Explainable Vision Transformer with Self-Supervised Learning to Predict Alzheimer’s Disease Progression Using 18F-FDG PET
Abstract
:1. Introduction
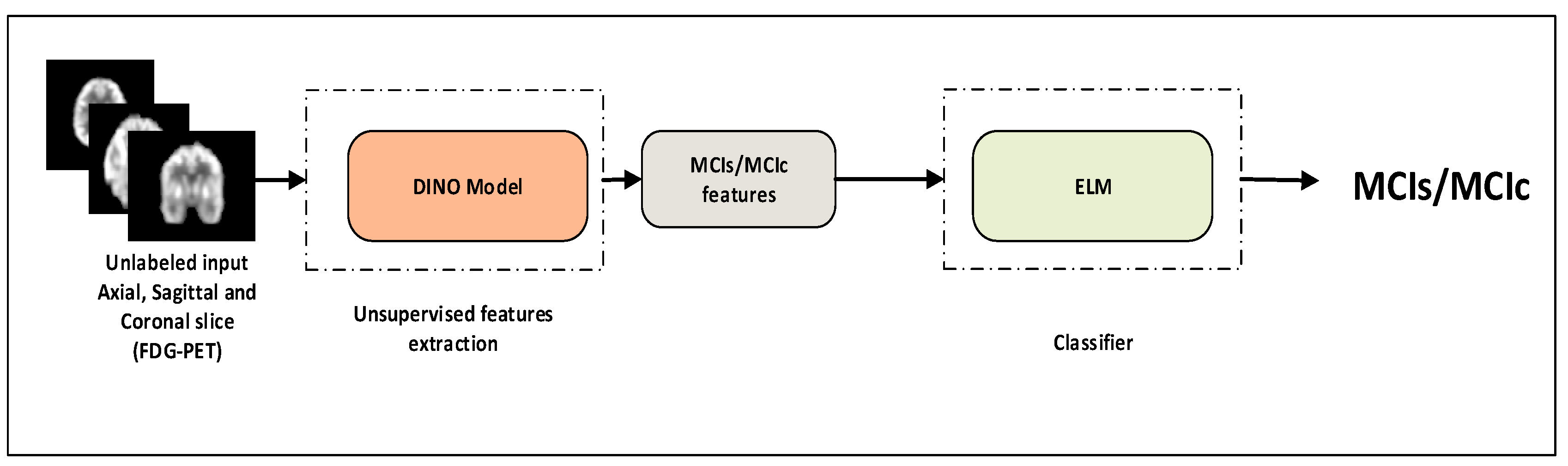
- A transformer model is suggested for the identification of MCI progression. The model expands upon the ViT backbone by utilizing 18F-FDG-PET and self-supervised learning to tackle the issue of MCI progression and disease identification.
- To address the issue of inadequate data in the field of brain imaging, we suggested a cross-domain transfer learning technique. We used ViT as the backbone with DINO.
- In the MCI recognition, experimental data show that the proposed method can achieve more competitive outcomes than current models. The model accuracy levels with the ADNI dataset were 92.31%, which is higher than the baseline’s ViT approach. Finally, we visualized important metabolic brain regions, which can assist the physician for proper analysis of MCI.
2. Materials and Methods
2.1. Dataset
2.2. FDG-PET Image Acquisition and Preprocessing
2.3. Self-Supervised Learning
2.4. Vision Transformer (ViT)
2.5. 18F-FDG-PET Feature Learning with ViT-Dino
2.6. Classifiers
2.7. Training Setup
2.8. Evaluation Matrixs
3. Results
3.1. Classification Performance on 18F-FDG-PET
3.2. Ablation Study
3.3. Performance Comparison with State-of-Art Methods
3.4. Pathological Attention Regions on FDG-PET by ViT DINO
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ottoy, J.; Niemantsverdriet, E.; Verhaeghe, J.; De Roeck, E.; Struyfs, H.; Somers, C.; Wyffels, L.; Ceyssens, S.; Van Mossevelde, S.; Van den Bossche, T.; et al. Association of short-term cognitive decline and MCI-to-AD dementia conversion with CSF, MRI, amyloid- and 18F-FDG-PET imaging. NeuroImage Clin. 2019, 22, 101771. [Google Scholar] [CrossRef] [PubMed]
- World Alzheimer Report 2021: Journey through the Diagnosis of Dementia. Available online: https://www.alzint.org/u/World-Alzheimer-Report-2021.pdf (accessed on 10 September 2023).
