331–L344. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Lin, C.J.; Yates, C.R.; Prasad, G.L. Metabolomics Analysis Identified Reduced Levels of Xenobiotics, Oxidative Stress, and Improved Vitamin Metabolism in Smokers Switched to Vuse Electronic Nicotine Delivery System. Nicotine Tob. Res. 2020, 23, 1133–1142. [Google Scholar] [CrossRef]
- Wang, Q.; Ji, X.; Rahman, I. Dysregulated Metabolites Serve as Novel Biomarkers for Metabolic Diseases Caused by E-Cigarette Vaping and Cigarette Smoking. Metabolites 2021, 11, 345. [Google Scholar] [CrossRef]
- Hsiao, Y.-C.; Matulewicz, R.S.; Sherman, S.E.; Jaspers, I.; Weitzman, M.L.; Gordon, T.; Liu, C.-W.; Yang, Y.; Lu, K.; Bjurlin, M.A. Untargeted Metabolomics to Characterize the Urinary Chemical Landscape of E-Cigarette Users. Chem. Res. Toxicol. 2023, 36, 630–642. [Google Scholar] [CrossRef]
- Alqahtani, S.; Cooper, B.; Spears, C.A.; Wright, C.; Shannahan, J. Electronic Nicotine Delivery System-Induced Alterations in Oral Health via Saliva Assessment. Exp. Biol. Med. 2020, 245, 1319–1325. [Google Scholar] [CrossRef]
- Ren, X.; Lin, L.; Sun, Q.; Li, T.; Sun, M.; Sun, Z.; Duan, J. Metabolomics-Based Safety Evaluation of Acute Exposure to Electronic Cigarettes in Mice. Sci. Total Environ. 2022, 839, 156392. [Google Scholar] [CrossRef]
- Madison, M.C.; Landers, C.T.; Gu, B.-H.; Chang, C.-Y.; Tung, H.-Y.; You, R.; Hong, M.J.; Baghaei, N.; Song, L.-Z.; Porter, P.; et al. Electronic Cigarettes Disrupt Lung Lipid Homeostasis and Innate Immunity Independent of Nicotine. J. Clin. Investig. 2019, 129, 4290–4304. [Google Scholar] [CrossRef]
- Moshensky, A.; Du, M.; Shin, J.; Advani, I.; Gunge, D.; Mathew, D.; Alkolla, R.; Du, A.; Javier, C.; Ma, L.; et al. Vaping-Induced Metabolomic Signatures in the Circulation of Mice Are Driven by Device Type, e-Liquid, Exposure Duration and Sex. ERJ Open Res. 2021, 7, 00229–02021. [Google Scholar] [CrossRef]
- Aug, A.; Altraja, S.; Kilk, K.; Porosk, R.; Soomets, U.; Altraja, A. E-Cigarette Affects the Metabolome of Primary Normal Human Bronchial Epithelial Cells. PLoS ONE 2015, 10, e0142053. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.R.; Jarrell, Z.R.; Orr, M.; Liu, K.H.; Go, Y.-M.; Jones, D.P. Metabolome-Wide Association Study of Flavorant Vanillin Exposure in Bronchial Epithelial Cells Reveals Disease-Related Perturbations in Metabolism. Environ. Int. 2021, 147, 106323. [Google Scholar] [CrossRef] [PubMed]
- Jarrell, Z.R.; Smith, M.R.; He, X.; Orr, M.; Jones, D.P.; Go, Y.-M. Firsthand and Secondhand Exposure Levels of Maltol-Flavored Electronic Nicotine Delivery System Vapors Disrupt Amino Acid Metabolism. Toxicol. Sci. 2021, 182, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Titz, B.; Boué, S.; Phillips, B.; Talikka, M.; Vihervaara, T.; Schneider, T.; Nury, C.; Elamin, A.; Guedj, E.; Peck, M.J.; et al. Effects of Cigarette Smoke, Cessation, and Switching to Two Heat-Not-Burn Tobacco Products on Lung Lipid Metabolism in C57BL/6 and Apoe-/- Mice-An Integrative Systems Toxicology Analysis. Toxicol. Sci. 2016, 149, 441–457. [Google Scholar] [CrossRef] [PubMed]
- Titz, B.; Szostak, J.; Sewer, A.; Phillips, B.; Nury, C.; Schneider, T.; Dijon, S.; Lavrynenko, O.; Elamin, A.; Guedj, E.; et al. Multi-Omics Systems Toxicology Study of Mouse Lung Assessing the Effects of Aerosols from Two Heat-Not-Burn Tobacco Products and Cigarette Smoke. Comput. Struct. Biotechnol. J. 2020, 18, 1056–1073. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, F.; Titz, B.; Sewer, A.; Lo Sasso, G.; Scotti, E.; Schlage, W.K.; Mathis, C.; Leroy, P.; Majeed, S.; Torres, L.O.; et al. Comparative Systems Toxicology Analysis of Cigarette Smoke and Aerosol from a Candidate Modified Risk Tobacco Product in Organotypic Human Gingival Epithelial Cultures: A 3-Day Repeated Exposure Study. Food Chem. Toxicol. 2017, 101, 15–35. [Google Scholar] [CrossRef]
- Anthérieu, S.; Garat, A.; Beauval, N.; Soyez, M.; Allorge, D.; Garçon, G.; Lo-Guidice, J.-M. Comparison of Cellular and Transcriptomic Effects between Electronic Cigarette Vapor and Cigarette Smoke in Human Bronchial Epithelial Cells. Toxicol. In Vitro 2017, 45, 417–425. [Google Scholar] [CrossRef]
- León, Z.; García-Cañaveras, J.C.; Donato, M.T.; Lahoz, A. Mammalian Cell Metabolomics: Experimental Design and Sample Preparation. Electrophoresis 2013, 34, 2762–2775. [Google Scholar] [CrossRef]
- Lenski, M.; Maallem, S.; Zarcone, G.; Garçon, G.; Lo-Guidice, J.-M.; Anthérieu, S.; Allorge, D. Prediction of a Large-Scale Database of Collision Cross-Section and Retention Time Using Machine Learning to Reduce False Positive Annotations in Untargeted Metabolomics. Metabolites 2023, 13, 282. [Google Scholar] [CrossRef]
- R Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022.
