Feature Papers in Advanced Computational Technologies for Biosignal Processing
Share This Topical Collection
Editors
 Dr. Shi Qiu
Dr. Shi Qiu
 Dr. Shi Qiu
Dr. Shi Qiu
E-Mail
Website
Collection Editor
Xi’an Institute of Optics and Precision Mechanics, Chinese Academy of Sciences, Xi’an, China
Interests: medical imaging; pattern recognition; signal and information processing
 Prof. Dr. Behnaz Ghoraani
Prof. Dr. Behnaz Ghoraani
 Prof. Dr. Behnaz Ghoraani
Prof. Dr. Behnaz Ghoraani
E-Mail
Website
Collection Editor
Department of Electrical Engineering and Computer Science, Florida Atlantic University, Boca Raton, FL 33431-0991, USA
Interests: biosignal processing; gait analysis; cardiovascular engineering; speech data analysis
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
Biosignal processing comprises an interdisciplinary field involving the acquisition, analysis, and interpretation of biological signals that originate from physiological mechanisms. It has become increasingly important in modern medicine and bioengineering, with applications ranging from use in diagnostics and therapeutic interventions to the development of prosthetic devices and brain–computer interfaces. The perspective of biosignal processing involves a deep understanding of the underlying physiological mechanisms that produce the signals, as well as advanced signal processing techniques that can extract useful information from noisy and complex signals. This field is poised to play an important role in advancing our understanding of the human body and developing new technologies that can improve human health and quality of life.
The aim of this Topical Collection, entitled “Feature Papers in Advanced Computational Technologies for Biosignal Processing”, is to collect high-quality research articles, short communications, and review articles in the fields of computational biosignal processing. We welcome scholars and engineers from academia and industry all around the world to contribute papers to highlight the scientific discoveries and cutting-edge technologies in this field.
Potential topics include, but are not limited to:
- electrophysiological signal (ECG, EEG, EMG, MEG) processing
- AI and machine learning for biomedical data analysis
- medical imaging and image analysis
- wearable sensor network and integration
- biodata mining
- computational methods and programs in biomedicine
- digital twins in healthcare
- simulation and modeling of physiological mechanisms
- biomedical speech and acoustic signal analysis
- computational biomechanics
Dr. Yunfeng Wu
Dr. Shi Qiu
Dr. Behnaz Ghoraani
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Bioengineering is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- electrophysiological signal (ECG, EEG, EMG, MEG) processing
- AI and machine learning for biomedical data analysis
- medical imaging and image analysis
- wearable sensor network and integration
- biodata mining
Published Papers (4 papers)
Open AccessArticle
Adaptive Filtering with Fitted Noise Estimate (AFFiNE): Blink Artifact Correction in Simulated and Real P300 Data
by
Kevin E. Alexander, Justin R. Estepp and Sherif M. Elbasiouny
Viewed by 1120
Abstract
(1) Background: The electroencephalogram (EEG) is frequently corrupted by ocular artifacts such as saccades and blinks. Methods for correcting these artifacts include independent component analysis (ICA) and recursive-least-squares (RLS) adaptive filtering (-AF). Here, we introduce a new method, AFFiNE, that applies Bayesian adaptive
[...] Read more.
(1) Background: The electroencephalogram (EEG) is frequently corrupted by ocular artifacts such as saccades and blinks. Methods for correcting these artifacts include independent component analysis (ICA) and recursive-least-squares (RLS) adaptive filtering (-AF). Here, we introduce a new method, AFFiNE, that applies Bayesian adaptive regression spline (BARS) fitting to the adaptive filter’s reference noise input to address the known limitations of both ICA and RLS-AF, and then compare the performance of all three methods. (2) Methods: Artifact-corrected P300 morphologies, topographies, and measurements were compared between the three methods, and to known truth conditions, where possible, using real and simulated blink-corrupted event-related potential (ERP) datasets. (3) Results: In both simulated and real datasets, AFFiNE was successful at removing the blink artifact while preserving the underlying P300 signal in all situations where RLS-AF failed. Compared to ICA, AFFiNE resulted in either a practically or an observably comparable error. (4) Conclusions: AFFiNE is an ocular artifact correction technique that is implementable in online analyses; it can adapt to being non-stationarity and is independent of channel density and recording duration. AFFiNE can be utilized for the removal of blink artifacts in situations where ICA may not be practically or theoretically useful.
Full article
►▼
Show Figures
Open AccessArticle
Multi-Shared-Task Self-Supervised CNN-LSTM for Monitoring Free-Body Movement UPDRS-III Using Wearable Sensors
by
Mustafa Shuqair, Joohi Jimenez-Shahed and Behnaz Ghoraani
Viewed by 1620
Abstract
The Unified Parkinson’s Disease Rating Scale (UPDRS) is used to recognize patients with Parkinson’s disease (PD) and rate its severity. The rating is crucial for disease progression monitoring and treatment adjustment. This study aims to advance the capabilities of PD management by developing
[...] Read more.
The Unified Parkinson’s Disease Rating Scale (UPDRS) is used to recognize patients with Parkinson’s disease (PD) and rate its severity. The rating is crucial for disease progression monitoring and treatment adjustment. This study aims to advance the capabilities of PD management by developing an innovative framework that integrates deep learning with wearable sensor technology to enhance the precision of UPDRS assessments. We introduce a series of deep learning models to estimate UPDRS Part III scores, utilizing motion data from wearable sensors. Our approach leverages a novel Multi-shared-task Self-supervised Convolutional Neural Network–Long Short-Term Memory (CNN-LSTM) framework that processes raw gyroscope signals and their spectrogram representations. This technique aims to refine the estimation accuracy of PD severity during naturalistic human activities. Utilizing 526 min of data from 24 PD patients engaged in everyday activities, our methodology demonstrates a strong correlation of 0.89 between estimated and clinically assessed UPDRS-III scores. This model outperforms the benchmark set by single and multichannel CNN, LSTM, and CNN-LSTM models and establishes a new standard in UPDRS-III score estimation for free-body movements compared to recent state-of-the-art methods. These results signify a substantial step forward in bioengineering applications for PD monitoring, providing a robust framework for reliable and continuous assessment of PD symptoms in daily living settings.
