Assessing Receptor Activation in 2D and 3D Cultured Hepatocytes: Responses to a Single Compound and a Complex Mixture
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture Maintenance
2.1.1. 2D Monolayer Cell Culture
2.1.2. 3D Spheroid Cell Culture
2.2. Preparation of NAFCs
2.3. Cell Viability
2.4. Cell Culture Treatments
2.5. Real-Time Quantitative PCR
2.6. Apoptosis: Caspase 3/7
2.7. Statistical Analysis
3. Results
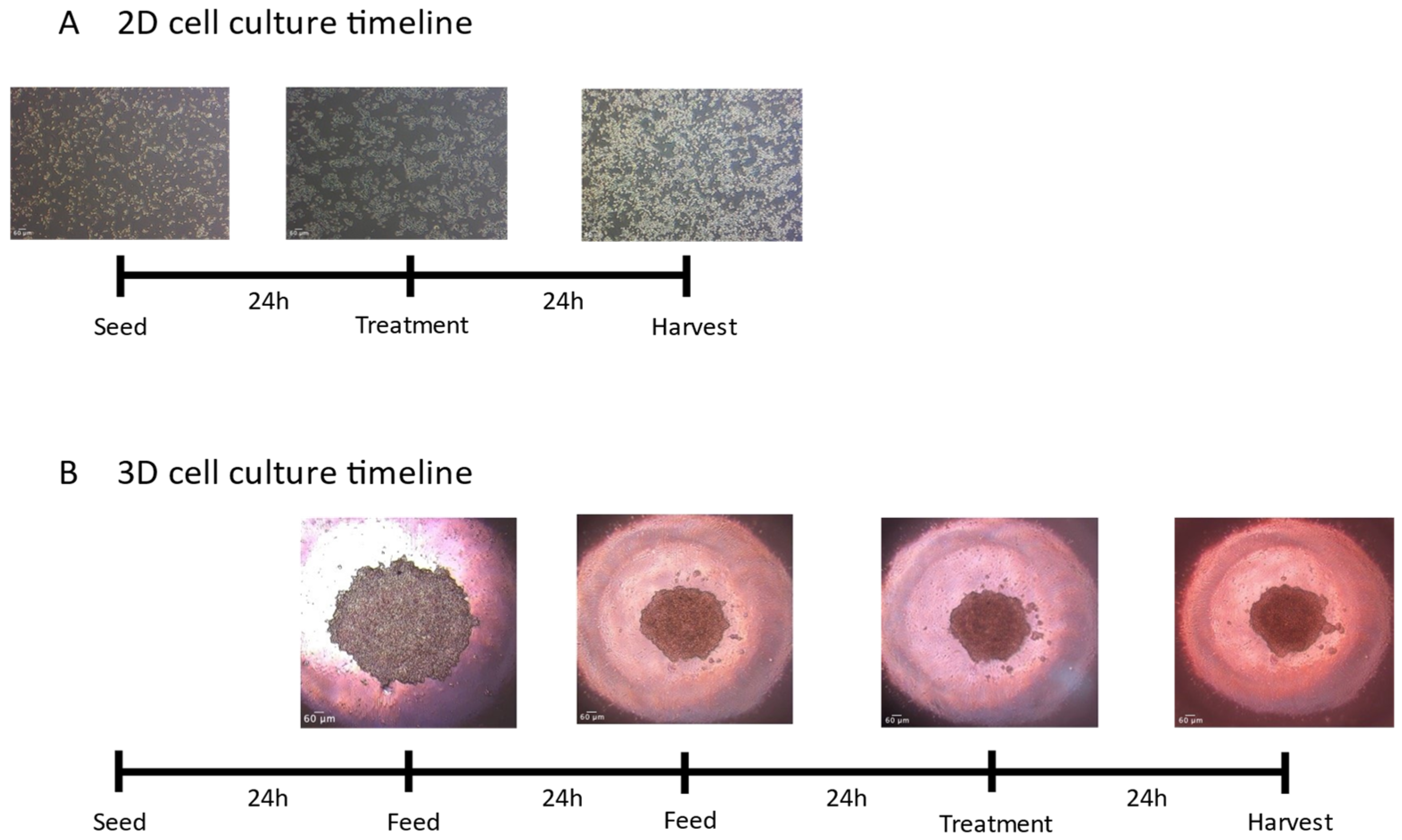
3.1. Morphology
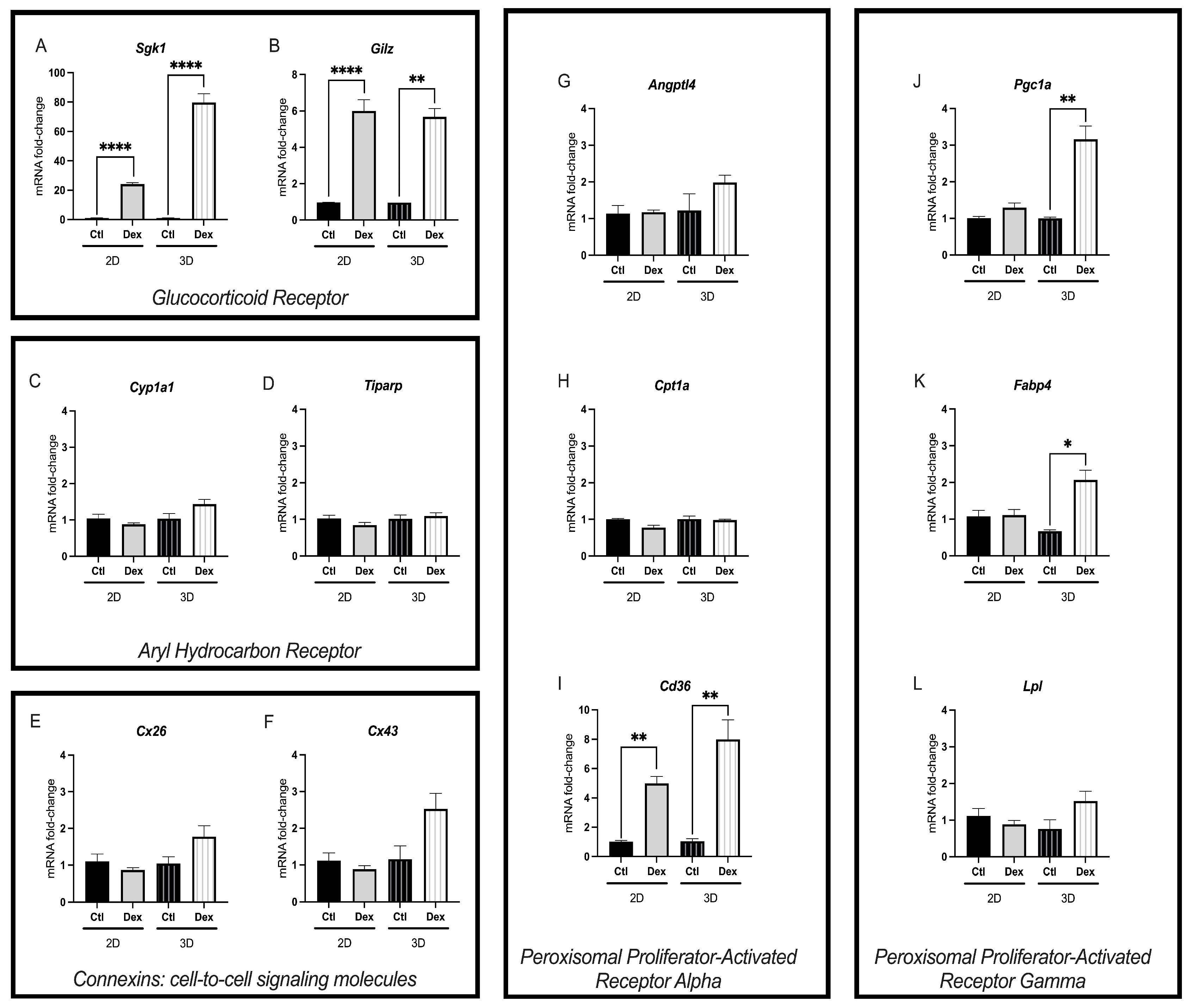
3.2. D-Cultured McA-RH7777s Exhibited Significantly Higher Fold Change of Altered Genes Following Dexamethasone Exposure
3.3. NAFCs Did Not Significantly Affect Cell-Viability in 2D or 3D Cultures
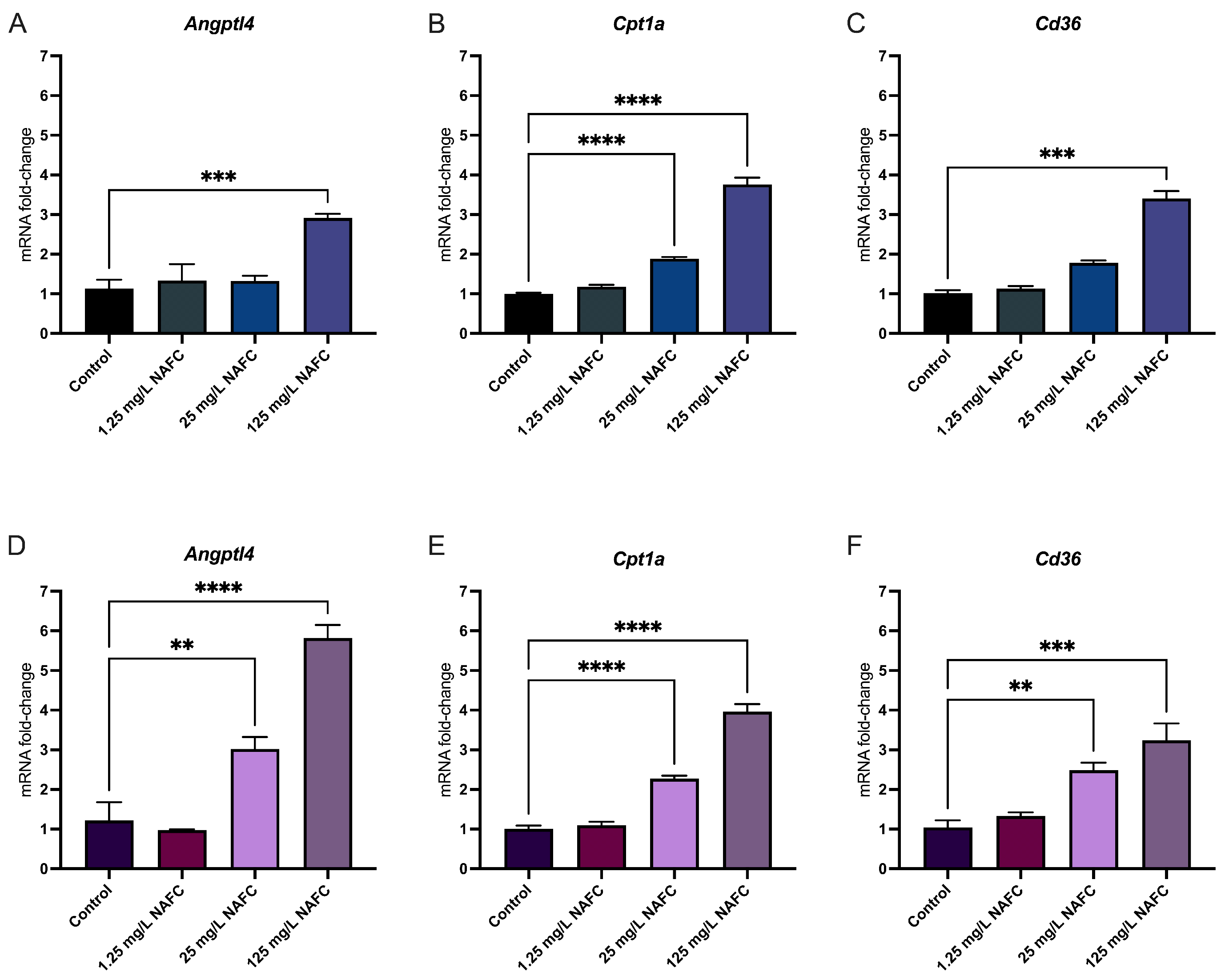
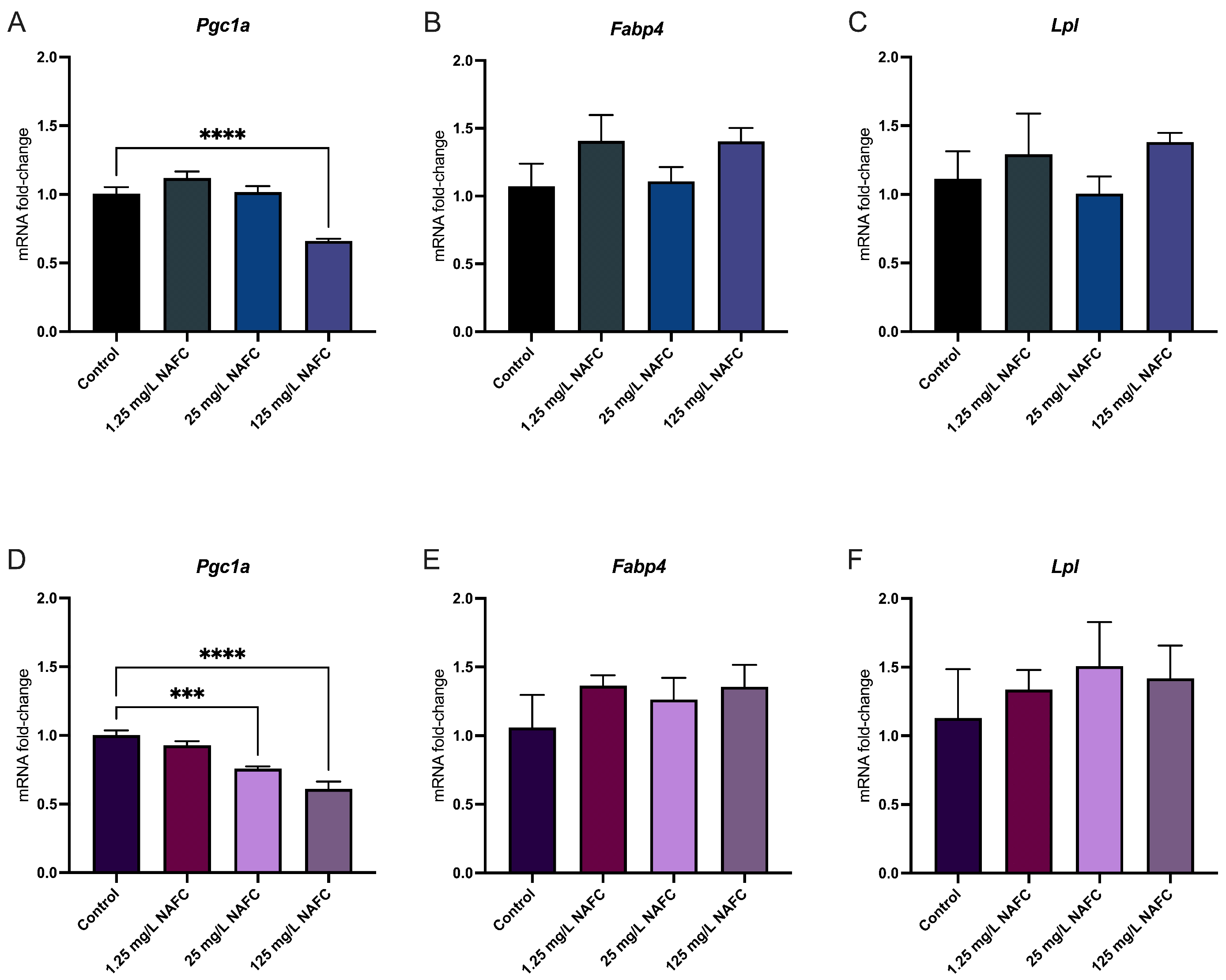
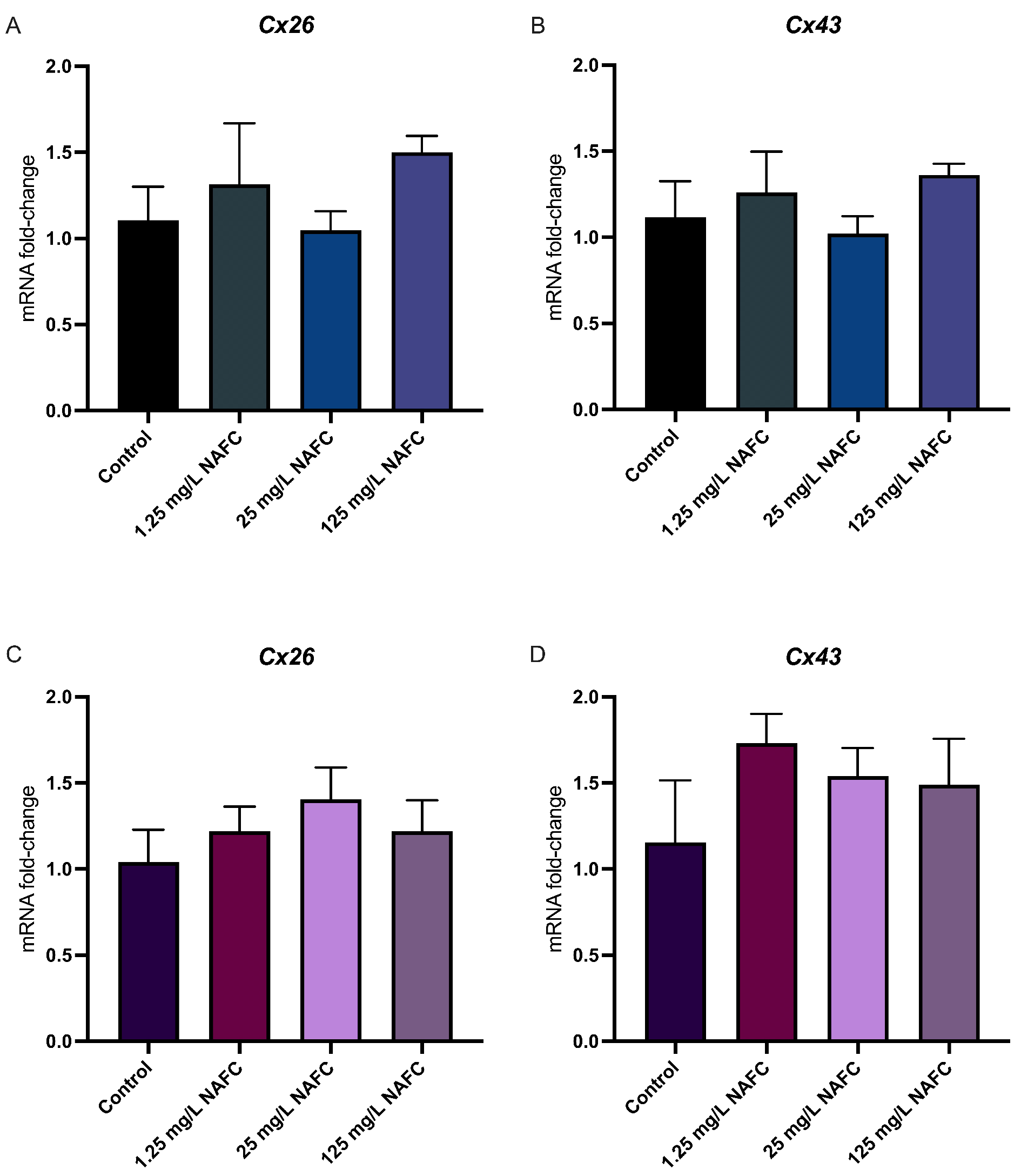
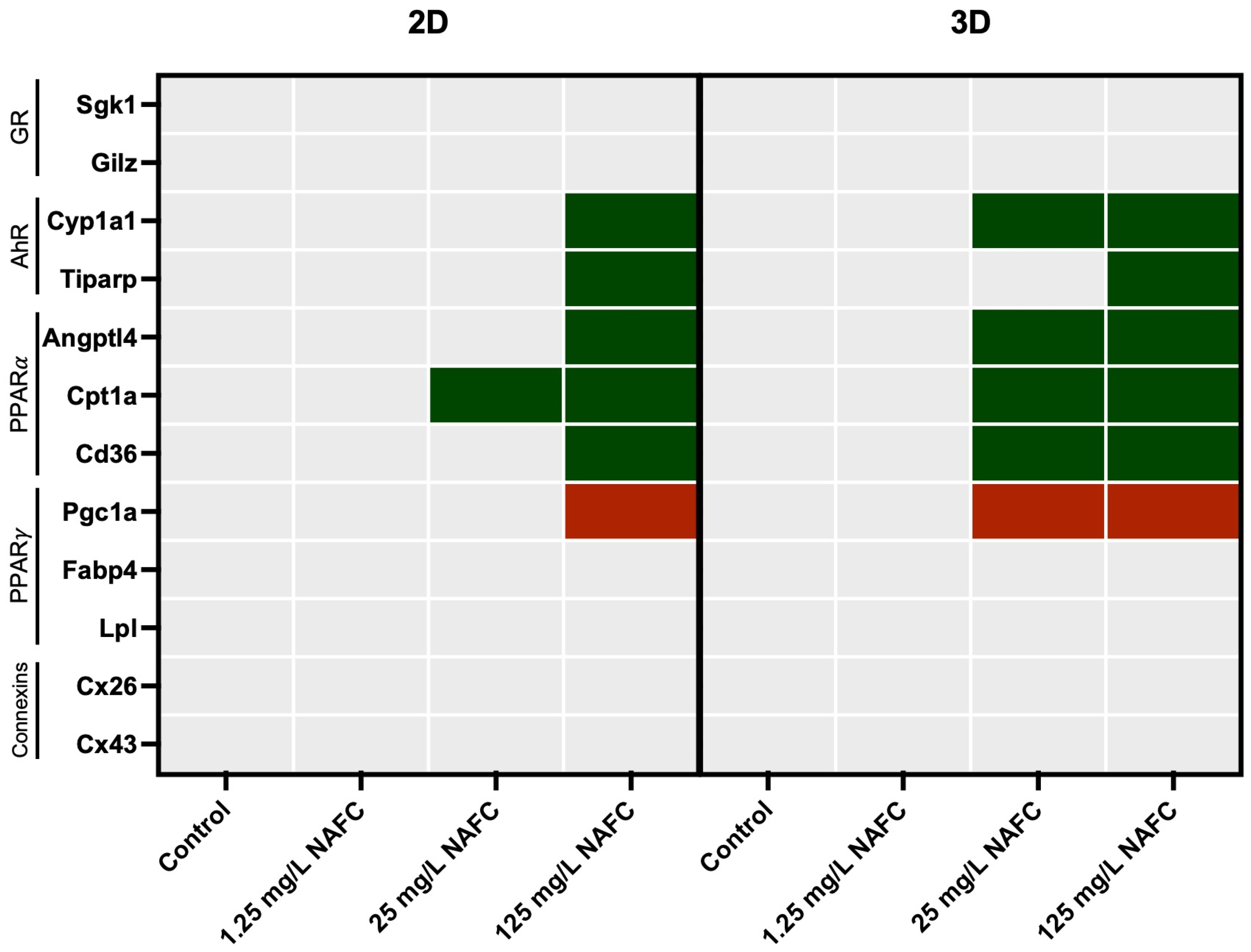
3.4. Exposure to NAFCs Elicits Differential Sensitivity in mRNA Expression of Genes Involved in Receptor Activation between 2D and 3D Cultured McA-RH7777s
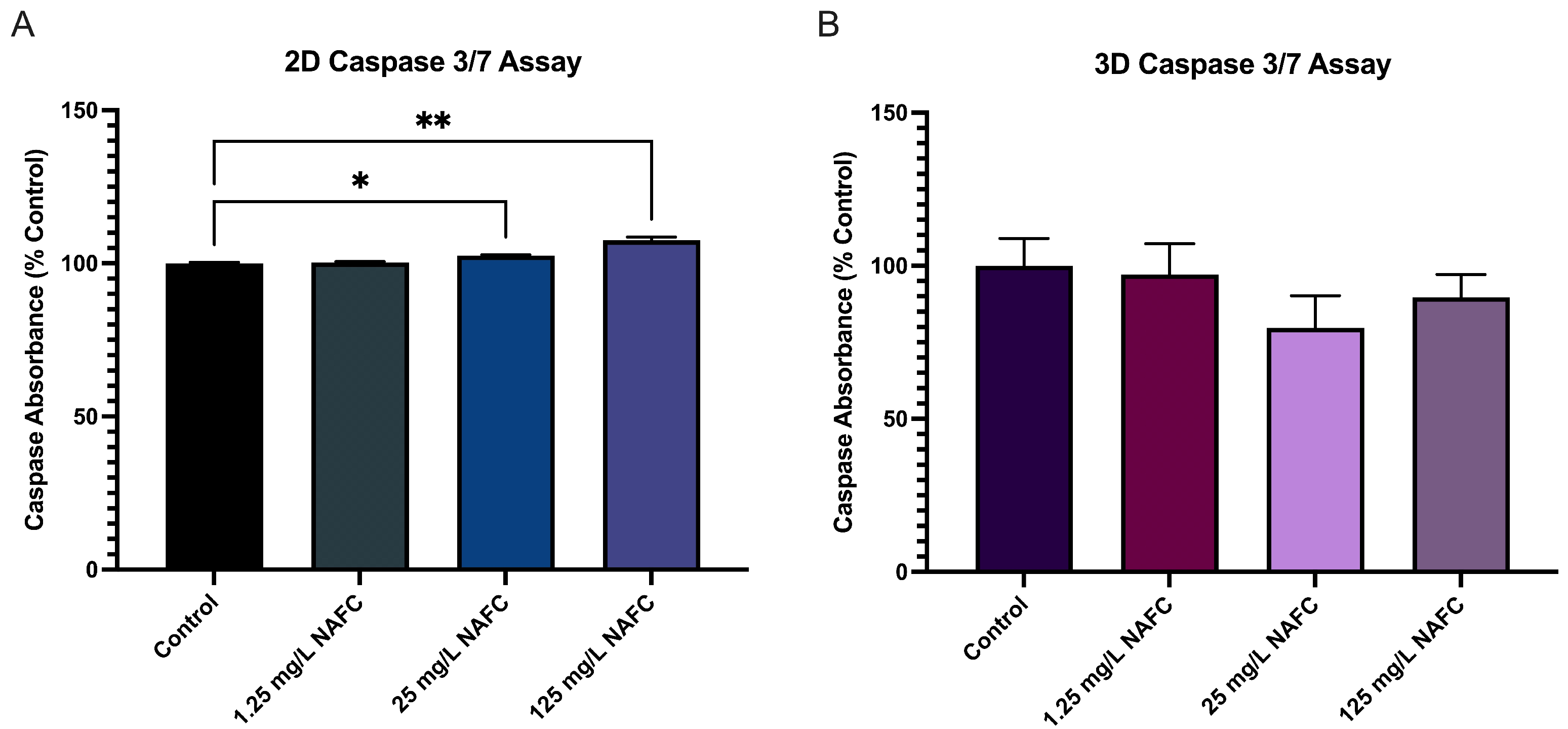
3.5. 3D Cultured McA-RH7777s Are More Resistant to NAFC-Induced Caspase 3/7 Activity
4. Discussion
4.1. Dexamethasone: A Model Corticosteroid
4.2. Naphthenic Acid Fraction Components: An Environmental Mixture
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pampaloni, F.; Stelzer, E.H. Three-Dimensional Cell Cultures in Toxicology. Biotechnol. Genet. Eng. Rev. 2009, 26, 117–138. [Google Scholar] [CrossRef]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning from 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- de Boo, J.; Hendriksen, C. Reduction Strategies in Animal Research: A Review of Scientific Approaches at the Intra-experimental, Supra-experimental and Extra-experimental Levels. Altern. Lab. Anim. 2005, 33, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Franco, N.; Olsson, I. Scientists and the 3Rs: Attitudes to animal use in biomedical research and the effect of mandatory training in laboratory animal science. Lab. Anim. 2014, 48, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Government of Canada. Bill S-5, Strengthening Environmental Protection for a Healthier Canada Act—Summary of Amendments. Available online: https://www.canada.ca/en/services/environment/pollution-waste-management/strengthening-canadian-environmental-protection-act-1999/bill-c-28-strengthening-environmental-protection-healthier-canada-act-summary-amendments.html (accessed on 8 December 2023).
- Lelièvre, S.A.; Kwok, T.; Chittiboyina, S. Architecture in 3D cell culture: An essential feature for in vitro toxicology. Toxicol. In Vitro 2017, 45, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Stock, K.; Estrada, M.F.; Vidic, S.; Gjerde, K.; Rudisch, A.; Santo, V.E.; Barbier, M.; Blom, S.; Arundkar, S.C.; Selvam, I.; et al. Capturing tumor complexity in vitro: Comparative analysis of 2D and 3D tumor models for drug discovery. Sci. Rep. 2016, 6, 28951. [Google Scholar] [CrossRef]
- Fang, Y.; Eglen, R.M. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov. Adv. Sci. Drug Discov. 2017, 22, 456–472. [Google Scholar] [CrossRef]
- Soldatow, V.Y.; LeCluyse, E.L.; Griffith, L.G.; Rusyn, I. In vitro models for liver toxicity testing. Toxicol. Res. 2013, 2, 23–39. [Google Scholar] [CrossRef]
- Sutherland, R.M.; Inch, W.R.; McCredie, J.A.; Kruuv, J. A Multi-component Radiation Survival Curve Using an in Vitro Tumour Model. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1970, 18, 491–495. [Google Scholar] [CrossRef]
- Sutherland, R.M.; McCredie, J.A.; Inch, W.R. Growth of multicell spheroids in tissue culture as a model of nodular carcinomas. J. Natl. Cancer Inst. 1971, 46, 113–120. [Google Scholar]