- Benjamini, Y.; Yekutieli, D. The Control of the False Discovery Rate in Multiple Testing under Dependency. Ann. Statist. 2001, 29, 1165–1188. [Google Scholar] [CrossRef]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A Public Repository for Sharing Mass Spectral Data for Life Sciences. J. Mass Spectrom 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed Minimum Reporting Standards for Chemical Analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Stabbert, R.; Schäfer, K.-H.; Biefel, C.; Rustemeier, K. Analysis of Aromatic Amines in Cigarette Smoke. Rapid Commun Mass Spectrom 2003, 17, 2125–2132. [Google Scholar] [CrossRef] [PubMed]
- Bie, Z.; Lu, W.; Zhu, Y.; Chen, Y.; Ren, H.; Ji, L. Rapid Determination of Six Carcinogenic Primary Aromatic Amines in Mainstream Cigarette Smoke by Two-Dimensional Online Solid Phase Extraction Combined with Liquid Chromatography Tandem Mass Spectrometry. J. Chromatogr. A 2017, 1482, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Liu, Q.; Cai, W.; Shao, X. Analysis of Scopoletin and Caffeic Acid in Tobacco by GC–MS After a Rapid Derivatization Procedure. Chroma 2009, 69, 743–748. [Google Scholar] [CrossRef]
- Pfau, W.; Skog, K. Exposure to Beta-Carbolines Norharman and Harman. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2004, 802, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Vivarelli, F.; Canistro, D.; Cirillo, S.; Elias, R.J.; Granata, S.; Mussoni, M.; Burattini, S.; Falcieri, E.; Turrini, E.; Fimognari, C.; et al. Unburned Tobacco Cigarette Smoke Alters Rat Ultrastructural Lung Airways and DNA. Nicotine Tob. Res. 2021, 23, 2127–2134. [Google Scholar] [CrossRef]
- Nichols, W.K.; Mehta, R.; Skordos, K.; Macé, K.; Pfeifer, A.M.A.; Carr, B.A.; Minko, T.; Burchiel, S.W.; Yost, G.S. 3-Methylindole-Induced Toxicity to Human Bronchial Epithelial Cell Lines. Toxicol. Sci. 2003, 71, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Weems, J.M.; Lamb, J.G.; D’Agostino, J.; Ding, X.; Yost, G.S. Potent Mutagenicity of 3-Methylindole Requires Pulmonary Cytochrome P450-Mediated Bioactivation: A Comparison to the Prototype Cigarette Smoke Mutagens B(a)P and NNK. Chem. Res. Toxicol. 2010, 23, 1682–1690. [Google Scholar] [CrossRef] [PubMed]
- Weems, J.M.; Cutler, N.S.; Moore, C.; Nichols, W.K.; Martin, D.; Makin, E.; Lamb, J.G.; Yost, G.S. 3-Methylindole Is Mutagenic and a Possible Pulmonary Carcinogen. Toxicol. Sci. 2009, 112, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Abraham, N.G.; Kappas, A. Pharmacological and Clinical Aspects of Heme Oxygenase. Pharmacol. Rev. 2008, 60, 79–127. [Google Scholar] [CrossRef] [PubMed]
- Even, B.; Fayad-Kobeissi, S.; Gagliolo, J.-M.; Motterlini, R.; Boczkowski, J.; Foresti, R.; Dagouassat, M. Heme Oxygenase-1 Induction Attenuates Senescence in Chronic Obstructive Pulmonary Disease Lung Fibroblasts by Protecting against Mitochondria Dysfunction. Aging Cell 2018, 17, e12837. [Google Scholar] [CrossRef]
- Song, Y.; Li, R.; Zhang, Y.; Wei, J.; Chen, W.; Chung, C.K.A.; Cai, Z. Mass Spectrometry-Based Metabolomics Reveals the Mechanism of Ambient Fine Particulate Matter and Its Components on Energy Metabolic Reprogramming in BEAS-2B Cells. Sci. Total Environ. 2019, 651, 3139–3150. [Google Scholar] [CrossRef]
- Ma, J.; Zhong, M.; Xiong, Y.; Gao, Z.; Wu, Z.; Liu, Y.; Hong, X. Emerging Roles of Nucleotide Metabolism in Cancer Development: Progress and Prospect. Aging 2021, 13, 13349–13358. [Google Scholar] [CrossRef]
- De Vitto, H.; Arachchige, D.B.; Richardson, B.C.; French, J.B. The Intersection of Purine and Mitochondrial Metabolism in Cancer. Cells 2021, 10, 2603. [Google Scholar] [CrossRef]
- Drazic, A.; Winter, J. The Physiological Role of Reversible Methionine Oxidation. Biochim. Biophys. Acta 2014, 1844, 1367–1382. [Google Scholar] [CrossRef]
- Solanki, H.S.; Babu, N.; Jain, A.P.; Bhat, M.Y.; Puttamallesh, V.N.; Advani, J.; Raja, R.; Mangalaparthi, K.K.; Kumar, M.M.; Prasad, T.S.K.; et al. Cigarette Smoke Induces Mitochondrial Metabolic Reprogramming in Lung Cells. Mitochondrion 2018, 40, 58–70. [Google Scholar] [CrossRef]
- Ishikawa, S.; Matsumura, K.; Kitamura, N.; Takanami, Y.; Ito, S. Multi-Omics Analysis: Repeated Exposure of a 3D Bronchial Tissue Culture to Whole-Cigarette Smoke. Toxicol. Vitr. 2019, 54, 251–262. [Google Scholar] [CrossRef]
- McElroy, J.P.; Carmella, S.G.; Heskin, A.K.; Tang, M.K.; Murphy, S.E.; Reisinger, S.A.; Jensen, J.A.; Hatsukami, D.K.; Hecht, S.S.; Shields, P.G. Effects of Cessation of Cigarette Smoking on Eicosanoid Biomarkers of Inflammation and Oxidative Damage. PLoS ONE 2019, 14, e0218386. [Google Scholar] [CrossRef]
- Lundström, S.L.; Balgoma, D.; Wheelock, Å.M.; Haeggström, J.Z.; Dahlén, S.-E.; Wheelock, C.E. Lipid Mediator Profiling in Pulmonary Disease. Curr. Pharm. Biotechnol. 2011, 12, 1026–1052. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.-Y.; Chen, G.G. Cigarette Smoking, Cyclooxygenase-2 Pathway and Cancer. Biochim. Biophys. Acta 2011, 1815, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Engel, K.M.; Schiller, J.; Galuska, C.E.; Fuchs, B. Phospholipases and Reactive Oxygen Species Derived Lipid Biomarkers in Healthy and Diseased Humans and Animals—A Focus on Lysophosphatidylcholine. Front. Physiol. 2021, 12, 732319. [Google Scholar] [CrossRef] [PubMed]
- ISO 20768:2018; Vapour Products—Routine Analytical Vaping Machine—Definitions and Standard Conditions. ISO: Geneva, Switzerland, 2018.
- ISO 3308:2012; Routine Analytical Cigarette-Smoking Machine—Definitions and Standard Conditions. ISO: Geneva, Switzerland, 2012.