- 2023 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2023, 19, alz.13016. [CrossRef]
- Silveira, M.; Marques, J. Boosting Alzheimer Disease Diagnosis Using PET Images. In Proceedings of the 2010 20th International Conference on Pattern Recognition, Istanbul, Turkey, 23–26 August 2010; pp. 2556–2559. [Google Scholar]
- Hampel, H.; Hardy, J.; Blennow, K.; Chen, C.; Perry, G.; Kim, S.H.; Villemagne, V.L.; Aisen, P.; Vendruscolo, M.; Iwatsubo, T.; et al. The Amyloid-β Pathway in Alzheimer’s Disease. Mol. Psychiatry 2021, 26, 5481–5503. [Google Scholar] [CrossRef]
- Jack, C.R.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimer’s Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef]
- Yu, W.; Lei, B.; Shen, Y.; Wang, S.; Liu, Y.; Feng, Z.; Hu, Y.; Ng, M.K. Morphological feature visualization of Alzheimer’s disease via Multidirectional Perception GAN. arXiv 2021, arXiv:2111.12886. [Google Scholar] [CrossRef]
- Mecocci, P.; Boccardi, V. The impact of aging in dementia: It is time to refocus attention on the main risk factor of dementia. Ageing Res. Rev. 2021, 65, 101210. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Cheng, D.; Yan, W. Alzheimer’s Disease Neuroimaging Initiative Classification of Alzheimer’s Disease by Combination of Convolutional and Recurrent Neural Networks Using FDG-PET Images. Front. Neuroinform. 2018, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Nesteruk, M.; Nesteruk, T.; Styczyńska, M.; Mandecka, M.; Barczak, A.; Barcikowska, M. Combined use of biochemical and volumetric biomarkers to assess the risk of conversion of mild cognitive impairment to Alzheimer’s disease. Folia Neuropathol. 2016, 54, 369–374. [Google Scholar] [CrossRef]
- Caminiti, S.P.; Ballarini, T.; Sala, A.; Cerami, C.; Presotto, L.; Santangelo, R.; Fallanca, F.; Vanoli, E.G.; Gianolli, L.; Iannaccone, S.; et al. FDG-PET and CSF biomarker accuracy in prediction of conversion to different dementias in a large multicentre MCI cohort. NeuroImage Clin. 2018, 18, 167–177. [Google Scholar] [CrossRef]
- Nuvoli, S.; Tanda, G.; Stazza, M.L.; Madeddu, G.; Spanu, A. Qualitative and Quantitative Analyses of Brain 18Fluoro-Deoxy-Glucose Positron Emission Tomography in Primary Progressive Aphasia. Dement. Geriatr. Cogn. Disord. 2020, 48, 250–260. [Google Scholar] [CrossRef]
- Liu, F.; Wee, C.-Y.; Chen, H.; Shen, D. Inter-modality relationship constrained multi-modality multi-task feature selection for Alzheimer’s Disease and mild cognitive impairment identification. NeuroImage 2014, 84, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Xia, Y.; Cai, T.W.; Feng, D.D. Semi-supervised manifold learning with affinity regularization for Alzheimer’s disease identification using positron emission tomography imaging. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 2251–2254. [Google Scholar]
- Shen, D.; Wu, G.; Suk, H.-I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef]
- Shaffer, J.L.; Petrella, J.R.; Sheldon, F.C.; Choudhury, K.R.; Calhoun, V.D.; Coleman, R.E.; Doraiswamy, P.M. Predicting Cognitive Decline in Subjects at Risk for Alzheimer Disease by Using Combined Cerebrospinal Fluid, MR Imaging, and PET Biomarkers. Radiology 2013, 266, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Massa, F.; Chincarini, A.; Bauckneht, M.; Raffa, S.; Peira, E.; Arnaldi, D.; Pardini, M.; Pagani, M.; Orso, B.; Donegani, M.I.; et al. Added value of semiquantitative analysis of brain FDG-PET for the differentiation between MCI-Lewy bodies and MCI due to Alzheimer’s disease. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 1263–1274. [Google Scholar] [CrossRef] [PubMed]