- Hamilton, G.A.; Westmorel, C.; George, A.E. Effects of medium composition on the morphology and function of rat hepatocytes cultured as spheroids and monolayers. In Vitro Cell. Dev. Biol. Anim. 2001, 37, 656–667. [Google Scholar] [CrossRef]
- Sharin, T.; Crump, D.; O’Brien, J.M. Evaluation of the Aryl Hydrocarbon Receptor Response in LMH 3D Spheroids. Environ. Toxicol. Chem. 2020, 39, 1693–1701. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q. Three-dimensional culture of hepatocytes for prediction of drug-induced hepatotoxicity. Expert Opin. Drug Metab. Toxicol. 2010, 6, 733–746. [Google Scholar] [CrossRef]
- Guillouzo, A. Liver cell models in in vitro toxicology. Environ. Health Perspect. 1998, 106, 511–532. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.S.; Friend, J.R.; Wu, F.J.; Sielaff, T.; Peshwa, M.V.; Lazar, A.; Nyberg, S.L.; Remmel, R.P.; Cerra, F.B. Development of a bioartificial liver employing xenogeneic hepatocytes. Cytotechnology 1997, 23, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Maurice, M.; Rogier, E.; Cassio, D.; Feldmann, G. Formation of plasma membrane domains in rat hepatocytes and hepatoma cell lines in culture. J. Cell Sci. 1988, 90, 79–92. [Google Scholar] [CrossRef]
- Jigorel, E.; Le Vee, M.; Boursier-Neyret, C.; Bertrand, M.; Fardel, O. Functional expression of sinusoidal drug transporters in primary human and rat hepatocytes. Drug Metab. Dispos. 2005, 33, 1418–1422. [Google Scholar] [CrossRef]
- Knop, E.; Bader, A.; Böker, K.; Pichlmayr, R.; Sewing, K.-F. Ultrastructural and functional differentiation of hepatocytes under long-term culture conditions: Differentiation of Hepatocytes. Anat. Rec. 1995, 242, 337–349. [Google Scholar] [CrossRef]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-Dimensional Cell Culture Systems and Their Applications in Drug Discovery and Cell-Based Biosensors. ASSAY Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef]
- Gunness, P.; Mueller, D.; Shevchenko, V.; Heinzle, E.; Ingelman-Sundberg, M.; Noor, F. 3D Organotypic Cultures of Human HepaRG Cells: A Tool for In Vitro Toxicity Studies. Toxicol. Sci. 2013, 133, 67–78. [Google Scholar] [CrossRef]
- Ramaiahgari, S.C.; den Braver, M.W.; Herpers, B.; Terpstra, V.; Commandeur, J.N.M.; van de Water, B.; Price, L.S. A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver-like properties for repeated dose high-throughput toxicity studies. Arch. Toxicol. 2014, 88, 1083–1095. [Google Scholar] [CrossRef]
- Skardal, A.; Devarasetty, M.; Soker, S.; Hall, A.R. In situ patterned micro 3D liver constructs for parallel toxicology testing in a fluidic device. Biofabrication 2015, 7, 031001. [Google Scholar] [CrossRef]
- Bierwolf, J.; Lutgehetmann, M.; Feng, K.; Erbes, J.; Deichmann, S.; Toronyi, E.; Stieglitz, C.; Nashan, B.; Ma, P.X.; Pollok, J.M. Primary rat hepatocyte culture on 3D nanofibrous polymer scaffolds for toxicology and pharmaceutical research. Biotechnol. Bioeng. 2011, 108, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.G.; Funk, J.; Robbins, J.B.; Crogan-Grundy, C.; Presnell, S.C.; Singer, T.; Roth, A.B. Bioprinted 3D Primary Liver Tissues Allow Assessment of Organ-Level Response to Clinical Drug Induced Toxicity In Vitro. PLoS ONE 2016, 11, e0158674. [Google Scholar] [CrossRef] [PubMed]
- Ingelman-Sundberg, M.; Lauschke, V.M. 3D human liver spheroids for translational pharmacology and toxicology. Basic Clin. Pharmacol. Toxicol. 2022, 130, 5–15. [Google Scholar] [CrossRef]
- Kammerer, S. Three-Dimensional Liver Culture Systems to Maintain Primary Hepatic Properties for Toxicological Analysis In Vitro. Int. J. Mol. Sci. 2021, 22, 10214. [Google Scholar] [CrossRef]