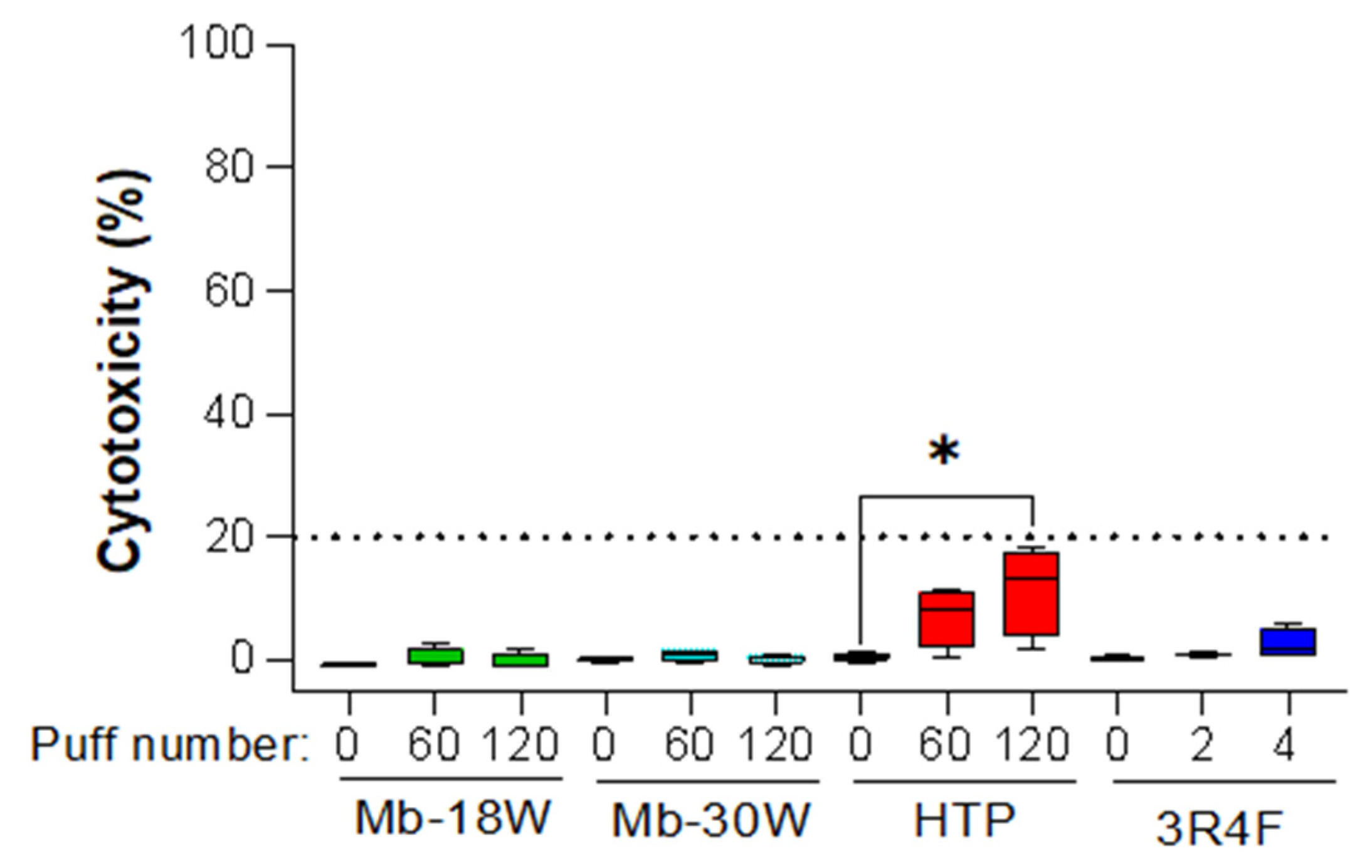
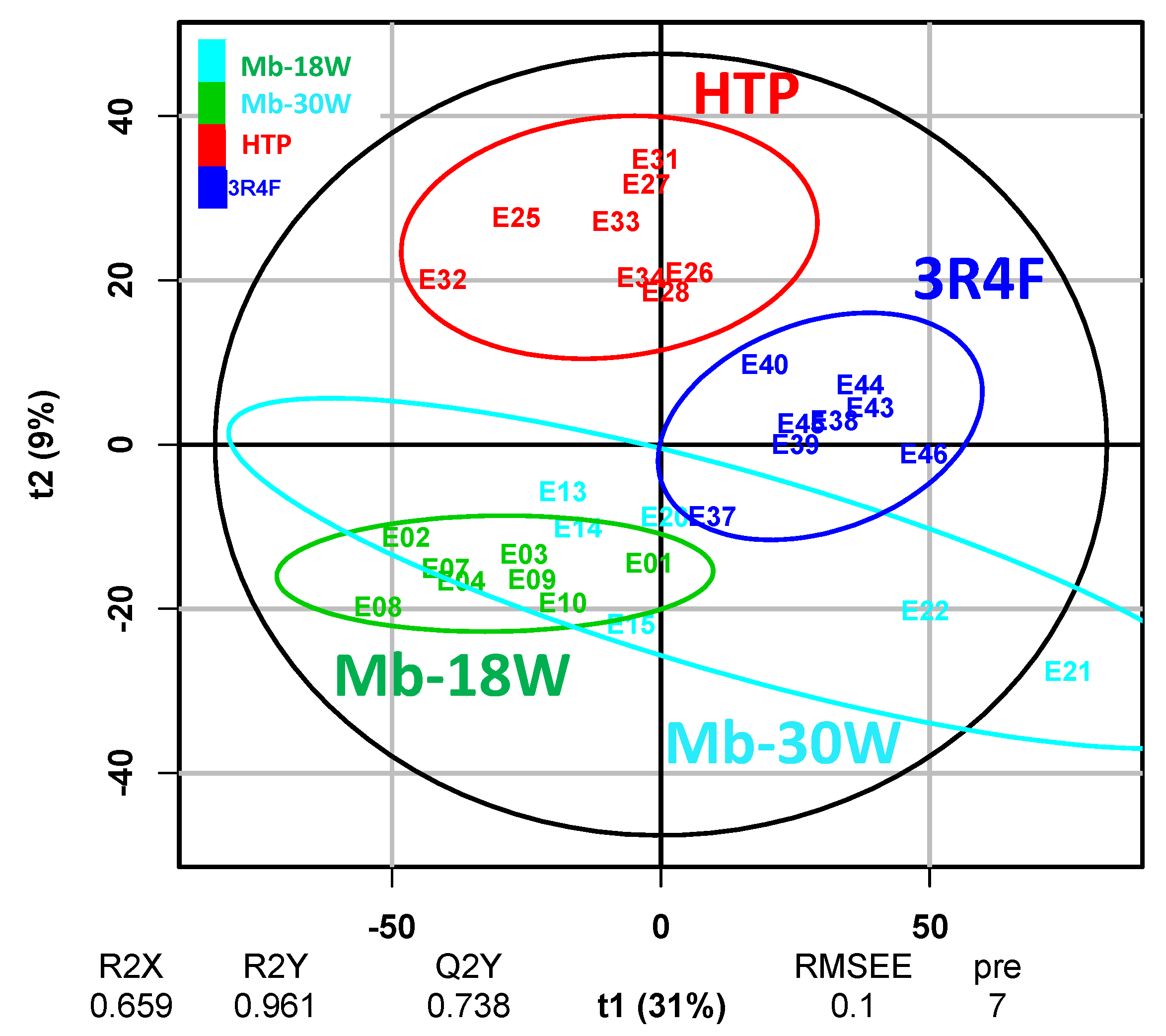
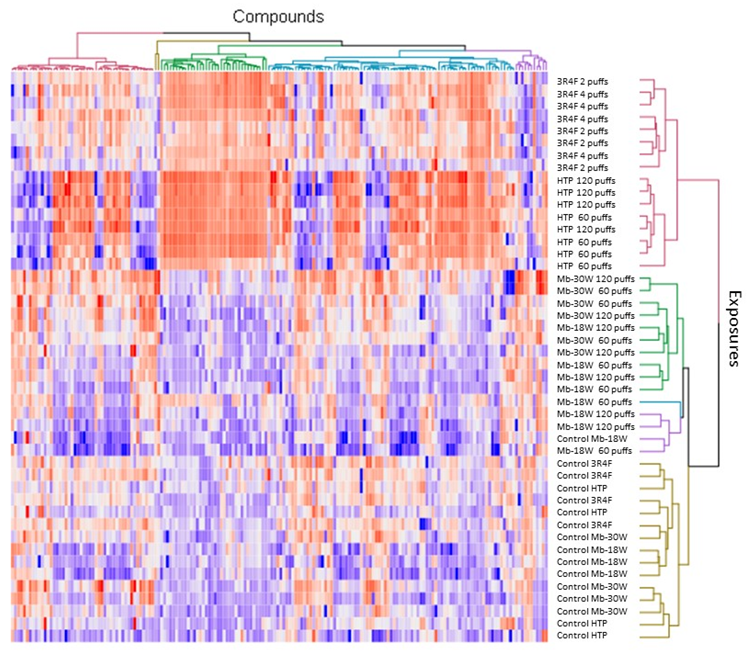
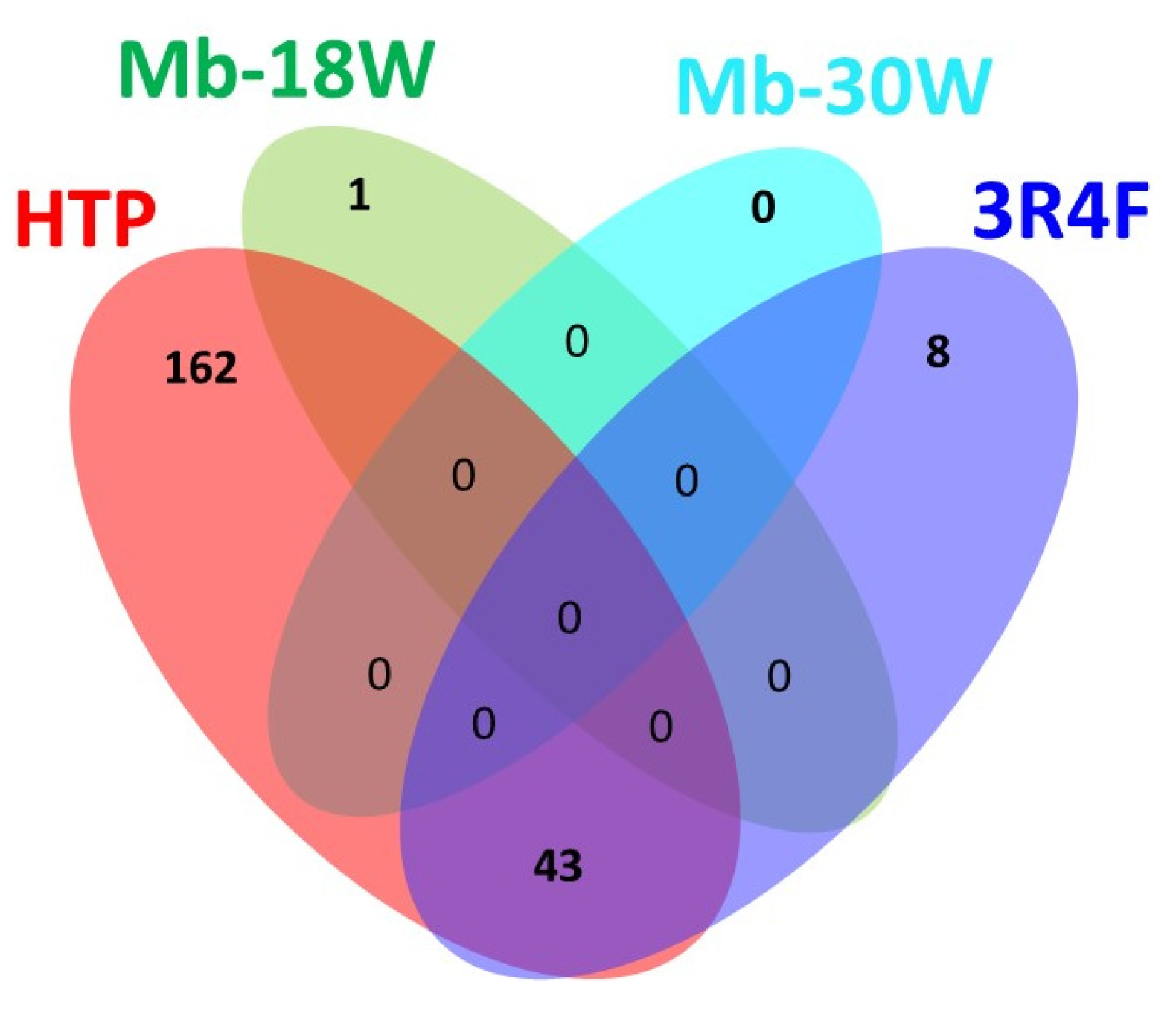
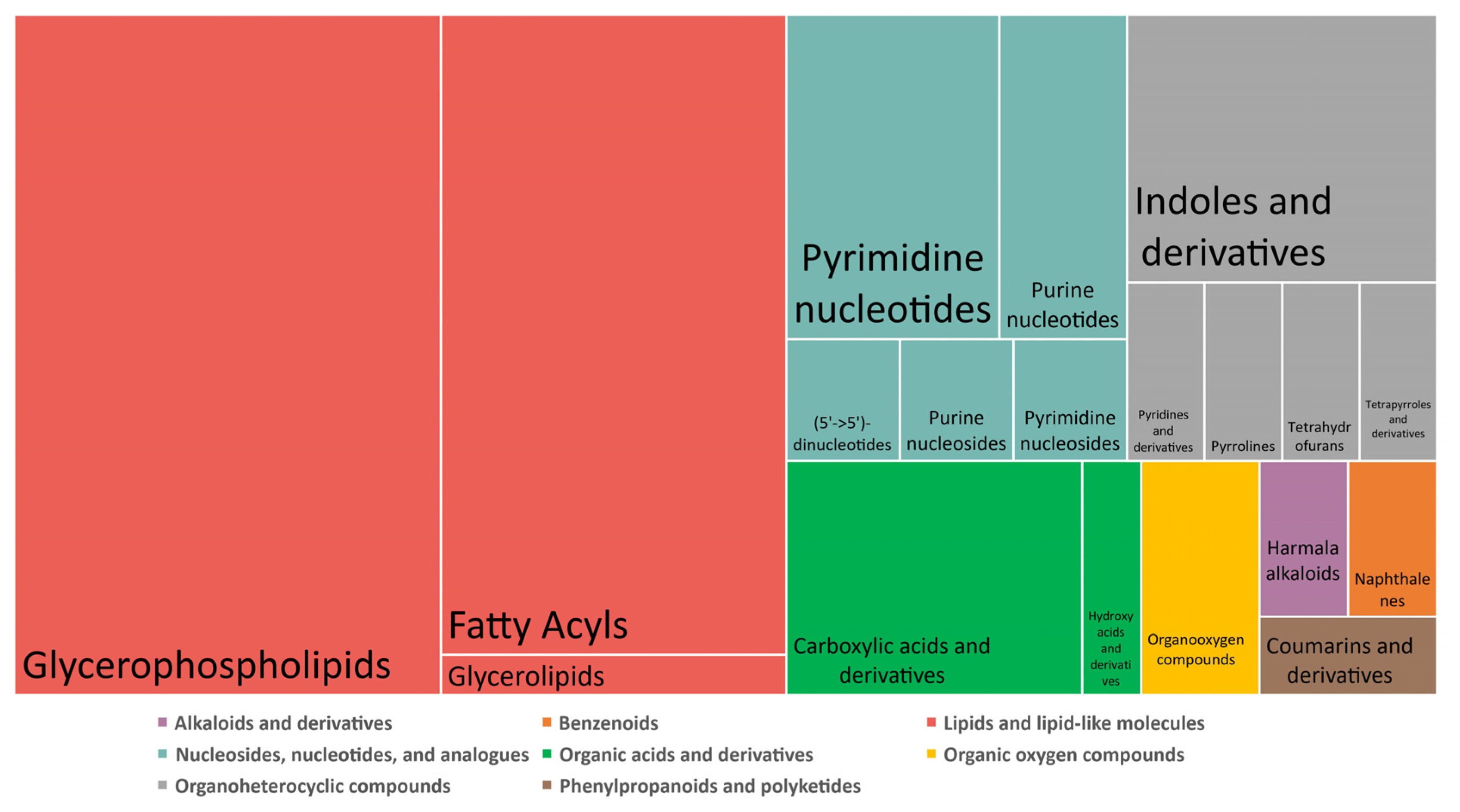
| Biological Question | Exposure Device Duration | Model Sample Type Sample Size | Deregulated Metabolite(s)/Metabolic Pathway(s) * | Conclusions | Independent Study | Ref |
|---|---|---|---|---|---|---|
| Cessation of vaping in regular heavy e-cigarette users | 4th e-cig generation Short-term (5 days) | Human Urine, plasma n = 30 | Serum: no difference. Urine: A specific metabolomic signature characterized the stop-session, including 3-hydroxyisovalerate (↘), pyruvate (↘), trimethylamine oxide (↗), hippurate (↗), and N-phenylacetyl-glycine (↗). | In regular e-cig users, short-term vaping cessation shifted baseline urine metabolome | Yes | [11] |
| Switch from cigarette to e-cig | 1st e-cig generation Short-term (5 days) | Human Urine, plasma n = 75 | ↘ xenobiotic exposure (nicotine and its metabolites, other cigarette smoke constituents). Improved vitamin metabolism and ↘ oxidative stress. | Less toxic environment for consumers of e-cigs and potential health benefits compared to people who smoke cigarettes | No | [12] |
| Effects of chronic e-cig vaping and cigarette smoking | No information on e-cig exposure device Long-term (>2 years) | Human Plasma n = 24 | E-cig vaping deregulated TCA cycle-related metabolites, while cigarette smoking altered sphingolipid metabolism. | Specific metabolic signatures could serve as potential systemic biomarkers for early pathogenesis of cardiopulmonary diseases | Yes | [13] |
| Long-term effects of e-cigs compared to tobacco | No information on e-cig exposure device Long-term effects (>6 months) | Human Urine n = 117 | Metabolomic signature of 839 and 396 features for people who smoke and vape, respectively, including 12% of common metabolites. ↗ acylcarnitines and acylglycines in vapers, suggesting higher lipid peroxidation. Trend of ↗ in cancer-related biomarkers (Me-Fapy) in people who vape. | Deregulation of markers of inflammatory status and fatty acid oxidation in people who vape, as well as a trend of elevated cancer-related biomarkers. | Yes | [14] |
| Effects of e-cigs on oral health | 4th e-cig generation Long-term (one month to 2 years) | Human Saliva n = 30 | Perturbation of 368 metabolites in vapors. ↗ prostaglandins, ↗ leukotrienes (arachidonic acid metabolism). Alterations in immune signaling metabolites (gangliosides, ceramides, angiotensin). | Potential biomarkers of periodontal disease in vapors. | Yes | [15] |
| Acute exposure to e-cigs | 4th e-cig generation Short-term (1 h to 8 h) | Mouse Serum n = 40 | Deregulation of 26 to 50 metabolites after exposure. The type of compound changed over time. Total of 24 metabolic pathways affected, mainly regulated amino acid metabolism, further affected the TCA cycle. | Highlight specific metabolic signatures of e-cigs acute exposure that are potentially beneficial for disease prevention | Yes | [16] |
