Prenatal Arsenic Exposure on DNA Methylation of C18ORF8 and ADAMTS9 Genes of Newborns from the POSGRAD Birth Cohort Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Population
2.2. Data Collection
2.2.1. Analysis of the Methylation of the C18ORF8 and ADAMTS9 Genes
2.2.2. Prenatal As Exposure Assessment
2.2.3. Other Variables of Interest
Maternal Information
Newborn Information
2.3. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Podgorski, J.; Berg, M. Global Threat of Arsenic in Groundwater. Science 2020, 368, 845–850. [Google Scholar] [CrossRef] [PubMed]
- Alarcón-Herrera, M.T.; Bundschuh, J.; Nath, B.; Nicolli, H.B.; Gutierrez, M.; Reyes-Gomez, V.M.; Nuñez, D.; Martín-Dominguez, I.R.; Sracek, O. Co-Occurrence of Arsenic and Fluoride in Groundwater of Semi-Arid Regions in Latin America: Genesis, Mobility and Remediation. J. Hazard. Mater. 2013, 262, 960–969. [Google Scholar] [CrossRef] [PubMed]
- Tovar Sánchez, E.; Mussali Galante, P.; Martínez Pacheco, M.; Ortiz Hernández, M.L.; Sánchez Salinas, E.; Olvera Velona, A. Relationship between Genotoxic Damage and Arsenic Blood Concentrations in Individuals Residing in an Arsenic Contaminated Area in Morelos, México. Rev. Int. Contam. Ambient. 2016, 32, 101–117. [Google Scholar]
- Garrido-Hoyos, S.; Romero-Velazquez, L. Synthesis of Minerals with Iron Oxide and Hydroxide Contents as a Sorption Medium to Remove Arsenic from Water for Human Consumption. Int. J. Environ. Res. Public Health 2015, 13, 69. [Google Scholar] [CrossRef] [PubMed]
- Mussali-Galante, P.; Tovar-Sánchez, E.; Valverde, M.; Valencia-Cuevas, L.; Rojas, E. Evidence of Population Genetic Effects in Peromyscus Melanophrys Chronically Exposed to Mine Tailings in Morelos, Mexico. Environ. Sci. Pollut. Res. Int. 2013, 20, 7666–7679. [Google Scholar] [CrossRef] [PubMed]
- Tchounwou, P.B.; Yedjou, C.G.; Udensi, U.K.; Pacurari, M.; Stevens, J.J.; Patlolla, A.K.; Noubissi, F.; Kumar, S. State of the Science Review of the Health Effects of Inorganic Arsenic: Perspectives for Future Research. Environ. Toxicol. 2019, 34, 188–202. [Google Scholar] [CrossRef] [PubMed]
- International Agency for Researc on Cancer Arsenic an Arsenic Compound. Available online: http://www.inchem.org/documents/iarc/suppl7/arsenic.html (accessed on 9 June 2020).
- Organización Mundial de la Salud Arsénico. 2018. Available online: https://www.who.int/es/news-room/fact-sheets/detail/arsenic (accessed on 4 April 2020).
- Quiñones_Valenzuela, A.; Gosset-Osuna, G.; Carboney-Castellanos, A. Arsénico y Salud. Salud Pública México 2014, 21, 187–197. [Google Scholar]
- Organización Mundial de la Salud Cancer. Available online: https://www.who.int/es/news-room/fact-sheets/detail/cancer (accessed on 30 July 2021).
- Organización Mundial de la Salud Cancer Colorectal. Available online: https://www.paho.org/hq/index.php?option=com_content&view=article&id=11761:colorectal-cancer&Itemid=41765&lang=es (accessed on 9 July 2020).