- Lauschke, V.M.; Shafagh, R.Z.; Hendriks, D.F.G.; Ingelman-Sundberg, M. 3D Primary Hepatocyte Culture Systems for Analyses of Liver Diseases, Drug Metabolism, and Toxicity: Emerging Culture Paradigms and Applications. Biotechnol. J. 2019, 14, 1800347. [Google Scholar] [CrossRef] [PubMed]
- Serras, A.S.; Rodrigues, J.S.; Cipriano, M.; Rodrigues, A.V.; Oliveira, N.G.; Miranda, J.P. A Critical Perspective on 3D Liver Models for Drug Metabolism and Toxicology Studies. Front. Cell Dev. Biol. 2021, 9, 626805. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Ooka, M.; Margolis, R.J.; Xia, M. Liver three-dimensional cellular models for high-throughput chemical testing. Cell Rep. Methods 2023, 3, 100432. [Google Scholar] [CrossRef]
- Messner, S.; Agarkova, I.; Moritz, W.; Kelm, J.M. Multi-cell type human liver microtissues for hepatotoxicity testing. Arch. Toxicol. 2013, 87, 209–213. [Google Scholar] [CrossRef]
- Wrzesinski, K.; Magnone, M.C.; Hansen, L.V.; Kruse, M.E.; Bergauer, T.; Bobadilla, M.; Gubler, M.; Mizrahi, J.; Zhang, K.; Andreasen, C.M.; et al. HepG2/C3A 3D spheroids exhibit stable physiological functionality for at least 24 days after recovering from trypsinisation. Toxicol. Res. 2013, 2, 163–172. [Google Scholar] [CrossRef]
- Lee, B.H.; Kim, M.H.; Lee, J.H.; Seliktar, D.; Cho, N.-J.; Tan, L.P. Modulation of Huh7.5 spheroid formation and functionality using modified PEG-based hydrogels of different stiffness. PLoS ONE 2015, 10, e0118123. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Hori, Y.; Yamamoto, T.; Urashima, T.; Ohara, Y.; Tanaka, H. 3D spheroid cultures improve the metabolic gene expression profiles of HepaRG cells. Biosci. Rep. 2015, 35, e00208. [Google Scholar] [CrossRef] [PubMed]
- Fey, S.J.; Wrzesinski, K. Determination of drug toxicity using 3D spheroids constructed from an immortal human hepatocyte cell line. Toxicol. Sci. Off. J. Soc. Toxicol. 2012, 127, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Sakai, Y.; Yamagami, S.; Nakazawa, K. Comparative Analysis of Gene Expression in Rat Liver Tissue and Monolayer- and Spheroid-Cultured Hepatocytes. Cells Tissues Organs 2009, 191, 281–288. [Google Scholar] [CrossRef]
- Abu-Absi, S.F.; Friend, J.R.; Hansen, L.K.; Hu, W.-S. Structural Polarity and Functional Bile Canaliculi in Rat Hepatocyte Spheroids. Exp. Cell Res. 2002, 274, 56–67. [Google Scholar] [CrossRef]
- Brophy, C.M.; Luebke-Wheeler, J.L.; Amiot, B.P.; Khan, H.; Remmel, R.P.; Rinaldo, P.; Nyberg, S.L. Rat hepatocyte spheroids formed by rocked technique maintain differentiated hepatocyte gene expression and function. Hepatology 2009, 49, 578–586. [Google Scholar] [CrossRef]
- Landry, J.; Bernier, D.; Ouellet, C.; Goyette, R.; Marceau, N. Spheroidal aggregate culture of rat liver cells: Histotypic reorganization, biomatrix deposition, and maintenance of functional activities. J. Cell Biol. 1985, 101, 914–923. [Google Scholar] [CrossRef]
- Kratschmar, D.V.; Messner, S.; Moritz, W.; Odermatt, A. Characterization of a Rat Multi-Cell Type 3D-Liver Microtissue System. J. Tissue Sci. Eng. 2013, 4, 1000130. [Google Scholar] [CrossRef]
- Frank, R.A.; Kavanagh, R.; Burnison, B.K.; Headley, J.V.; Peru, K.M.; Der Kraak, G.V.; Solomon, K.R. Diethylaminoethyl-cellulose clean-up of a large volume naphthenic acid extract. Chemosphere 2006, 64, 1346–1352. [Google Scholar] [CrossRef]
- Marentette, J.R.; Frank, R.A.; Bartlett, A.J.; Gillis, P.L.; Hewitt, L.M.; Peru, K.M.; Headley, J.V.; Brunswick, P.; Shang, D.; Parrott, J.L. Toxicity of naphthenic acid fraction components extracted from fresh and aged oil sands process-affected waters, and commercial naphthenic acid mixtures, to fathead minnow (Pimephales promelas) embryos. Aquat. Toxicol. 2015, 164, 108–117. [Google Scholar] [CrossRef]