| Long-term effects of e-cigs with or without nicotine | 2nd e-cig generation Long-term (4 months) | Mouse Bronchoalveolar lavage n = 9 | Independently of nicotine presence, altered lung lipid homeostasis in alveolar macrophages and epithelial cells. Aberrant phospholipids in alveolar macrophages and increased surfactant-associated phospholipids in the airway. Downregulation of innate immunity against viral pathogens in resident macrophages. | Alterations in lipid homeostasis and immune impairment are independent of nicotine, thereby warranting more extensive investigations on the vehicle solvents used in e-cigs | Yes | [17] |
| Effects of the type of e-cig consumption | 2nd, 3rd, 4th e-cig generation 4 to 12 weeks | Mouse Plasma n = 6 | Different alterations in metabolomic profiles depending on the e-cig generation, chemical compounds, duration of exposure, and gender. These signatures have been associated with cardiovascular diseases and can serve as predictors of chronic kidney diseases. | Each e-cig generation and each e-liquid are likely to lead to their own set of health effects. | Yes | [18] |
| Effects of e-liquid compared to cigarette smoke condensate | E-liquid or cigarette smoke condensate Short-term (1 h to 13 h) | HBEC * (at ALI) Intracellular content n = 3 | E-liquid and cigarette smoke condensate affected 24% and 35% of the metabolome, respectively, with biphasic fluctuations: first maximum after 5 h, second maximum after 13 h. Alterations in amino acids, energy, β-oxidation of fatty acid metabolism. | E-liquid profoundly alters the metabolome of HBEC in a manner which is comparable and partially overlapping with the effects of cigarette smoke condensate | Yes | [19] |
| Effects of e-cig vanillin (flavorant) | e-liquid on cells Short-term (18 h) | BEAS-2B cell line Intracellular content n = 3 | Vanillin perturbed specific energy, amino acid, antioxidant, and sphingolipid pathways previously associated with human disease such as lung disease including asthma, idiopathic pulmonary fibrosis, and acute respiratory distress syndrome. | Vanillin could drive the lung metabolic microenvironment to a more pathogenic state. | Yes | [20] |
| Effects of e-cig maltol (flavorant) | 3rd e-cig generation Short-term (1 h) | BEAS-2B cell line Intracellular content n = 3 | Perturbation of oxidative stress with e-liquids with or without maltol. Deregulation of amino acid metabolism specifically with maltol. Many effects of firsthand exposure were also observed with secondhand exposure. | Flavorants in e-liquids impact lung metabolism after both firsthand and secondhand exposure. | Yes | [21] |
| Switch from cigarettes to HTPs or smoke cessation | HTP Long-term (2–8 months) | Mouse Lung intracellular content n = 8 | ↗ candidate surfactant lipids, ↗ inflammatory eicosanoids, ↗ ceramide classes after cigarette exposure that were absent in mice from the cessation group and the switching group to HTPs. | Benefits of tobacco cessation or switching to an HTP for lipidomic lung profile. | No | [22] |
| Switch from cigarettes to HTPs or smoke cessation | HTP Long-term (6 months) | Mouse Lung intracellular content n = 9 | Substantial effects of 3R4F exposure: ↗ inflammatory and oxidative stress responses, ↗ metabolites with immunoregulatory roles (itaconate, polyamines, quinolinate), ↗ metabolites of oxidative stress response (heme–biliverdin–bilirubin pathway). HTP aerosol exposure was associated with fewer to absent effects. | Benefits of tobacco cessation or switching to an HTP for metabolic lung profile | No | [23] |