- Kile, M.L.; Baccarelli, A.; Hoffman, E.; Tarantini, L.; Quamruzzaman, Q.; Rahman, M.; Mahiuddin, G.; Mostofa, G.; Hsueh, Y.-M.; Wright, R.O.; et al. Prenatal Arsenic Exposure and DNA Methylation in Maternal and Umbilical Cord Blood Leukocytes. Environ. Health Perspect. 2012, 120, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Miao, Z.; Wu, L.; Lu, M.; Meng, X.; Gao, B.; Qiao, X.; Zhang, W.; Xue, D. Analysis of the Transcriptional Regulation of Cancer-Related Genes by Aberrant DNA Methylation of the Cis-Regulation Sites in the Promoter Region during Hepatocyte Carcinogenesis Caused by Arsenic. Oncotarget 2015, 6, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Chanda, S.; Dasgupta, U.B.; GuhaMazumder, D.; Gupta, M.; Chaudhuri, U.; Lahiri, S.; Das, S.; Ghosh, N.; Chatterjee, D. DNA Hypermethylation of Promoter of Gene P53 and P16 in Arsenic-Exposed People with and without Malignancy. Toxicol. Sci. 2006, 89, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Arechederra, M.; Daian, F.; Yim, A.; Bazai, S.K.; Richelme, S.; Dono, R.; Saurin, A.J.; Habermann, B.H.; Maina, F. Hypermethylation of Gene Body CpG Islands Predicts High Dosage of Functional Oncogenes in Liver Cancer. Nat. Commun. 2018, 9, 3164. [Google Scholar] [CrossRef] [PubMed]
- Kile, M.L.; Houseman, E.A.; Baccarelli, A.A.; Quamruzzaman, Q.; Rahman, M.; Mostofa, G.; Cardenas, A.; Wright, R.O.; Christiani, D.C. Effect of Prenatal Arsenic Exposure on DNA Methylation and Leukocyte Subpopulations in Cord Blood. Epigenetics 2014, 9, 774–782. [Google Scholar] [CrossRef] [PubMed]
- Koestler, D.C.; Avissar-Whiting, M.; Houseman, E.A.; Karagas, M.R.; Marsit, C.J. Differential DNA Methylation in Umbilical Cord Blood of Infants Exposed to Low Levels of Arsenic in Utero. Environ. Health Perspect. 2013, 121, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Mesa-Cornejo, V.M.; Barros-Núñez, P.; Medina-Lozano, C. Methylation of DNA: A marker for the diagnosis and prognosis in cancer. Gac. Médica México 2006, 142, 81–82. [Google Scholar]
- Martin, E.M.; Fry, R.C. Environmental Influences on the Epigenome: Exposure—Associated DNA Methylation in Human Populations. Annu. Rev. Public Health 2018, 39, 309–333. [Google Scholar] [CrossRef] [PubMed]
- Rojas, D.; Rager, J.E.; Smeester, L.; Bailey, K.A.; Drobná, Z.; Rubio-Andrade, M.; Stýblo, M.; García-Vargas, G.; Fry, R.C. Prenatal Arsenic Exposure and the Epigenome: Identifying Sites of 5-Methylcytosine Alterations That Predict Functional Changes in Gene Expression in Newborn Cord Blood and Subsequent Birth Outcomes. Toxicol. Sci. 2015, 143, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Montes-Castro, N.; Alvarado-Cruz, I.; Torres-Sánchez, L.; García-Aguiar, I.; Barrera-Hernández, A.; Escamilla-Núñez, C.; Del Razo, L.M.; Quintanilla-Vega, B. Prenatal Exposure to Metals Modified DNA Methylation and the Expression of Antioxidant- and DNA Defense-Related Genes in Newborns in an Urban Area. J. Trace Elem. Med. Biol. 2019, 55, 110–120. [Google Scholar] [CrossRef] [PubMed]
- OGG1 8-Oxoguanine DNA Glycosylase [Homo Sapiens (Human)]-Gene-NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/4968 (accessed on 2 August 2021).