- Bartlett, A.J.; Frank, R.A.; Gillis, P.L.; Parrott, J.L.; Marentette, J.R.; Brown, L.R.; Hooey, T.; Vanderveen, R.; McInnis, R.; Brunswick, P.; et al. Toxicity of naphthenic acids to invertebrates: Extracts from oil sands process-affected water versus commercial mixtures. Environ. Pollut. 2017, 227, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Björnsson, E.S.; Vucic, V.; Stirnimann, G.; Robles-Díaz, M. Role of Corticosteroids in Drug-Induced Liver Injury. A Systematic Review. Front. Pharmacol. 2022, 13, 820724. [Google Scholar] [CrossRef] [PubMed]
- Flynn, B.P.; Birnie, M.T.; Kershaw, Y.M.; Pauza, A.G.; Kim, S.; Baek, S.; Rogers, M.F.; Paterson, A.R.; Stavreva, D.A.; Murphy, D.; et al. Corticosterone pattern-dependent glucocorticoid receptor binding and transcriptional regulation within the liver. PLoS Genet. 2021, 17, e1009737. [Google Scholar] [CrossRef]
- Wang, S.-H.; Liang, C.-T.; Liu, Y.-W.; Huang, M.-C.; Huang, S.-C.; Hong, W.-F.; Su, J.-G.J. Crosstalk between activated forms of the aryl hydrocarbon receptor and glucocorticoid receptor. Toxicology 2009, 262, 87–97. [Google Scholar] [CrossRef]
- Rando, G.; Tan, C.K.; Khaled, N.; Montagner, A.; Leuenberger, N.; Bertrand-Michel, J.; Paramalingam, E.; Guillou, H.; Wahli, W. Glucocorticoid receptor-PPARα axis in fetal mouse liver prepares neonates for milk lipid catabolism. eLife 2016, 5, e11853. [Google Scholar] [CrossRef]
- Hasan, A.U.; Ohmori, K.; Hashimoto, T.; Kamitori, K.; Yamaguchi, F.; Rahman, A.; Tokuda, M.; Kobori, H. PPARγ activation mitigates glucocorticoid receptor-induced excessive lipolysis in adipocytes via homeostatic crosstalk. J. Cell. Biochem. 2018, 119, 4627–4635. [Google Scholar] [CrossRef]
- Yamamoto, A.; Kakuta, H.; Sugimoto, Y. Involvement of glucocorticoid receptor activation on anti-inflammatory effect induced by peroxisome proliferator-activated receptor γ agonist in mice. Int. Immunopharmacol. 2014, 22, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Marentette, J.R.; Frank, R.A.; Hewitt, L.M.; Gillis, P.L.; Bartlett, A.J.; Brunswick, P.; Shang, D.; Parrott, J.L. Sensitivity of walleye (Sander vitreus) and fathead minnow (Pimephales promelas) early-life stages to naphthenic acid fraction components extracted from fresh oil sands process-affected waters. Environ. Pollut. 2015, 207, 59–67. [Google Scholar] [CrossRef]
- Clemente, J.S.; Prasad, N.G.N.; MacKinnon, M.D.; Fedorak, P.M. A statistical comparison of naphthenic acids characterized by gas chromatography–mass spectrometry. Chemosphere 2003, 50, 1265–1274. [Google Scholar] [CrossRef]
- Quagraine, E.K.; Peterson, H.G.; Headley, J.V. In Situ Bioremediation of Naphthenic Acids Contaminated Tailing Pond Waters in the Athabasca Oil Sands Region—Demonstrated Field Studies and Plausible Options: A Review. J. Environ. Sci. Health Part A 2005, 40, 685–722. [Google Scholar] [CrossRef] [PubMed]
- Vander Meulen, I.J.; Schock, D.M.; Parrott, J.L.; Mundy, L.J.; Pauli, B.D.; Peru, K.M.; McMartin, D.W.; Headley, J.V. Characterization of naphthenic acid fraction compounds in water from Athabasca oil sands wetlands by Orbitrap high-resolution mass spectrometry. Sci. Total Environ. 2021, 780, 146342. [Google Scholar] [CrossRef] [PubMed]
- Jamshed, L.; Perono, G.A.; Yacoub, L.R.; Gutgesell, R.M.; Frank, R.A.; Hewitt, L.M.; Thomas, P.J.; Holloway, A.C. The effects of oil sands process-affected water naphthenic acid fraction components on GDF15 secretion in extravillous trophoblast cells. Toxicol. Appl. Pharmacol. 2022, 441, 115970. [Google Scholar] [CrossRef] [PubMed]