| Effects of HTPs compared to tobacco | HTP Short-term (3 days) | Human gingival epithelial cells Intracellular content n = 5 | 13 metabolites perturbed after HTP exposure vs. 181 for cigarettes. Reduction in the metabolic impact in HTP aerosol-exposed samples with respect to cigarettes. | Exposure to HTP aerosol had a lower impact on the pathophysiology of human gingival organotypic cultures than cigarette smoke | No | [24] |
| Type of Exposure | D0 vs. D1 | D0 vs. D2 | Common Compounds |
|---|---|---|---|
| 3R4F | 46 | 51 | 46 |
| HTP | 198 | 204 | 197 |
| Mb-18W | 1 | 1 | 1 |
| Mb-30W | 0 | 0 | 0 |
| ESI | Peak | Confidence Level | HMDB | Name | HTP Log2(FC) D0 vs. | 3R4F Log2(FC) D0 vs. | Mb-18W Log2(FC) D0 vs. | Mb-30W Log2(FC) D0 vs. | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D1 | D2 | D1 | D2 | D1 | D2 | D1 | D2 | |||||
| NEG | 0.93_565.0441m/z | 2 | HMDB0000286 | Uridine diphosphate glucose | −1.3 | −1.7 | −0.3 | −1.1 | −0.1 | −0.5 | 0.1 | 0 |
| NEG | 0.97_607.0776n | 2 | HMDB0000290 | Uridine diphosphate-N-acetylglucosamine | −1.1 | −1.2 | −0.1 | −0.7 | −0.3 | −0.3 | −0.7 | −0.7 |
| NEG | 0.97_628.0517m/z | 2 | HMDB0000290 | Uridine diphosphate-N-acetylglucosamine | −2.3 | −2 | −0.2 | −1.1 | −0.3 | −0.3 | −1.3 | −1.2 |
| NEG | 1.00_482.9586m/z | 1 | HMDB00285 | Uridine triphosphate | −0.8 | −1.2 | −0.5 | −1.5 | 0 | −0.4 | 0.3 | 0 |
| NEG | 1.00_506.9926n | 2 | HMDB0001440 | dGTP | −0.5 | −1 | −0.4 | −0.8 | −0.2 | −0.4 | 0.3 | 0 |
| NEG | 1.04_191.0547m/z | 1 | HMDB03072 | Quinic acid | 2.8 | 3 | 1.1 | 2.3 | 1.2 | 1 | −0.4 | 0 |
| NEG | 1.08_427.0267n | 1 | HMDB00061 | Adenosine 3′,5′-diphosphate | 1.1 | 1.5 | 0.4 | 1.1 | 0.3 | 0.1 | 0 | 0 |
| NEG | 1.08_604.0656m/z | 1 | HMDB01163 | Guanosine diphosphate mannose | −1.2 | −1.4 | −0.3 | −0.6 | −0.5 | −0.4 | −0.9 | −0.5 |
| NEG | 1.09_429.0553m/z | 2 | HMDB0060067 | CMP-2-aminoethylphosphonate | −0.6 | −1.3 | −0.3 | −0.5 | 0 | 0 | 0.5 | 0.1 |
| NEG | 1.59_742.0631m/z | 1 | HMDB00217 | NADP | −0.4 | −0.7 | −0.1 | −0.3 | 0.1 | −0.4 | 0 | −0.3 |
| NEG | 1.60_347.0374m/z | 1 | HMDB00175 | Inosine 5′-monophosphate | 3.6 | 3.7 | 1.5 | 3.6 | 0.4 | 0 | 1.4 | 1 |
| NEG | 1.64_148.0420m/z | 1 | HMDB00696 | Methionine | −1.2 | −1.2 | −0.2 | −0.4 | −0.5 | −0.5 | −0.2 | −0.8 |
| NEG | 2.35_321.0676n | 2 | HMDB0013220 | Beta-citryl-l-glutamic acid | −1 | −1.2 | −0.5 | −0.6 | −0.3 | −0.5 | −0.1 | −0.3 |
| NEG | 2.96_612.1481n | 2 | HMDB0003337 | Oxidized glutathione | 0.3 | 0.7 | 0.2 | 1.2 | 0.1 | 0.1 | 0.1 | 0.2 |
| NEG | 14.11_498.2602m/z | 2 | HMDB0011519 | LysoPE 20:5 | 2.3 | 2.7 | 0.8 | 0.3 | 0.3 | 0.6 | 0.5 | 0.4 |
| NEG | 14.98_526.2911m/z | 2 | HMDB0011525 | LysoPE 22:5 | 2.4 | 2.9 | 0.7 | 0.4 | 0.2 | 0.3 | 0.2 | 0.1 |
| NEG | 15.47_506.3213m/z | 2 | HMDB0011512 | LysoPE 20:1 | 0.9 | 1.4 | 0.2 | 0.5 | 0.4 | 0.3 | 0.1 | −0.1 |
| NEG | 15.69_506.3213m/z | 2 | HMDB0011512 | LysoPE 20:1 | 0.9 | 1.8 | 0 | 0.3 | 0.4 | 0.2 | 0.3 | 0.2 |