- Lei, B.; Yu, S.; Zhao, X.; Frangi, A.F.; Tan, E.-L.; Elazab, A.; Wang, T.; Wang, S. Diagnosis of early Alzheimer’s disease based on dynamic high order networks. Brain Imaging Behav. 2021, 15, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Q.; Lei, B.; Shen, Y.; Liu, Y.; Feng, Z.; Wang, S. Multimodal Representations Learning and Adversarial Hypergraph Fusion for Early Alzheimer’s Disease Prediction. arXiv 2021, arXiv:2107.09928. [Google Scholar]
- Hu, S.; Yu, W.; Chen, Z.; Wang, S. Medical Image Reconstruction Using Generative Adversarial Network for Alzheimer Disease Assessment with Class-Imbalance Problem. In Proceedings of the 2020 IEEE 6th International Conference on Computer and Communications (ICCC), Chengdu, China, 11–14 December 2020; pp. 1323–1327. [Google Scholar]
- Aggarwal, R.; Sounderajah, V.; Martin, G.; Ting, D.S.W.; Karthikesalingam, A.; King, D.; Ashrafian, H.; Darzi, A. Diagnostic accuracy of deep learning in medical imaging: A systematic review and meta-analysis. NPJ Digit. Med. 2021, 4, 65. [Google Scholar] [CrossRef]
- Varoquaux, G.; Cheplygina, V. Machine learning for medical imaging: Methodological failures and recommendations for the future. NPJ Digit. Med. 2022, 5, 48. [Google Scholar] [CrossRef]
- Salahuddin, Z.; Woodruff, H.C.; Chatterjee, A.; Lambin, P. Transparency of deep neural networks for medical image analysis: A review of interpretability methods. Comput. Biol. Med. 2022, 140, 105111. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. arXiv 2019, arXiv:1801.04381. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Li, J.; Wei, Y.; Wang, C.; Hu, Q.; Liu, Y.; Xu, L. 3-D CNN-Based Multichannel Contrastive Learning for Alzheimer’s Disease Automatic Diagnosis. IEEE Trans. Instrum. Meas. 2022, 71, 1–11. [Google Scholar] [CrossRef]
- Zhu, W.; Sun, L.; Huang, J.; Han, L.; Zhang, D. Dual Attention Multi-Instance Deep Learning for Alzheimer’s Disease Diagnosis With Structural MRI. IEEE Trans. Med. Imaging 2021, 40, 2354–2366. [Google Scholar] [CrossRef] [PubMed]
- Acquarelli, J.; van Laarhoven, T.; Postma, G.J.; Jansen, J.J.; Rijpma, A.; van Asten, S.; Heerschap, A.; Buydens, L.M.C.; Marchiori, E. Convolutional neural networks to predict brain tumor grades and Alzheimer’s disease with MR spectroscopic imaging data. PLoS ONE 2022, 17, e0268881. [Google Scholar] [CrossRef]
- Kang, W.; Lin, L.; Zhang, B.; Shen, X.; Wu, S. Multi-model and multi-slice ensemble learning architecture based on 2D convolutional neural networks for Alzheimer’s disease diagnosis. Comput. Biol. Med. 2021, 136, 104678. [Google Scholar] [CrossRef]
- Park, S.W.; Yeo, N.Y.; Kim, Y.; Byeon, G.; Jang, J.-W. Deep learning application for the classification of Alzheimer’s disease using 18F-flortaucipir (AV-1451) tau positron emission tomography. Sci. Rep. 2023, 13, 8096. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. arXiv 2021, arXiv:2010.11929. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is All you Need. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Volume 30. [Google Scholar]