- Monostory, K.; Kőhalmy, K.; Prough, R.A.; Kóbori, L.; Vereczkey, L. The effect of synthetic glucocorticoid, dexamethasone on CYP1A1 inducibility in adult rat and human hepatocytes. FEBS Lett. 2005, 579, 229–235. [Google Scholar] [CrossRef]
- Gomez, A.; Bindesbøll, C.; Satheesh, S.V.; Grimaldi, G.; Hutin, D.; MacPherson, L.; Ahmed, S.; Tamblyn, L.; Cho, T.; Nebb, H.I.; et al. Characterization of TCDD-inducible poly-ADP-ribose polymerase (TIPARP/ARTD14) catalytic activity. Biochem. J. 2018, 475, 3827–3846. [Google Scholar] [CrossRef]
- Xu, A.; Lam, M.C.; Chan, K.W.; Wang, Y.; Zhang, J.; Hoo, R.L.C.; Xu, J.Y.; Chen, B.; Chow, W.-S.; Tso, A.W.K.; et al. Angiopoietin-like protein 4 decreases blood glucose and improves glucose tolerance but induces hyperlipidemia and hepatic steatosis in mice. Proc. Natl. Acad. Sci. USA 2005, 102, 6086–6091. [Google Scholar] [CrossRef]
- Staiger, H.; Haas, C.; Machann, J.; Werner, R.; Weisser, M.; Schick, F.; Machicao, F.; Stefan, N.; Fritsche, A.; Häring, H.-U. Muscle-Derived Angiopoietin-Like Protein 4 Is Induced by Fatty Acids via Peroxisome Proliferator–Activated Receptor (PPAR)-δ and Is of Metabolic Relevance in Humans. Diabetes 2009, 58, 579–589. [Google Scholar] [CrossRef]
- Singh, A.K.; Chaube, B.; Zhang, X.; Sun, J.; Citrin, K.M.; Canfrán-Duque, A.; Aryal, B.; Rotllan, N.; Varela, L.; Lee, R.G.; et al. Hepatocyte-specific suppression of ANGPTL4 improves obesity-associated diabetes and mitigates atherosclerosis in mice. J. Clin. Investig. 2021, 131, e140989. [Google Scholar] [CrossRef]
- Lichtenstein, L.; Berbée, J.F.P.; van Dijk, S.J.; van Dijk, K.W.; Bensadoun, A.; Kema, I.P.; Voshol, P.J.; Müller, M.; Rensen, P.C.N.; Kersten, S. Angptl4 Upregulates Cholesterol Synthesis in Liver via Inhibition of LPL- and HL-Dependent Hepatic Cholesterol Uptake. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 2420–2427. [Google Scholar] [CrossRef]
- Yu, C.-Y.; Mayba, O.; Lee, J.V.; Tran, J.; Harris, C.; Speed, T.P.; Wang, J.-C. Genome-Wide Analysis of Glucocorticoid Receptor Binding Regions in Adipocytes Reveal Gene Network Involved in Triglyceride Homeostasis. PLoS ONE 2010, 5, e15188. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Y.; Ortlund, E.A. First High-Resolution Crystal Structures of the Glucocorticoid Receptor Ligand-Binding Domain–Peroxisome Proliferator-Activated γ Coactivator 1-α Complex with Endogenous and Synthetic Glucocorticoids. Mol. Pharmacol. 2019, 96, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Furuhashi, M.; Saitoh, S.; Shimamoto, K.; Miura, T. Fatty Acid-Binding Protein 4 (FABP4): Pathophysiological Insights and Potent Clinical Biomarker of Metabolic and Cardiovascular Diseases. Clin. Med. Insights Cardiol. 2014, 8, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.; Kong, P.; Zhang, S.; Zhang, L.; Cao, Y.; Duan, X.; Sun, T.; Tao, Z.; Liu, W. SGK1 in Human Cancer: Emerging Roles and Mechanisms. Front. Oncol. 2021, 10, 608722. [Google Scholar] [CrossRef] [PubMed]
- Ronchetti, S.; Migliorati, G.; Riccardi, C. GILZ as a Mediator of the Anti-Inflammatory Effects of Glucocorticoids. Front. Endocrinol. 2015, 6, 170. [Google Scholar] [CrossRef] [PubMed]
- Nataraja, C.; Dankers, W.; Flynn, J.; Lee, J.P.W.; Zhu, W.; Vincent, F.B.; Gearing, L.J.; Ooi, J.; Pervin, M.; Cristofaro, M.A.; et al. GILZ Regulates the Expression of Pro-Inflammatory Cytokines and Protects Against End-Organ Damage in a Model of Lupus. Front. Immunol. 2021, 12, 652800. [Google Scholar] [CrossRef]
- Cannarile, L.; Delfino, D.V.; Adorisio, S.; Riccardi, C.; Ayroldi, E. Implicating the Role of GILZ in Glucocorticoid Modulation of T-Cell Activation. Front. Immunol. 2019, 10, 1823. [Google Scholar] [CrossRef]