| POS | 0.96_404.0019n | 2 | HMDB0000295 | Uridine 5′-diphosphate | −1.3 | −1.2 | −0.4 | −0.5 | −0.3 | −0.1 | −0.4 | −1.1 |
| POS | 0.98_489.1138m/z | 1 | HMDB0001413 | Cytidine 5′-diphosphocholine | 5.5 | 6.9 | 0.7 | 3.9 | 0.7 | 1.7 | −0.2 | 1 |
| POS | 0.99_506.9954n | 2 | HMDB0001440 | dGTP | −0.6 | −1.1 | −0.1 | −0.4 | −0.3 | −0.3 | 0 | 0 |
| POS | 1.07_192.0265n | 1 | HMDB0000193 | Isocitric acid | −1 | −1.3 | −0.4 | −0.3 | −0.3 | −0.1 | 0 | −0.1 |
| POS | 1.07_321.0689n | 2 | HMDB0013220 | Beta-citryl-l-glutamic acid | −1.3 | −1.4 | −0.6 | −0.7 | −0.4 | −0.2 | −0.3 | −0.5 |
| POS | 1.07_612.1507n | 2 | HMDB0003337 | Oxidized glutathione | 0.3 | 0.5 | 0 | 1.4 | 0 | 0.2 | 0.1 | 0 |
| POS | 1.11_250.0931m/z | 2 | HMDB0000085 | Deoxyguanosine | −1.4 | −1.5 | −0.6 | −0.8 | −0.1 | 0 | 0 | −0.3 |
| POS | 2.01_227.0902n | 1 | HMDB00014 | Deoxycytidine | −0.5 | −1 | −0.2 | −0.2 | −0.1 | 0 | 0.3 | 0.1 |
| POS | 2.29_321.0690n | 2 | HMDB0013220 | Beta-citryl-l-glutamic acid | −1.3 | −1.5 | −0.5 | −0.7 | −0.4 | −0.1 | 0.1 | −0.5 |
| POS | 2.98_612.1497n | 2 | HMDB0003337 | Oxidized glutathione | 0.4 | 0.9 | 0.2 | 1.7 | 0 | 0.3 | 0.4 | 0.2 |
| POS | 3.63_132.0806m/z | 2 | HMDB0000466 | 3-Methylindole | 4.8 | 6.6 | 1.9 | 2.3 | 4 | 4.5 | 4.3 | 3.3 |
| POS | 3.82_229.1781n | 2 | HMDB0041947 | N1,N8-diacetylspermidine | 4.6 | 6.8 | 2.8 | 6.5 | 4.2 | 4.4 | 1.5 | 2.8 |
| POS | 3.83_230.1859m/z | 2 | HMDB0041947 | N1,N8-diacetylspermidine | 3.1 | 4.9 | 1.7 | 4.2 | 1.5 | 1.7 | 1.1 | 1.9 |
| POS | 4.22_221.1280m/z | 2 | HMDB0002096 | 3-indolebutyric acid | 8.6 | 9.6 | 6.2 | 8.7 | 2.1 | 0.9 | 0 | 0.4 |
| POS | 4.53_143.0734n | 2 | HMDB0243964 | 1-naphthylamine | 15.3 | 15.6 | 9.9 | 11.9 | 5 | 7.5 | 4.5 | 6.6 |
| POS | 5.62_163.1228m/z | 1 | HMDB0001934 | Nicotine | 16.5 | 17.2 | 11.9 | 14.3 | 7.6 | 8.9 | 11.9 | 12.8 |
| POS | 6.54_191.1176m/z | 2 | HMDB0004369 | N-methylserotonin | 10.5 | 11.3 | 6.5 | 9 | 2.3 | 3.9 | −0.3 | 1.2 |
| POS | 6.59_190.0840n | 2 | HMDB0000325 | 3-hydroxysuberic acid | 6.6 | 6.8 | 1.9 | 3.2 | −0.1 | −1 | 1.7 | 3.2 |
| POS | 7.26_187.0629n | 2 | HMDB0000734 | Indoleacrylic acid | −0.9 | −0.8 | −0.2 | −0.2 | −0.3 | −0.2 | 0 | −0.5 |
| POS | 8.18_246.1697m/z | 2 | HMDB0000688 | Isovalerylcarnitine | −0.9 | −1.4 | −0.7 | −1.3 | −0.5 | −0.3 | −0.2 | −0.6 |
| POS | 8.53_169.0760m/z | 2 | HMDB0012897 | Beta-carboline | 9.5 | 10.2 | 7.6 | 8.8 | 1.7 | 2 | 0.6 | 0.1 |
| POS | 8.82_183.0914m/z | 2 | HMDB0035196 | Harman | 6.6 | 7 | 6.4 | 7.5 | 0.1 | 0.6 | −0.6 | −0.5 |
| POS | 9.00_215.1177m/z | 2 | HMDB0001389 | Melatonin | 11.1 | 12.4 | 5.1 | 7.5 | 3.5 | 4.9 | 6.9 | 7.1 |
| POS | 9.22_260.1851m/z | 2 | HMDB0000705 | Hexanoylcarnitine | −1 | −0.9 | −0.6 | −0.7 | −0.1 | 0 | 0.5 | 0.2 |
| POS | 9.34_193.0494m/z | 2 | HMDB0034344 | Scopoleptin | 13.1 | 13.4 | 6.9 | 9.5 | −0.6 | −1 | −0.2 | 0.1 |
| POS | 11.43_236.2004m/z | 2 | HMDB0036823 | Theaspirane | −2.4 | −3.2 | −2 | −2.5 | −1 | −1 | −0.8 | −1.3 |
| POS | 12.54_262.1781n | 2 | HMDB0032297 | Glyceryl 5-hydroxydecanoate | 0.6 | 1.6 | −0.1 | 1 | 1.9 | 2.9 | 0.6 | 1 |
| POS | 12.78_391.1874m/z | 3 | - | Eicosanoid | 11.3 | 11.8 | 6.6 | 8.9 | −0.5 | 1 | −0.9 | −0.6 |