- Hoang, G.M.; Kim, U.-H.; Kim, J.G. Vision transformers for the prediction of mild cognitive impairment to Alzheimer’s disease progression using mid-sagittal sMRI. Front. Aging Neurosci. 2023, 15, 1102869. [Google Scholar] [CrossRef]
- Lyu, Y.; Yu, X.; Zhu, D.; Zhang, L. Classification of Alzheimer’s Disease via Vision Transformer: Classification of Alzheimer’s Disease via Vision Transformer. In Proceedings of the 15th International Conference on PErvasive Technologies Related to Assistive Environments, Corfu, Greece, 29 June–1 July 2022; Association for Computing Machinery: New York, NY, USA, 2022; pp. 463–468. [Google Scholar]
- Sarraf, S.; Sarraf, A.; DeSouza, D.D.; Anderson, J.A.E.; Kabia, M. The Alzheimer’s Disease Neuroimaging Initiative OViTAD: Optimized Vision Transformer to Predict Various Stages of Alzheimer’s Disease Using Resting-State fMRI and Structural MRI Data. Brain Sci. 2023, 13, 260. [Google Scholar] [CrossRef]
- Yin, Y.; Jin, W.; Bai, J.; Liu, R.; Zhen, H. SMIL-DeiT:Multiple Instance Learning and Self-supervised Vision Transformer network for Early Alzheimer’s disease classification. In Proceedings of the 2022 International Joint Conference on Neural Networks (IJCNN), Padua, Italy, 18–23 July 2022; pp. 1–6. [Google Scholar]
- Zhu, J.; Tan, Y.; Lin, R.; Miao, J.; Fan, X.; Zhu, Y.; Liang, P.; Gong, J.; He, H. Efficient self-attention mechanism and structural distilling model for Alzheimer’s disease diagnosis. Comput. Biol. Med. 2022, 147, 105737. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Khalvati, F. Introducing Vision Transformer for Alzheimer’s Disease classification task with 3D input. arXiv 2022, arXiv:2210.01177. [Google Scholar]
- Grill, J.-B.; Strub, F.; Altché, F.; Tallec, C.; Richemond, P.H.; Buchatskaya, E.; Doersch, C.; Pires, B.A.; Guo, Z.D.; Azar, M.G.; et al. Bootstrap your own latent: A new approach to self-supervised Learning. arXiv 2020, arXiv:2006.07733. [Google Scholar]
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A Simple Framework for Contrastive Learning of Visual Representations. arXiv 2020, arXiv:2002.05709. [Google Scholar]
- He, K.; Fan, H.; Wu, Y.; Xie, S.; Girshick, R. Momentum Contrast for Unsupervised Visual Representation Learning. arXiv 2020, arXiv:1911.05722. [Google Scholar]
- Caron, M.; Touvron, H.; Misra, I.; Jégou, H.; Mairal, J.; Bojanowski, P.; Joulin, A. Emerging Properties in Self-Supervised Vision Transformers. arXiv 2021, arXiv:2104.14294. [Google Scholar]
- Chen, L.; Bentley, P.; Mori, K.; Misawa, K.; Fujiwara, M.; Rueckert, D. Self-supervised learning for medical image analysis using image context restoration. Med. Image Anal. 2019, 58, 101539. [Google Scholar] [CrossRef]
- Tang, Y.; Yang, D.; Li, W.; Roth, H.; Landman, B.; Xu, D.; Nath, V.; Hatamizadeh, A. Self-Supervised Pre-Training of Swin Transformers for 3D Medical Image Analysis. arXiv 2022, arXiv:2111.14791. [Google Scholar]
- Dong, Q.-Y.; Li, T.-R.; Jiang, X.-Y.; Wang, X.-N.; Han, Y.; Jiang, J.-H. Glucose metabolism in the right middle temporal gyrus could be a potential biomarker for subjective cognitive decline: A study of a Han population. Alzheimer’s Res. Ther. 2021, 13, 74. [Google Scholar] [CrossRef]
- SPM—Statistical Parametric Mapping. Available online: https://www.fil.ion.ucl.ac.uk/spm/ (accessed on 11 January 2023).