- Mittelbrunn, M.; Sánchez-Madrid, F. Intercellular communication: Diverse structures for exchange of genetic information. Nat. Rev. Mol. Cell Biol. 2012, 13, 328–335. [Google Scholar] [CrossRef]
- Bruzzone, R.; White, T.W.; Paul, D.L. Connections with Connexins: The Molecular Basis of Direct Intercellular Signaling. Eur. J. Biochem. 1996, 238, 1–27. [Google Scholar] [CrossRef]
- Stout, R.F.; Snapp, E.L.; Spray, D.C. Connexin Type and Fluorescent Protein Fusion Tag Determine Structural Stability of Gap Junction Plaques. J. Biol. Chem. 2015, 290, 23497–23514. [Google Scholar] [CrossRef]
- Crespo Yanguas, S.; Willebrords, J.; Maes, M.; da Silva, T.C.; Veloso Alves Pereira, I.; Cogliati, B.; Zaidan Dagli, M.L.; Vinken, M. Connexins and pannexins in liver damage. EXCLI J. 2016, 15, 177–186. [Google Scholar] [CrossRef]
- Li, Y.; Yang, L.; Tao, R.; Shang, Y.; Sun, M.; Peng, S.; Zhao, G.; Zhao, Y. The Expression of Connexin 26 Regulates the Radiosensitivity of Hepatocellular Carcinoma Cells through a Mitogen-Activated Protein Kinases Signal Pathway. Int. J. Mol. Sci. 2022, 23, 14644. [Google Scholar] [CrossRef]
- Cogliati, B.; Da Silva, T.C.; Aloia, T.P.A.; Chaible, L.M.; Real-Lima, M.A.; Sanches, D.S.; Matsuzaki, P.; Hernandez-Blazquez, F.J.; Dagli, M.L.Z. Morphological and molecular pathology of CCL4-induced hepatic fibrosis in connexin43-deficient mice. Microsc. Res. Tech. 2011, 74, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Quax, R.A.; Manenschijn, L.; Koper, J.W.; Hazes, J.M.; Lamberts, S.W.J.; van Rossum, E.F.C.; Feelders, R.A. Glucocorticoid sensitivity in health and disease. Nat. Rev. Endocrinol. 2013, 9, 670–686. [Google Scholar] [CrossRef]
- Hazra, A.; Pyszczynski, N.A.; DuBois, D.C.; Almon, R.R.; Jusko, W.J. Modeling of corticosteroid effects on hepatic low-density lipoprotein receptors and plasma lipid dynamics in rats. Pharm. Res. 2008, 25, 769–780. [Google Scholar] [CrossRef]
- Scheving, L.A.; Buchanan, R.; Krause, M.A.; Zhang, X.; Stevenson, M.C.; Russell, W.E. Dexamethasone modulates ErbB tyrosine kinase expression and signaling through multiple and redundant mechanisms in cultured rat hepatocytes. Am. J. Physiol.-Gastrointest. Liver Physiol. 2007, 293, G552–G559. [Google Scholar] [CrossRef][Green Version]
- Ayyar, V.S.; Almon, R.R.; DuBois, D.C.; Sukumaran, S.; Qu, J.; Jusko, W.J. Functional proteomic analysis of corticosteroid pharmacodynamics in rat liver: Relationship to hepatic stress, signaling, energy regulation, and drug metabolism. J. Proteom. 2017, 160, 84–105. [Google Scholar] [CrossRef] [PubMed]
- Steger, D.J.; Grant, G.R.; Schupp, M.; Tomaru, T.; Lefterova, M.I.; Schug, J.; Manduchi, E.; Stoeckert, C.J.; Lazar, M.A. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 2010, 24, 1035–1044. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Zhang, L.; Ma, X.; Hu, W.; Chen, Y.; Yu, M.; Wang, Q.; Li, X.; Yin, Z.; Zhu, Y.; et al. Tamoxifen inhibits macrophage FABP4 expression through the combined effects of the GR and PPARγ pathways. Biochem. J. 2013, 454, 467–477. [Google Scholar] [CrossRef]
- Brennan-Speranza, T.C.; Henneicke, H.; Gasparini, S.J.; Blankenstein, K.I.; Heinevetter, U.; Cogger, V.C.; Svistounov, D.; Zhang, Y.; Cooney, G.J.; Buttgereit, F.; et al. Osteoblasts mediate the adverse effects of glucocorticoids on fuel metabolism. J. Clin. Investig. 2012, 122, 4172–4189. [Google Scholar] [CrossRef]
- Rafacho, A.; Roma, L.P.; Taboga, S.R.; Boschero, A.C.; Bosqueiro, J.R. Dexamethasone-induced insulin resistance is associated with increased connexin 36 mRNA and protein expression in pancreatic rat islets. Can. J. Physiol. Pharmacol. 2007, 85, 536–545. [Google Scholar] [CrossRef]