| POS | 12.85_253.1335m/z | 2 | HMDB0037554 | Rollipyrrole | 11.7 | 10.3 | 10.9 | 11.5 | −5.9 | −5.7 | 3.7 | −0.1 |
| POS | 12.93_377.2081m/z | 3 | - | Eicosanoid | 11.1 | 10.9 | 10 | 10.8 | 4.9 | 5.7 | −9.3 | −0.7 |
| POS | 13.03_391.1889m/z | 3 | - | Eicosanoid | 5.3 | 6.4 | 2 | 4.1 | 0 | 0.5 | 0 | 0 |
| POS | 13.15_391.1875m/z | 3 | - | Eicosanoid | 9.2 | 9.6 | 2.7 | 5.7 | 0.9 | 0.9 | 2.1 | 1 |
| POS | 13.17_377.2074m/z | 3 | - | Eicosanoid | 12.1 | 11 | 8.4 | 8.3 | 11.4 | 2.1 | 2.8 | 2.2 |
| POS | 13.36_336.2287n | 3 | - | Eicosanoid | 9.5 | 10.4 | 6.3 | 8.2 | 0.4 | 1.7 | 0.4 | 1 |
| POS | 13.37_465.2850n | 2 | HMDB0010380 | LysoPC(14:1) | 2.1 | 2.8 | 0 | 0.5 | 0.4 | 0.8 | 0.2 | 0.7 |
| POS | 13.52_377.2078m/z | 3 | - | Eicosanoid | 11.4 | 11.4 | 6.6 | 9.3 | 3 | 2.4 | −1.1 | 0.9 |
| POS | 13.61_583.2540m/z | 2 | HMDB01008 | Biliverdin | 1.7 | 0.3 | 1.5 | 2.1 | −0.3 | 0.4 | 0 | 0.1 |
| POS | 13.86_467.3004n | 2 | HMDB0010379 | LysoPC(14:0/0:0) | 1.3 | 1.6 | 0.3 | 0.4 | 0.2 | 0.5 | 0.3 | 0.4 |
| POS | 13.97_359.1982m/z | 3 | - | Eicosanoid | 9.3 | 9.2 | 3.2 | 5 | 0.1 | −0.6 | 1.7 | 1.5 |
| POS | 14.07_499.2692n | 2 | HMDB0011489 | LysoPE(0:0/20:5) | 2.4 | 3 | 0.4 | 0.5 | 0.3 | 0.9 | 0.6 | 1.3 |
| POS | 14.12_541.3164n | 2 | HMDB0010397 | LysoPC(20:5) | 5.5 | 7.1 | 0.8 | 1.5 | 1.1 | 1.5 | 2.3 | 0.9 |
| POS | 14.18_373.1770m/z | 3 | - | Eicosanoid | 14.9 | 13.9 | 4.1 | 5.3 | −5.1 | −10.7 | 0.2 | 5.9 |
| POS | 14.24_493.3160n | 2 | HMDB0010383 | LysoPC(16:1/0:0) | 1.2 | 1.7 | 0 | 0.1 | 0 | 0.2 | 0 | 0.2 |
| POS | 14.27_518.3212m/z | 2 | HMDB0010382 | LysoPC(16:0) | 2.4 | 3.5 | −0.3 | 0.4 | 1 | 1.6 | 1.4 | 0.7 |
| POS | 14.42_359.1980m/z | 3 | - | Eicosanoid | 17.1 | 16 | 10.5 | 11.5 | 1.5 | −4.6 | 0 | 0 |
| POS | 14.66_322.2497n | 3 | - | Eicosanoid | 9.4 | 8.8 | 0 | 2.3 | −1 | 5.5 | 2.1 | 4.3 |
| POS | 14.68_519.3316n | 2 | HMDB0010386 | LysoPC(18:2) | 2 | 2.9 | 0 | 0.4 | 0 | 0.5 | 0.3 | 0.5 |
| POS | 14.72_543.3316n | 2 | HMDB0010395 | LysoPC(20:4) | 2.9 | 4.2 | 0.1 | 0.7 | 0.4 | 0.6 | 1.1 | 0.4 |
| POS | 14.83_508.3388m/z | 2 | HMDB0012108 | LPC 17:1 | 1.9 | 2.5 | −0.3 | 0.4 | 0.9 | 1.6 | −0.1 | 0.5 |
| POS | 14.85_510.3983m/z | 2 | HMDB0072866 | MG(10:0/0:0/0:0) | −3.2 | −7.3 | −1.5 | −4.4 | −0.9 | −0.1 | 1.3 | −2.1 |
| POS | 14.96_519.3317n | 2 | HMDB0010386 | LysoPC(18:2) | 1 | 1.5 | −0.3 | −0.1 | 0.2 | 0.3 | 0.2 | 0.4 |
| POS | 15.00_569.3475n | 2 | HMDB0010403 | LysoPC(22:5) | 4.6 | 6.5 | 0.4 | 1.3 | 0.9 | 1 | 1.8 | 1.3 |
| POS | 15.07_495.3320n | 2 | HMDB0010382 | LysoPC(16:0) | 0.7 | 1.1 | 0.1 | 0.1 | 0.1 | 0.3 | 0.5 | 0.3 |
| POS | 15.21_584.3104m/z | 2 | HMDB0010393 | LysoPC(20:3) | 2.6 | 3.4 | −0.6 | −0.3 | 0.9 | 1.1 | 1 | 0.2 |
| POS | 15.22_547.3573m/z | 2 | HMDB0094688 | 1-stearoylglycerophosphocholine | 3.1 | 4.4 | −0.1 | 0.1 | 0.6 | 1.1 | 1.5 | 0.9 |
| POS | 15.24_453.2849n | 2 | HMDB0011503 | LysoPE(16:0/0:0) | −1.7 | −2 | −0.8 | −1.2 | 0 | 0.6 | 0.2 | 0.5 |
| POS | 15.27_318.2190n | 3 | - | Eicosanoid | 11.1 | 11.1 | 4.6 | 7.3 | 0.5 | −0.5 | 0.8 | 1.3 |
| POS | 15.43_521.3476n | 2 | HMDB0002815 | LysoPC(18:1) | 0.8 | 1.2 | 0 | 0.2 | 0 | 0.1 | 0.2 | 0.3 |
| POS | 15.43_545.3462n | 2 | HMDB0010393 | LysoPC(20:3) | 1.9 | 2.6 | −0.2 | 0 | 0.6 | 0.6 | 0.7 | 0.4 |