- Gonzalez-Escamilla, G.; Lange, C.; Teipel, S.; Buchert, R.; Grothe, M.J. PETPVE12: An SPM toolbox for Partial Volume Effects correction in brain PET—Application to amyloid imaging with AV45-PET. NeuroImage 2017, 147, 669–677. [Google Scholar] [CrossRef]
- Caron, M.; Misra, I.; Mairal, J.; Goyal, P.; Bojanowski, P.; Joulin, A. Unsupervised Learning of Visual Features by Contrasting Cluster Assignments. arXiv 2021, arXiv:2006.09882. [Google Scholar]
- Qureshi, M.N.I.; Min, B.; Jo, H.J.; Lee, B. Multiclass Classification for the Differential Diagnosis on the ADHD Subtypes Using Recursive Feature Elimination and Hierarchical Extreme Learning Machine: Structural MRI Study. PLoS ONE 2016, 11, e0160697. [Google Scholar] [CrossRef]
- Zhang, W.; Shen, H.; Ji, Z.; Meng, G.; Wang, B. Identification of Mild Cognitive Impairment Using Extreme Learning Machines Model. In Intelligent Computing Theories and Methodologies; Huang, D.-S., Jo, K.-H., Hussain, A., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 589–600. [Google Scholar]
- Self-Supervised Vision Transformers with DINO. Meta Research. Available online: https://github.com/facebookresearch/dino#self-supervised-vision-transformers-with-dino (accessed on 29 August 2023).
- Loshchilov, I.; Hutter, F. Decoupled Weight Decay Regularization. arXiv 2019, arXiv:1711.05101. [Google Scholar]
- rwightman/pytorch-image-models: v0.6.11 Release. Available online: https://zenodo.org/records/7140899 (accessed on 10 September 2023).
- Nozadi, S.H.; Kadoury, S.; Alzheimer’s Disease Neuroimaging Initiative. Classification of Alzheimer’s and MCI Patients from Semantically Parcelled PET Images: A Comparison between AV45 and FDG-PET. Int. J. Biomed. Imaging 2018, 2018, e1247430. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.; Stocks, J.; Heywood, A.; Jung, Y.; Jenkins, L.; Hill, V.; Katsaggelos, A.; Popuri, K.; Rosen, H.; Beg, M.F.; et al. Transfer learning for predicting conversion from mild cognitive impairment to dementia of Alzheimer’s type based on a three-dimensional convolutional neural network. Neurobiol. Aging 2021, 99, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Liu, Y.; Wu, H.; Wang, J.; Chen, L.; Chen, C.L.P. Broad learning for early diagnosis of Alzheimer’s disease using FDG-PET of the brain. Front. Neurosci. 2023, 17, 1137567. [Google Scholar] [CrossRef]
- Choi, H.; Jin, K.H. Predicting cognitive decline with deep learning of brain metabolism and amyloid imaging. Behav. Brain Res. 2018, 344, 103–109. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, Y.; Zhou, L.; Yuan, H.; Shen, D. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. NeuroImage 2011, 55, 856–867. [Google Scholar] [CrossRef]
- Aggleton, J.P.; Nelson, A.J.D. Why do lesions in the rodent anterior thalamic nuclei cause such severe spatial deficits? Neurosci. Biobehav. Rev. 2015, 54, 131–144. [Google Scholar] [CrossRef]
- Role of the Medial Prefrontal Cortex in Cognition, Ageing and Dementia|Brain Communications|Oxford Academic. Available online: https://academic.oup.com/braincomms/article/3/3/fcab125/6296836 (accessed on 28 June 2023).