- Padhi, S.S.; Roy, S.; Kar, M.; Saha, A.; Roy, S.; Adhya, A.; Baisakh, M.; Banerjee, B. Role of CDKN2A/P16 Expression in the Prognostication of Oral Squamous Cell Carcinoma. Oral Oncol. 2017, 73, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, V.; Castellví-Bel, S.; Balaguer, F.; Pellisé, M.; Ocaña, T.; Castells, A. Epigenética Del Cáncer. Gastroenterol. Hepatol. 2008, 31, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Tang, J.; Feng, Y.; Li, S.; Xiang, Q.; He, X.; Ren, G.; Peng, W.; Xiang, T. ADAMTS9 Is Silenced by Epigenetic Disruption in Colorectal Cancer and Inhibits Cell Growth and Metastasis by Regulating Akt/P53 Signaling. Cell. Physiol. Biochem. 2017, 44, 1370–1380. [Google Scholar] [CrossRef] [PubMed]
- Hammond, R.K.; Pahl, M.C.; Su, C.; Cousminer, D.L.; Leonard, M.E.; Lu, S.; Doege, C.A.; Wagley, Y.; Hodge, K.M.; Lasconi, C.; et al. Biological Constraints on GWAS SNPs at Suggestive Significance Thresholds Reveal Additional BMI Loci. eLife 2021, 10, e62206. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Ni, Z.; Chen, W.; Zhou, Y. The ADAMTS9 Gene Is Associated with Mandibular Retrusion in a Chinese Population. Gene 2020, 749, 144701. [Google Scholar] [CrossRef] [PubMed]
- Bu, X.; Qin, A.; Luo, Z.; Hu, Y. Overexpression of the long non-coding RNA ADAMTS9-AS2 suppresses colorectal cancer proliferation and metastasis. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2019, 44, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, U.; Stein, A.D.; Parra-Cabrera, S.; Wang, M.; Imhoff-Kunsch, B.; Juárez-Márquez, S.; Rivera, J.; Martorell, R. Effects of Docosahexaenoic Acid Supplementation during Pregnancy on Gestational Age and Size at Birth: Randomized, Double-Blind, Placebo-Controlled Trial in Mexico. Food Nutr. Bull. 2010, 31, S108–S116. [Google Scholar] [CrossRef] [PubMed]
- Castro-Rodríguez, J.A.; Krause, B.J.; Uauy, R.; Casanello, P. Epigenética En Enfermedades Alérgicas y Asma. Rev. Chil. Pediatría 2016, 87, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Mehta, R.S.; Song, M.; Bezawada, N.; Wu, K.; Garcia-Albeniz, X.; Morikawa, T.; Fuchs, C.S.; Ogino, S.; Giovannucci, E.L.; Chan, A.T. A Prospective Study of Macrophage Inhibitory Cytokine-1 (MIC-1/GDF15) and Risk of Colorectal Cancer. J. Natl. Cancer Inst. 2014, 106, dju016. [Google Scholar] [CrossRef] [PubMed]
- Instituto Nacional del Cáncer Nrf2. Available online: https://www.cancer.gov/espanol/publicaciones/diccionarios/diccionario-cancer/def/nrf2 (accessed on 2 December 2021).
- Cheng, T.F.; Choudhuri, S.; Muldoon-Jacobs, K. Epigenetic Targets of Some Toxicologically Relevant Metals: A Review of the Literature. J. Appl. Toxicol. 2012, 32, 643–653. [Google Scholar] [CrossRef] [PubMed]
- Sage, A.P.; Minatel, B.C.; Ng, K.W.; Stewart, G.L.; Dummer, T.J.B.; Lam, W.L.; Martinez, V.D. Oncogenomic Disruptions in Arsenic-Induced Carcinogenesis. Oncotarget 2017, 8, 25736–25755. [Google Scholar] [CrossRef] [PubMed]
- RMC1 Regulator of MON1-CCZ1 [Homo Sapiens (Human)]. Available online: https://www.ncbi.nlm.nih.gov/gene/29919 (accessed on 19 June 2021).
- Uniprot C18ORF8. Available online: https://www.uniprot.org/uniprot/G3GIT6 (accessed on 19 June 2021).