- Liu, X.; Qin, J.; Chang, M.; Wang, S.; Li, Y.; Pei, X.; Wang, Y. Enhanced differentiation of human pluripotent stem cells into pancreatic endocrine cells in 3D culture by inhibition of focal adhesion kinase. Stem Cell Res. Ther. 2020, 11, 488. [Google Scholar] [CrossRef] [PubMed]
- Clemente, J.S.; Fedorak, P.M. A review of the occurrence, analyses, toxicity, and biodegradation of naphthenic acids. Chemosphere 2005, 60, 585–600. [Google Scholar] [CrossRef]
- Li, C.; Fu, L.; Stafford, J.; Belosevic, M.; Gamal El-Din, M. The toxicity of oil sands process-affected water (OSPW): A critical review. Sci. Total Environ. 2017, 601–602, 1785–1802. [Google Scholar] [CrossRef]
- Gutgesell, R.M.; Jamshed, L.; Frank, R.A.; Hewitt, L.M.; Thomas, P.J.; Holloway, A.C. Naphthenic acid fraction components from oil sands process-affected water from the Athabasca Oil Sands Region impair murine osteoblast differentiation and function. J. Appl. Toxicol. 2022, 42, 2005–2015. [Google Scholar] [CrossRef]
- Kassotis, C.D.; Nagel, S.C.; Stapleton, H.M. Unconventional oil and gas chemicals and wastewater-impacted water samples promote adipogenesis via PPARγ-dependent and independent mechanisms in 3T3-L1 cells. Sci. Total Environ. 2018, 640–641, 1601–1610. [Google Scholar] [CrossRef]
- Lister, A.; Nero, V.; Farwell, A.; Dixon, D.G.; Van Der Kraak, G. Reproductive and stress hormone levels in goldfish (Carassius auratus) exposed to oil sands process-affected water. Aquat. Toxicol. Amst. Neth. 2008, 87, 170–177. [Google Scholar] [CrossRef]
- Sérée, E.; Villard, P.-H.; Pascussi, J.-M.; Pineau, T.; Maurel, P.; Nguyen, Q.B.; Fallone, F.; Martin, P.-M.; Champion, S.; Lacarelle, B.; et al. Evidence for a new human CYP1A1 regulation pathway involving PPAR-α and 2 PPRE sites. Gastroenterology 2004, 127, 1436–1445. [Google Scholar] [CrossRef]
- Tan, M.J.; Teo, Z.; Sng, M.K.; Zhu, P.; Tan, N.S. Emerging Roles of Angiopoietin-like 4 in Human Cancer. Mol. Cancer Res. 2012, 10, 677–688. [Google Scholar] [CrossRef]
- Oike, Y.; Akao, M.; Kubota, Y.; Suda, T. Angiopoietin-like proteins: Potential new targets for metabolic syndrome therapy. Trends Mol. Med. 2005, 11, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Smati, S.; Régnier, M.; Fougeray, T.; Polizzi, A.; Fougerat, A.; Lasserre, F.; Lukowicz, C.; Tramunt, B.; Guillaume, M.; Burnol, A.-F.; et al. Regulation of hepatokine gene expression in response to fasting and feeding: Influence of PPAR-α and insulin-dependent signalling in hepatocytes. Diabetes Metab. 2020, 46, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chen, W.; Wang, Z.; Xin, M.; Gao, S.; Liu, W.; Wang, K.; Ma, J.; Yan, X.; Ren, Y. Profiles of transcriptome and metabolic pathways after hypobaric hypoxia exposure. Proteome Sci. 2022, 20, 16. [Google Scholar] [CrossRef]
- Fotschki, B.; Laparra, J.M.; Sójka, M. Raspberry Polyphenolic Extract Regulates Obesogenic Signals in Hepatocytes. Molecules 2018, 23, 2103. [Google Scholar] [CrossRef] [PubMed]
- Assunção, M.; Dehghan-Baniani, D.; Yiu, C.H.K.; Später, T.; Beyer, S.; Blocki, A. Cell-Derived Extracellular Matrix for Tissue Engineering and Regenerative Medicine. Front. Bioeng. Biotechnol. 2020, 8, 602009. [Google Scholar] [CrossRef]
- Salehi, M.S.; Neumann, I.D.; Jurek, B.; Pandamooz, S. Co-Stimulation of Oxytocin and Arginine-Vasopressin Receptors Affect Hypothalamic Neurospheroid Size. Int. J. Mol. Sci. 2021, 22, 8464. [Google Scholar] [CrossRef] [PubMed]