- Brewer, A.; Barton, B. Visual cortex in aging and Alzheimer’s disease: Changes in visual field maps and population receptive fields. Front. Psychol. 2014, 5, 74. [Google Scholar] [CrossRef]
- Liu, S.; Liu, S.; Cai, W.; Che, H.; Pujol, S.; Kikinis, R.; Feng, D.; Fulham, M.J. ADNI Multimodal Neuroimaging Feature Learning for Multiclass Diagnosis of Alzheimer’s Disease. IEEE Trans. Biomed. Eng. 2015, 62, 1132–1140. [Google Scholar] [CrossRef] [PubMed]








| Groups | Gender (M/F) | Education | Age (Years) | MoCA | MMSE | CDR | APOEƐ4 |
|---|---|---|---|---|---|---|---|
| MCI-s | 130/115 # | 16.3 ± 2.7 | 72.3 ± 7.5 * | 23.7 ± 2.4 * | 28.0 ± 1.7 * | 1.1 ± 0.5 * | 43.1% |
| MCI-c | 119/105 | 16.2 ± 2.1 | 74.1 ± 7.3 | 21.1 ± 2.7 | 26.3 ± 2.1 | 2.3 ± 1.0 | 74.0% |
| Model | Classifiers | ACC % (mean ± std) | SEN % (mean ± std) | SPE % (mean ± std) | PRE % (mean ± std) | Recall % (mean ± std) | F1-Score % (mean ± std) |
|---|---|---|---|---|---|---|---|
| ViT-S | 82.37 ± 1.29 | 75.51 ± 2.01 | 88.71 ± 1.03 | 83.87 ± 1.45 | 84.33 ± 3.02 | 83.30 ± 1.05 | |
| ViT-B | 81.75 ± 2.13 | 85.38 ± 3.14 | 79.85 ± 1.71 | 83.54 ± 2.52 | 82.47 ± 2.45 | 82.71 ± 1.71 | |
| ViT-L | 78.93 ± 1.07 | 67.83 ± 2.73 | 90.97 ± 1.07 | 81.75 ± 2.59 | 78.83 ± 3.74 | 79.34 ± 2.15 | |
| DINO ViT-B | KNN | 88.36 ± 1.91 | 81.71 ± 2.47 | 95.08 ± 1.32 | 89.15 ± 3.11 | 88.31 ± 2.14 | 88.25 ± 1.72 |
| SVM | 85.24 ± 3.73 | 92.92 ± 1.01 | 78.06 ± 3.45 | 86.01 ± 2.47 | 85.49 ± 1.75 | 85.21 ± 1.04 | |
| ELM | 92.31 ± 1.07 | 90.21 ± 3.37 | 95.50 ± 2.15 | 93.10 ± 1.88 | 92.95 ± 2.31 | 93.92 ± 1.33 |
| Model | Patch Size | Classifiers | ACC % (mean ± std) | SEN % (mean ± std) | SPE % (mean ± std) | PRE % (mean ± std) | Recall % (mean ± std) | F1-Score % (mean ± std) |
|---|---|---|---|---|---|---|---|---|
| DINO ViT-B | 8 | KNN | 87.49 ± 1.23 | 95.93 ± 3.11 | 79.61 ± 2.15 | 88.45 ± 1.33 | 87.77 ± 1.04 | 87.46 ± 1.87 |
| SVM | 86.47 ± 1.55 | 78.16 ± 2.45 | 94.23 ± 1.07 | 87.44 ± 1.58 | 86.2 ± 2.78 | 86.31 ± 1.95 | ||
| ELM | 91.56 ± 1.03 | 86.75 ± 1.47 | 96.06 ± 1.56 | 91.98 ± 1.19 | 91.4 ± 1.74 | 91.51 ± 1.51 | ||
| 16 | KNN | 88.36 ± 1.91 | 81.71 ± 2.47 | 95.08 ± 1.32 | 89.15 ± 3.11 | 88.31 ± 2.14 | 88.25 ± 1.72 | |