- Wang, X.; Baek, S.J.; Eling, T.E. The Diverse Roles of Nonsteroidal Anti-Inflammatory Drug Activated Gene (NAG-1/GDF15) in Cancer. Biochem. Pharmacol. 2013, 85, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Bansal, N.; Kumar, D.; Gupta, A.; Chandra, D.; Sankhwar, S.N.; Mandhani, A. Relevance of MIC-1 in the Era of PSA as a Serum Based Predictor of Prostate Cancer: A Critical Evaluation. Sci. Rep. 2017, 7, 16824. [Google Scholar] [CrossRef] [PubMed]
- Emmerson, P.J.; Duffin, K.L.; Chintharlapalli, S.; Wu, X. GDF15 and Growth Control. Front. Physiol. 2018, 9, 1712. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, Z.; Tian, H.; Li, Y.; Li, M.; Zhao, W.; Zhang, C.; Wang, T.; Liu, J.; Zhang, A.; et al. Circulating MIC-1/GDF15 Is a Complementary Screening Biomarker with CEA and Correlates with Liver Metastasis and Poor Survival in Colorectal Cancer. Oncotarget 2017, 8, 24892–24901. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.A.; Ward, R.L.; Buckhaults, P.; Liu, T.; Romans, K.E.; Hawkins, N.J.; Bauskin, A.R.; Kinzler, K.W.; Vogelstein, B.; Breit, S.N. MIC-1 Serum Level and Genotype: Associations with Progress and Prognosis of Colorectal Carcinoma. Clin. Cancer Res. 2003, 9, 2642–2650. [Google Scholar] [PubMed]
- Danta, M.; Barber, D.A.; Zhang, H.P.; Lee-Ng, M.; Baumgart, S.W.L.; Tsai, V.W.W.; Husaini, Y.; Saxena, M.; Marquis, C.P.; Errington, W.; et al. Macrophage Inhibitory Cytokine-1/Growth Differentiation Factor-15 as a Predictor of Colonic Neoplasia. Aliment. Pharmacol. Ther. 2017, 46, 347–354. [Google Scholar] [CrossRef] [PubMed]
- González-Becerra, K.; Ramos-Lopez, O.; Barrón-Cabrera, E.; Riezu-Boj, J.I.; Milagro, F.I.; Martínez-López, E.; Martínez, J.A. Fatty Acids, Epigenetic Mechanisms and Chronic Diseases: A Systematic Review. Lipids Health Dis. 2019, 18, 178. [Google Scholar] [CrossRef] [PubMed]