| POS | 15.65_502.3256m/z | 2 | HMDB0010407 | LysoPC(P-16:0) | 3.9 | 4.5 | 0.3 | 1.4 | 1.3 | 1.5 | −0.1 | 0.1 |
| POS | 15.66_479.3361n | 2 | HMDB0010407 | LysoPC(P-16:0) | 3.1 | 3.7 | −0.1 | 0.8 | 0.6 | 1 | 0 | 0.2 |
| POS | 15.79_548.3696m/z | 2 | HMDB0010392 | LysoPC(20:2) | 2.4 | 3.2 | 0 | 0.4 | 0.2 | 0.5 | 0.6 | 0.3 |
| POS | 15.96_547.3629n | 2 | HMDB0010392 | LysoPC(20:2) | 2.6 | 3.5 | −0.6 | −0.3 | 1.6 | 0.2 | 1.3 | 1.3 |
| POS | 15.99_548.3700m/z | 2 | HMDB0010392 | LysoPC(20:2) | 1.8 | 2.6 | −0.3 | −0.3 | 0.6 | 0.2 | 0.5 | 0.6 |
| POS | 16.80_549.3785n | 2 | HMDB0010391 | LysoPC(20:1) | 1.4 | 2.2 | 0.1 | 0.2 | 0.2 | 0.2 | 0.2 | 0.8 |
| POS | 16.90_508.3750m/z | 2 | HMDB0013122 | LysoPC(P-18:0) | 5.3 | 6 | 0.4 | 1.2 | 2.4 | 1.7 | 0 | 1.5 |
| Pathway | Total Number of Compounds in the Pathway | Hits | p-Value | Adjusted p-Value (FDR) | Pathway Impact Value Calculated Based on Pathway Topology Analysis |
|---|---|---|---|---|---|
| Amino sugar and nucleotide sugar metabolism | 37 | 3 | 0.009 | 0.45 | 0.15 |
| Pyrimidine metabolism | 39 | 3 | 0.010 | 0.45 | 0.07 |
| Purine metabolism | 65 | 3 | 0.042 | 0.93 | 0.13 |
| Glutathione metabolism | 28 | 2 | 0.044 | 0.93 | 0.02 |
| Glycerophospholipid metabolism | 36 | 2 | 0.070 | 0.98 | 0.03 |
| Phosphonate and phosphinate metabolism | 6 | 1 | 0.071 | 0.98 | 0.50 |
| Tryptophan metabolism | 41 | 2 | 0.087 | 0.98 | 0.02 |
| Ascorbate and aldarate metabolism | 8 | 1 | 0.094 | 0.98 | 0.00 |
| Nicotinate and nicotinamide metabolism | 15 | 1 | 0.169 | 1.00 | 0.00 |
| Starch and sucrose metabolism | 18 | 1 | 0.200 | 1.00 | 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lenski, M.; Zarcone, G.; Maallem, S.; Garçon, G.; Lo-Guidice, J.-M.; Allorge, D.; Anthérieu, S. Metabolomics Provides Novel Insights into the Potential Toxicity Associated with Heated Tobacco Products, Electronic Cigarettes, and Tobacco Cigarettes on Human Bronchial Epithelial BEAS-2B Cells. Toxics 2024, 12, 128. https://doi.org/10.3390/toxics12020128
Lenski M, Zarcone G, Maallem S, Garçon G, Lo-Guidice J-M, Allorge D, Anthérieu S. Metabolomics Provides Novel Insights into the Potential Toxicity Associated with Heated Tobacco Products, Electronic Cigarettes, and Tobacco Cigarettes on Human Bronchial Epithelial BEAS-2B Cells. Toxics. 2024; 12(2):128. https://doi.org/10.3390/toxics12020128
Chicago/Turabian StyleLenski, Marie, Gianni Zarcone, Saïd Maallem, Guillaume Garçon, Jean-Marc Lo-Guidice, Delphine Allorge, and Sébastien Anthérieu. 2024. "Metabolomics Provides Novel Insights into the Potential Toxicity Associated with Heated Tobacco Products, Electronic Cigarettes, and Tobacco Cigarettes on Human Bronchial Epithelial BEAS-2B Cells" Toxics 12, no. 2: 128. https://doi.org/10.3390/toxics12020128
APA StyleLenski, M., Zarcone, G., Maallem, S., Garçon, G., Lo-Guidice, J.-M., Allorge, D., & Anthérieu, S. (2024). Metabolomics Provides Novel Insights into the Potential Toxicity Associated with Heated Tobacco Products, Electronic Cigarettes, and Tobacco Cigarettes on Human Bronchial Epithelial BEAS-2B Cells. Toxics, 12(2), 128. https://doi.org/10.3390/toxics12020128