| SVM | 85.24 ± 3.73 | 92.92 ± 1.01 | 78.06 ± 3.45 | 86.01 ± 2.47 | 85.49 ± 1.75 | 85.21 ± 1.04 | ||
| ELM | 92.95 ± 1.07 | 90.21 ± 3.37 | 95.50 ± 2.15 | 93.10 ± 1.88 | 92.95 ± 2.31 | 93.92 ± 1.33 | ||
| 32 | KNN | 82.62 ± 3.01 | 93.52 ± 1.41 | 72.43 ± 4.75 | 84.15 ± 2.13 | 82.98 ± 1.71 | 82.58 ± 1.37 | |
| SVM | 81.31 ± 2.45 | 64.16 ± 5.78 | 97.33 ± 1.21 | 85.07 ± 2.04 | 80.74 ± 3.41 | 80.58 ± 1.78 | ||
| ELM | 86.84 ± 1.58 | 75.45 ± 3.71 | 97.47 ± 1.03 | 88.74 ± 1.23 | 86.64 ± 1.59 | 86.57 ± 1.79 |
| Study | Modality | Method | ACC | SEN | SPE |
|---|---|---|---|---|---|
| Nozadi et al. [55] | FDG-PET | RF | 72.5 | 79.2 | 69.9 |
| Bae et al. [56] | MRI | ResNet | 86.1 | 84 | 74.8 |
| Zhu et al. [28] | MRI | Dual attention multi-instance deep learning network | 80.2 | 77.1 | 82.6 |
| MRI | ViT-S | 83.27 | 85.07 | 81.48 | |
| Hoang et al. [34] | ViT-B | 80.67 | 79.1 | 82.22 | |
| ViT-L | 72.86 | 74.63 | 71.11 | ||
| Duan J et al. [57] | FDG-PET | CNN | - | 81.63 | 85.19 |
| Choi and Jin et al. [58] | FDG-PET | Deep Learning | 84.2 | 81.0 | 87.0 |
| Our | FDG-PET | DINO-ELM | 92.95 | 90.21 | 95.50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khatri, U.; Kwon, G.-R. Explainable Vision Transformer with Self-Supervised Learning to Predict Alzheimer’s Disease Progression Using 18F-FDG PET. Bioengineering 2023, 10, 1225. https://doi.org/10.3390/bioengineering10101225
Khatri U, Kwon G-R. Explainable Vision Transformer with Self-Supervised Learning to Predict Alzheimer’s Disease Progression Using 18F-FDG PET. Bioengineering. 2023; 10(10):1225. https://doi.org/10.3390/bioengineering10101225
Chicago/Turabian StyleKhatri, Uttam, and Goo-Rak Kwon. 2023. "Explainable Vision Transformer with Self-Supervised Learning to Predict Alzheimer’s Disease Progression Using 18F-FDG PET" Bioengineering 10, no. 10: 1225. https://doi.org/10.3390/bioengineering10101225
APA StyleKhatri, U., & Kwon, G.-R. (2023). Explainable Vision Transformer with Self-Supervised Learning to Predict Alzheimer’s Disease Progression Using 18F-FDG PET. Bioengineering, 10(10), 1225. https://doi.org/10.3390/bioengineering10101225