- Hussey, B.; Steel, R.P.; Gyimah, B.; Reynolds, J.C.; Taylor, I.M.; Lindley, M.R.; Mastana, S. DNA Methylation of Tumour Necrosis Factor (TNF) Alpha Gene Is Associated with Specific Blood Fatty Acid Levels in a Gender-Specific Manner. Mol. Genet. Genom. Med. 2021, 9, e1679. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, B.P.; Mitra, T.; Ganguly, S.; Das Sarkar, S.; Mahanty, A. Curcumin Has Protective Effect on the Eye Lens Against Arsenic Toxicity. Biol. Trace Elem. Res. 2021, 199, 3354–3359. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, S.J.; Zhou, J.; Peters, T.J.; Buckley, M.; Sutcliffe, B.; Oytam, Y.; Gibson, R.A.; McPhee, A.; Yelland, L.N.; Makrides, M.; et al. Effect of Prenatal DHA Supplementation on the Infant Epigenome: Results from a Randomized Controlled Trial. Clin. Epigenetics 2016, 8, 114. [Google Scholar] [CrossRef] [PubMed]
- Moradi Sarabi, M.; Mohammadrezaei Khorramabadi, R.; Zare, Z.; Eftekhar, E. Polyunsaturated Fatty Acids and DNA Methylation in Colorectal Cancer. World J. Clin. Cases 2019, 7, 4172–4185. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gao, D.; Zhang, G.; Zhang, X.; Li, Q.; Gao, Q.; Chen, R.; Xu, S.; Huang, L.; Zhang, Y.; et al. Exposure to Multiple Metals in Early Pregnancy and Gestational Diabetes Mellitus: A Prospective Cohort Study. Environ. Int. 2020, 135, 105370. [Google Scholar] [CrossRef] [PubMed]
- Soler-Blasco, R.; Murcia, M.; Lozano, M.; Sarzo, B.; Esplugues, A.; Vioque, J.; Lertxundi, N.; Marina, L.S.; Lertxundi, A.; Irizar, A.; et al. Urinary Arsenic Species and Methylation Efficiency during Pregnancy: Concentrations and Associated Factors in Spanish Pregnant Women. Environ. Res. 2021, 196, 100889. [Google Scholar] [CrossRef] [PubMed]
- Michel-Ramirez, G.; Recio-Vega, R.; Lantz, R.C.; Gandolfi, A.J.; Olivas-Calderon, E.; Chau, B.T.; Amistadi, M.K. Assessment of YAP Gene Polymorphisms and Arsenic Interaction in Mexican Women with Breast Cancer. J. Appl. Toxicol. 2020, 40, 342–351. [Google Scholar] [CrossRef] [PubMed]