Full article
►▼
Show Figures
Open AccessArticle
Explainable Vision Transformer with Self-Supervised Learning to Predict Alzheimer’s Disease Progression Using 18F-FDG PET
by
Uttam Khatri and Goo-Rak Kwon
Cited by 4 | Viewed by 3405
Abstract
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder that affects millions of people worldwide. Early and accurate prediction of AD progression is crucial for early intervention and personalized treatment planning. Although AD does not yet have a reliable therapy, several medications help slow
[...] Read more.
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder that affects millions of people worldwide. Early and accurate prediction of AD progression is crucial for early intervention and personalized treatment planning. Although AD does not yet have a reliable therapy, several medications help slow down the disease’s progression. However, more study is still needed to develop reliable methods for detecting AD and its phases. In the recent past, biomarkers associated with AD have been identified using neuroimaging methods. To uncover biomarkers, deep learning techniques have quickly emerged as a crucial methodology. A functional molecular imaging technique known as fluorodeoxyglucose positron emission tomography (18F-FDG-PET) has been shown to be effective in assisting researchers in understanding the morphological and neurological alterations to the brain associated with AD. Convolutional neural networks (CNNs) have also long dominated the field of AD progression and have been the subject of substantial research, while more recent approaches like vision transformers (ViT) have not yet been fully investigated. In this paper, we present a self-supervised learning (SSL) method to automatically acquire meaningful AD characteristics using the ViT architecture by pretraining the feature extractor using the self-distillation with no labels (DINO) and extreme learning machine (ELM) as classifier models. In this work, we examined a technique for predicting mild cognitive impairment (MCI) to AD utilizing an SSL model which learns powerful representations from unlabeled 18F-FDG PET images, thus reducing the need for large-labeled datasets. In comparison to several earlier approaches, our strategy showed state-of-the-art classification performance in terms of accuracy (92.31%), specificity (90.21%), and sensitivity (95.50%). Then, to make the suggested model easier to understand, we highlighted the brain regions that significantly influence the prediction of MCI development. Our methods offer a precise and efficient strategy for predicting the transition from MCI to AD. In conclusion, this research presents a novel Explainable SSL-ViT model that can accurately predict AD progress based on 18F-FDG PET scans. SSL, attention, and ELM mechanisms are integrated into the model to make it more predictive and interpretable. Future research will enable the development of viable treatments for neurodegenerative disorders by combining brain areas contributing to projection with observed anatomical traits.
Full article
►▼
Show Figures
Open AccessArticle
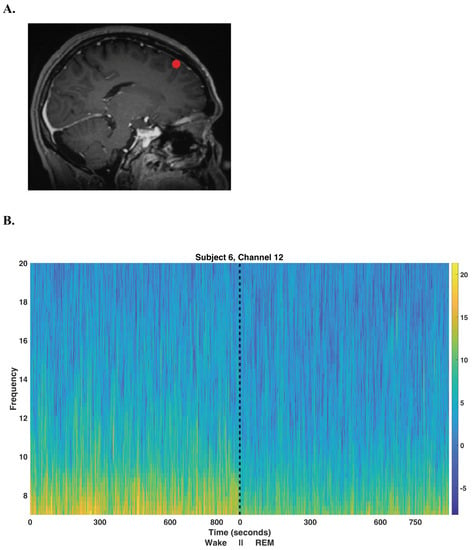
Unsupervised Multitaper Spectral Method for Identifying REM Sleep in Intracranial EEG Recordings Lacking EOG/EMG Data
by
Kyle Q. Lepage, Sparsh Jain, Andrew Kvavilashvili, Mark Witcher and Sujith Vijayan
Cited by 1 | Viewed by 2727
Abstract
A large number of human intracranial EEG (iEEG) recordings have been collected for clinical purposes, in institutions all over the world, but the vast majority of these are unaccompanied by EOG and EMG recordings which are required to separate Wake episodes from REM
[...] Read more.
A large number of human intracranial EEG (iEEG) recordings have been collected for clinical purposes, in institutions all over the world, but the vast majority of these are unaccompanied by EOG and EMG recordings which are required to separate Wake episodes from REM sleep using accepted methods. In order to make full use of this extremely valuable data, an accurate method of classifying sleep from iEEG recordings alone is required. Existing methods of sleep scoring using only iEEG recordings accurately classify all stages of sleep, with the exception that wake (W) and rapid-eye movement (REM) sleep are not well distinguished. A novel multitaper (Wake vs. REM) alpha-rhythm classifier is developed by generalizing K-means clustering for use with multitaper spectral eigencoefficients. The performance of this unsupervised method is assessed on eight subjects exhibiting normal sleep architecture in a hold-out analysis and is compared against a classical power detector. The proposed multitaper classifier correctly identifies
min of REM in one night of recorded sleep, while incorrectly labeling less than
of all labeled 30 s epochs for all but one subject (human rater reliability is estimated to be near
), and outperforms the equivalent statistical-power classical test. Hold-out analysis indicates that when using one night’s worth of data, an accurate generalization of the method on new data is likely. For the purpose of studying sleep, the introduced multitaper alpha-rhythm classifier further paves the way to making available a large quantity of otherwise unusable IEEG data.
Full article
►▼
Show Figures