- Totaro, A.; Panciera, T.; Piccolo, S. YAP/TAZ upstream signals and downstream responses. Nat. Cell Biol. 2018, 20, 888–899. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.M.; Pankov, R.; Cukierman, E. Dimensions and dynamics in integrin function. Braz. J. Med. Biol. Res. 2003, 36, 959–966. [Google Scholar] [CrossRef]
- Kim, D.; Koh, B.; Kim, K.R.; Kim, K.Y.; Jung, W.H.; Kim, H.Y.; Kim, S.; Rhee, S.D. Anticancer effect of XAV939 is observed by inhibiting lactose dehydrogenase A in a 3-dimensional culture of colorectal cancer cells. Oncol. Lett. 2019, 18, 4858–4864. [Google Scholar] [CrossRef]
- Keeratichamroen, S.; Sornprachum, T.; Ngiwsara, L.; Ornnork, N.; Svasti, J. p-STAT3 influences doxorubicin and etoposide resistance of A549 cells grown in an in vitro 3D culture model. Oncol. Rep. 2023, 49, 1–12. [Google Scholar] [CrossRef]
- Imamura, Y.; Mukohara, T.; Shimono, Y.; Funakoshi, Y.; Chayahara, N.; Toyoda, M.; Kiyota, N.; Takao, S.; Kono, S.; Nakatsura, T.; et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015, 33, 1837–1843. [Google Scholar] [CrossRef]



| Accession Number | Gene Name | Symbol | Forward Sequence (5′-3′) | Reverse Sequence (5′-3′) |
|---|---|---|---|---|
| NM_017101.1 | Peptidylprolyl isomerase A | Ppia | CCGCTGTCTCTTTTCGCC | GCTGTCTTTGGAACTTTGTCTGC |
| NM_012512.2 | beta-2-microglobulin | B2m | AATTCACACCCACCGAGACC | GCTCCTTCAGAGTGACGTGT |
| NM_012540.3 | Cytochrome P450 Family 1 Subfamily A Member 1 | Cyp1a1 | TTGGGGAGGTTACTGGTTCTG | GAGTTAGGGAGGTAACGGAGG |
| XM_039102285.1 | TCDD Inducible Poly(ADP-Ribose) Polymerase | Tiparp | GGTGGGATTGGGTTAAGCTCT | CAGTCCATCTATACACCTGCCC |
| XM_039079418.1 | Angiopoietin Like 4 | Angptl4 | CAGGACTGGGATGGCAATG | CCTCACCCCCCAAATGG |
| XM_006230695.3 | Carnitine Palmitoyltransferase 1A | Cpt1a | CACCCCAACCCATATCCAGG | TCCTCACGGTCTAATGTGCG |
| NM_031561.2 | CD36 Molecule | Cd36 | TCTCACACAACTCAGATACTGCT | GCACTTGCTTCTTGCCAACT |
| NM_001193568.1 | Serum/Glucocorticoid Regulated Kinase 1 | Sgk1 | GAAACGGTCTTGCAGTGACG | CTGACTGACAACTGGGGCAT |
| NM_031345.1 | TSC22 Domain Family Member 3 | Gilz | GGAGGTCCTAAAGGAGCAGATTC | GCGTCTTCAGGAGGGTATTCTC |
| NM_031347.1 | PPARG Coactivator 1 Alpha | Pgc1α | ATGGAGTGACATAGAGTGTGCT | CACCACTTCAATCCACCCAGA |
| NM_001173450.1 | Fatty Acid Binding Protein 4 | Fabp4 | GCCCAAGCTGGGATCTCA | CCCCCATGAAAAGTCTTCTAGTAAAA |
| NM_012598.2 | Lipoprotein Lipase | Lpl | AACCCCAGCAAGGCATACAG | AGGCAGAGCCCTTTCTCAAAT |
| NM_001004099.2 | Gap Junction Protein Beta 2 | Cx26 | GCGACCCACTTCTGACCAA | TCCCCTGTAAGGAGCAGGT |
| NM_012567.2 | Gap Junction Protein Alpha 1 | Cx43 | GCACTTTTCTTTCATTGGGGGA | GTCTGCTGTTTGGTACT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jamshed, L.; Jamshed, S.; Frank, R.A.; Hewitt, L.M.; Thomas, P.J.; Holloway, A.C. Assessing Receptor Activation in 2D and 3D Cultured Hepatocytes: Responses to a Single Compound and a Complex Mixture. Toxics 2024, 12, 631. https://doi.org/10.3390/toxics12090631
Jamshed L, Jamshed S, Frank RA, Hewitt LM, Thomas PJ, Holloway AC. Assessing Receptor Activation in 2D and 3D Cultured Hepatocytes: Responses to a Single Compound and a Complex Mixture. Toxics. 2024; 12(9):631. https://doi.org/10.3390/toxics12090631
Chicago/Turabian StyleJamshed, Laiba, Shanza Jamshed, Richard A. Frank, L. Mark Hewitt, Philippe J. Thomas, and Alison C. Holloway. 2024. "Assessing Receptor Activation in 2D and 3D Cultured Hepatocytes: Responses to a Single Compound and a Complex Mixture" Toxics 12, no. 9: 631. https://doi.org/10.3390/toxics12090631
APA StyleJamshed, L., Jamshed, S., Frank, R. A., Hewitt, L. M., Thomas, P. J., & Holloway, A. C. (2024). Assessing Receptor Activation in 2D and 3D Cultured Hepatocytes: Responses to a Single Compound and a Complex Mixture. Toxics, 12(9), 631. https://doi.org/10.3390/toxics12090631