- Gamboa-Loira, B.; Cebrián, M.E.; López-Carrillo, L. Arsenic Exposure in Northern Mexican Women. Salud Pública México 2020, 62, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Barats, A.; Renac, C.; Orani, A.M.; Durrieu, G.; Saint Martin, H.; Esteller, M.V.; Garrido Hoyos, S.E. Tracing Source and Mobility of Arsenic and Trace Elements in a Hydrosystem Impacted by Past Mining Activities (Morelos State, Mexico). Sci. Total Environ. 2020, 712, 135565. [Google Scholar] [CrossRef] [PubMed]
- Shao, B.; Feng, Y.; Zhang, H.; Yu, F.; Li, Q.; Tan, C.; Xu, H.; Ying, J.; Li, L.; Yang, D.; et al. The 3p14.2 Tumour Suppressor ADAMTS9 Is Inactivated by Promoter CpG Methylation and Inhibits Tumour Cell Growth in Breast Cancer. J. Cell. Mol. Med. 2018, 22, 1257–1271. [Google Scholar] [CrossRef] [PubMed]
- Khan Academy Resumen de La Transcripción. Available online: https://es.khanacademy.org/science/ap-biology/gene-expression-and-regulation/transcription-and-rna-processing/a/overview-of-transcription (accessed on 3 August 2021).
- Brázda, V.; Fojta, M. The Rich World of P53 DNA Binding Targets: The Role of Dna Structure. Int. J. Mol. Sci. 2019, 20, 5605. [Google Scholar] [CrossRef] [PubMed]
- Alvarado-Cruz, I.; Alegría-Torres, J.A.; Montes-Castro, N.; Jiménez-Garza, O.; Quintanilla-Vega, B. Environmental Epigenetic Changes, as Risk Factors for the Development of Diseases in Children: A Systematic Review. Ann. Glob. Health 2018, 84, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Poblete-Naredo, I.; Garcia-Salcedo, J.J.; Salinas-Aguirre, J.E.; Corona-Nuñez, R.O.; Albores, A. Capítulo 5: Casos de Estudio y Experiencias En Campo. Caso La Laguna. In Arsénico y Fluoruro en Agua: Riesgos y Perspectivas Desde la Sociedad Civil y la Academia en México; Secretaría de Gobernación: Ciudad de México, México, 2018; pp. 105–113. [Google Scholar]
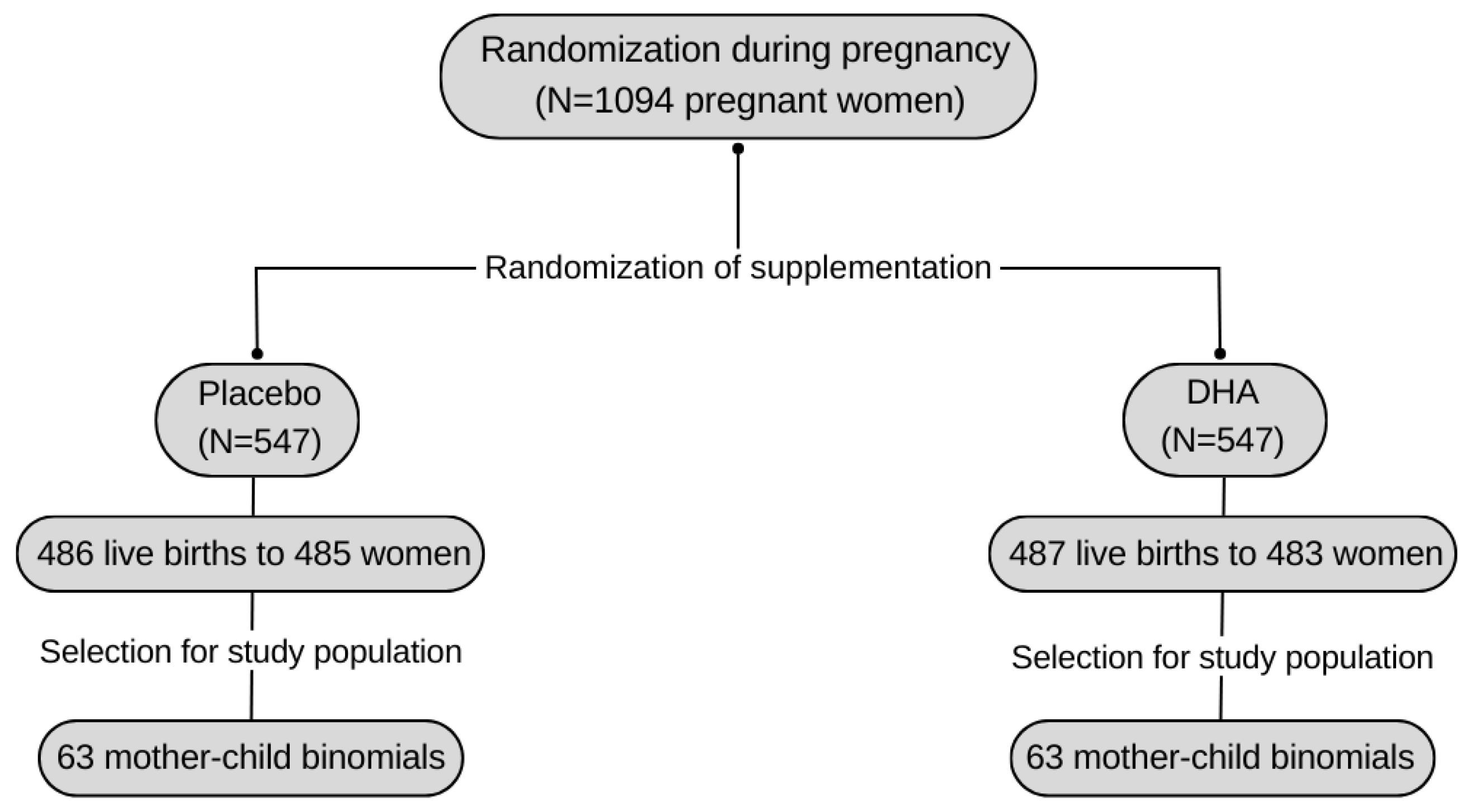


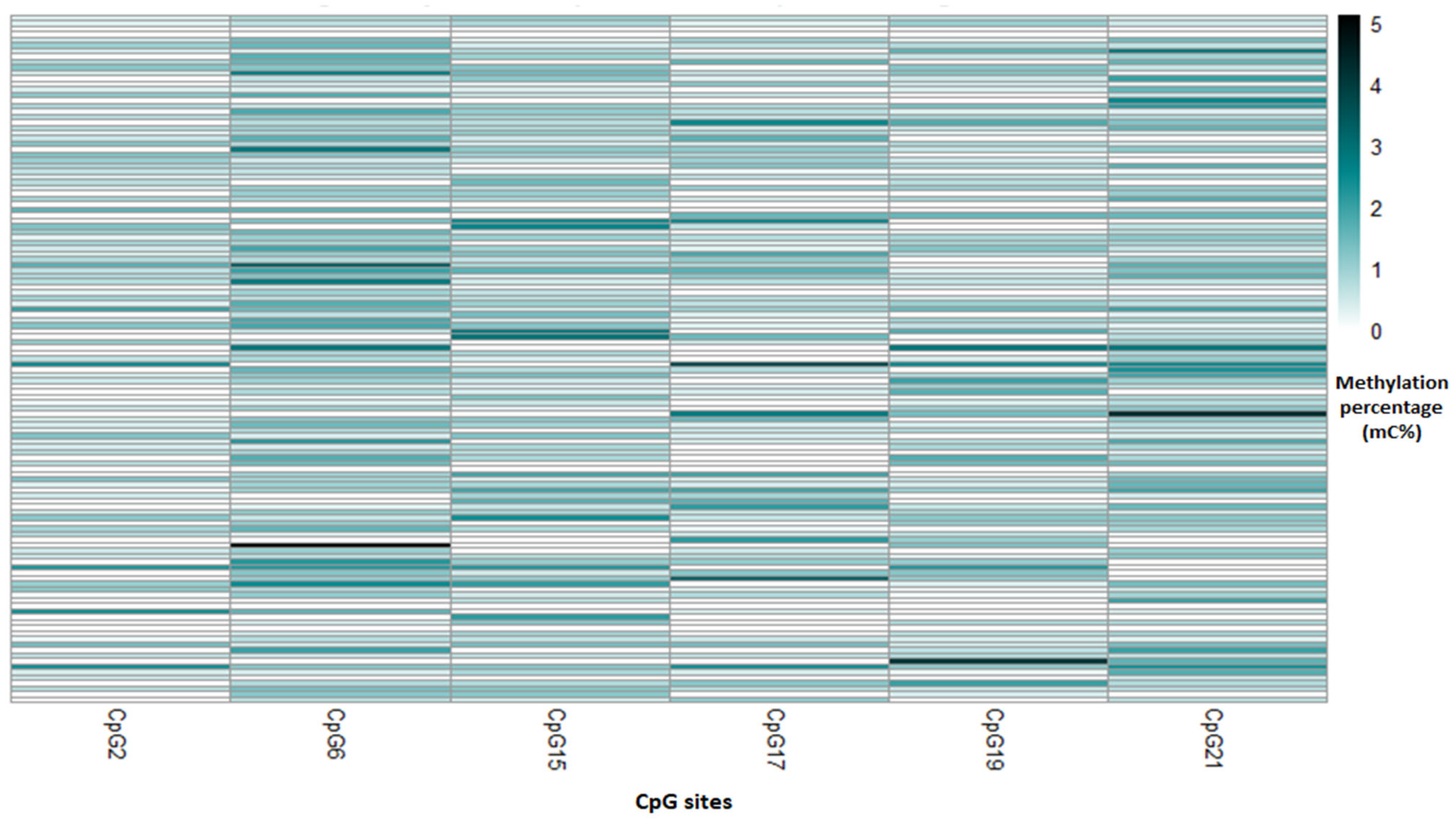

| Included Population | Not Included Population | ||
|---|---|---|---|
| Maternal Variables | (n = 126) | (n = 858) | |
| (18 to 22 Pregnancy Weeks) | Mean ± SD o n (%) | Mean ± SD o n (%) | p-Value |
| Maternal age (years) | 26 ± 4.738 | 26 ± 4.714 | 0.51 |
| Smoke habits | |||
| Yes | 1 (0.79) | 16 (1.88) | 0.39 |
| No Passive smoker | 125 (99.21) | 837 (98.12) | |
| Yes | 46 (38.98) | 197 (40.96) | 0.7 |
| No Supplement | 72 (61.02) | 284 (59.04) | |
| DHA | 63 (50) | 428 (49.94) | 0.99 |
| Placebo | 63 (50) | 429 (50.06) | |
| Folic acid | |||
| Yes | 107 (84.92) | 667 (77.73) | 0.22 |
| No | 19 (15.07) | 182 (22.26) | |
| BMI | 26.22 ± 4.47 | 26.01 ± 4.25 | 0.62 |
| Included population | Not included population | ||
| (n = 126) | (n = 858) | ||
| Newborn variables | Mean ± SD o n (%) | Mean ± SD o n (%) | p-value |
| Infant sex | |||
| Female | 68 (53.97) | 394 (45.92) | 0.19 |
| Male | 58 (46.03) | 464 (54.08) | |
| Premature (<37 weeks) | 6 (5.13) | 67 (9.38) | 0.13 |
| Gestational weight (g) | 3257.64 ± 404.91 | 3183 ± 486.34 | 0.11 |
| Mean TUA ± SD | 44.43 ± 38.17 | 38.56 ± 35.89 | 0.15 |
| Median (min, max) | 31 (1.5, 180) | 29 (0.15–190) | |
| Percentile 25% | 13 | 6.5 | |
| Percentile 75% | 70 | 59 | |
| Percentile 90% | 99 | 90 | |
| As in maternal urine adjusted by creatinine (µg g−1) | Study population | Cohort population | p-value |
| Mean TUA µg g−1 ± SD | 49.4 ± 73.77 | 49.31 ± 60.25 | 0.97 |
| Median (min, max) | 30.01 (1.34, 532.97) | 29.35 (0.216–532.96) | |
| Percentile 25% | 12.54 | 12.51 | |
| Percentile 75% | 52.11 | 56.86 |
| C18ORF8 Gene Methylation (%) | ADAMTS9 Gene Methylation (%) | ||
|---|---|---|---|
|
Mean SD *
First index I | 0.67 ± 0.28 |
Mean SD *
First index II | 1.45 ± 1.03 |
| Mean SD | 0.7 ± 0.43 | Mean SD | 0.59 ± 1.14 |
| Median (min, max) Second index III | 0.7 (0, 2.03) | Median (min, max) Second index IV | 0 (0, 5.36) |
| Mean SD | 0.78 ± 0.44 | Mean SD | 0.95 ± 1.8 |
| Median (min, max) | 0.70 (0, 2.51) | Median (min, max) | 0 (0, 8.61) |
| TUA gr−1 | ||||
|---|---|---|---|---|
| Crude | Adjusted | |||
| ß (95% CI) | p-Value | ß (95% CI) | p-Value | |
| C18ORF8 | ||||
| First Index | ||||
| (CpG2, CpG6, and | ||||
| CpG17) | 0.03 (−0.16, 0.21) | 0.76 | 0.05 (−0.14, 0.23) | 0.63 |
| Second Index | ||||
| (CpG15, CpG19, | ||||
| and CpG21) | −0.22 (−0.39, −0.05) | 0.01 * | −0.21 (−0.38, −0.04) | 0.02 * |
| ADAMTS9 | ||||
| First Index | ||||
| (CpG2 and CpG6) | 0.04 (−0.45, 0.53) | 0.86 | 0.06 (−0.43, 0.55) | 0.81 |
| Second Index | ||||
| (CpG1, CpG4, | ||||
| CpG5, CpG13, | ||||
| CpG14, and CpG20) | −0.52 (−1.38, 0.24) | 0.18 | −0.56 (−1.3, 0.21) | 0.15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lerma-Treviño, C.; Hernández-Cadena, L.; Acosta-Montes, J.O.; Hernández-Montes, G.; Alvarado-Cruz, I.; Romieu, I.; Barraza-Villarreal, A. Prenatal Arsenic Exposure on DNA Methylation of C18ORF8 and ADAMTS9 Genes of Newborns from the POSGRAD Birth Cohort Study. Toxics 2024, 12, 476. https://doi.org/10.3390/toxics12070476
Lerma-Treviño C, Hernández-Cadena L, Acosta-Montes JO, Hernández-Montes G, Alvarado-Cruz I, Romieu I, Barraza-Villarreal A. Prenatal Arsenic Exposure on DNA Methylation of C18ORF8 and ADAMTS9 Genes of Newborns from the POSGRAD Birth Cohort Study. Toxics. 2024; 12(7):476. https://doi.org/10.3390/toxics12070476
Chicago/Turabian StyleLerma-Treviño, Carolina, Leticia Hernández-Cadena, Jorge Octavio Acosta-Montes, Georgina Hernández-Montes, Isabel Alvarado-Cruz, Isabelle Romieu, and Albino Barraza-Villarreal. 2024. "Prenatal Arsenic Exposure on DNA Methylation of C18ORF8 and ADAMTS9 Genes of Newborns from the POSGRAD Birth Cohort Study" Toxics 12, no. 7: 476. https://doi.org/10.3390/toxics12070476







