Development and Evaluation of a Natural Language Processing System for Curating a Trans-Thoracic Echocardiogram (TTE) Database
Abstract
:1. Introduction
- Improved Data Extraction Using NLP. This study introduces and illustrates the usage of a GATE-based NLP system to extract comprehensive information from echocardiography reports. This approach substantially improves the efficiency while maintaining accuracy to extract meaningful insights from these reports by automating the data extraction process.
- Data-Driven Clinical Decision-Making. By utilising Big Data resources, this work contributes to the realisation of learning health systems. It provides the foundation for using data-driven [8] insights to help healthcare practitioners make precise diagnoses, identify at-risk patients, develop personalised treatment regimens and enhance medical research in complex patient populations. It enables healthcare workers to immediately identify critical parameters and red flags in preoperative and postoperative settings, such as changes in aortic diameters or valve haemodynamics. This feature can also help to expedite surgical prioritisation, patient monitoring and waiting list administration.
- Opportunities for Research and Audits. The NLP system’s capabilities go beyond treating specific patients. It paves the way to advanced audit and research projects in cardiac care. With the use of this technology, researchers can monitor patterns and examine large datasets, advancing our knowledge of heart conditions and therapeutic outcomes.
- Interpretable Integration of Data. The study addresses the challenge of integrating both continuous and discrete data, such as qualitative ratings and quantitative measurements, in echocardiogram reports. The structured database generated by the NLP system offers a clear and interpretable representation of this combined information, facilitating its practical use.
- Potential for Broader Healthcare Applications. While the study focuses on echocardiography reports, the concepts and methods explored here can potentially extend to other investigative modalities, including CT scans. The NLP system’s ability to integrate with risk modelling approaches further expands its potential impact on healthcare research and practice.
2. Related Work
3. Materials and Methods
3.1. Dataset
3.2. Data Exploration
3.3. Data Annotation Approach
3.4. Model Development
3.5. Statistical Analysis and Validation
3.6. Practical System Application
4. Results
4.1. Demographics
4.2. Aortic Regurgitation (AR) Level
4.3. LV Systolic Function
4.4. MV Regurgitation Level
4.5. AV+MV+PV+TV Stenosis
4.6. TR Level
5. Discussion
5.1. Technical Perspective
5.2. Relevance to Clinical Practice
5.2.1. Cardiac Surgery Perspective
5.2.2. Cardiologist Perspective
5.3. Limitations and Future Work
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thompson, J.; Hu, J.; Mudaranthakam, D.P.; Streeter, D.; Neums, L.; Park, M.; Koestler, D.C.; Gajewski, B.; Jensen, R.; Mayo, M.S. Relevant Word Order Vectorization for Improved Natural Language Processing in Electronic Health Records. Sci. Rep. 2019, 9, 9253. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, M.; Hu, S.; Shen, Y.; Lan, J.; Jiang, B.; de Bock, G.H.; Vliegenthart, R.; Chen, X.; Xie, X. Development and multicenter validation of chest X-ray radiography interpretations based on natural language processing. Commun. Med. 2021, 1, 43. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, J.H.; Choi, S.; Kim, J.-H.; Seok, J.; Joo, H.J. Validation of deep learning natural language processing algorithm for keyword extraction from pathology reports in electronic health records. Sci. Rep. 2020, 10, 20265. [Google Scholar] [CrossRef]
- Morgan, S.E.; Diederen, K.; Vértes, P.E.; Ip, S.H.Y.; Wang, B.; Thompson, B.; Demjaha, A.; De Micheli, A.; Oliver, D.; Liakata, M.; et al. Natural Language Processing markers in first episode psychosis and people at clinical high-risk. Transl. Psychiatry 2021, 11, 630. [Google Scholar] [CrossRef]
- Dickerson, L.K.; Rouhizadeh, M.; Korotkaya, Y.; Bowring, M.G.; Massie, A.B.; McAdams-Demarco, M.A.; Segev, D.L.; Cannon, A.; Guerrerio, A.L.; Chen, P.-H.; et al. Language impairment in adults with end-stage liver disease: Application of natural language processing towards patient-generated health records. NPJ Digit. Med. 2019, 2, 106. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, C.; Tao, D. GAN-MDF: A Method for Multi-fidelity Data Fusion in Digital Twins. arXiv 2023, arXiv:2106.14655. [Google Scholar]
- Liu, Z.; Yin, H.; Chai, Y.; Yang, S.X. A novel approach for multimodal medical image fusion. Expert Syst. Appl. 2014, 41, 7425–7435. [Google Scholar] [CrossRef]
- Gotz, D.; Borland, D. Data-Driven Healthcare: Challenges and Opportunities for Interactive Visualization. IEEE Comput. Graph. Appl. 2016, 36, 90–96. [Google Scholar] [CrossRef]
- Large-Scale Identification of Aortic Stenosis and Its Severity Using Natural Language Processing on Electronic Health Records—ScienceDirect. Available online: https://www.sciencedirect.com/science/article/pii/S2666693621000256 (accessed on 20 June 2023).
- Szekér, S.; Fogarassy, G.; Vathy-Fogarassy, Á. A general text mining method to extract echocardiography measurement results from echocardiography documents. Artif. Intell. Med. 2023, 143, 102584. [Google Scholar] [CrossRef]
- Nath, C.; Albaghdadi, M.S.; Jonnalagadda, S.R. A Natural Language Processing Tool for Large-Scale Data Extraction from Echocardiography Reports. PLoS ONE 2016, 11, e0153749. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Garvin, J.H.; Goldstein, M.K.; Hwang, T.S.; Redd, A.; Bolton, D.; Heidenreich, P.A.; Meystre, S.M. Extraction of left ventricular ejection fraction information from various types of clinical reports. J. Biomed. Inform. 2017, 67, 42–48. [Google Scholar] [CrossRef]
- Zheng, C.; Sun, B.C.; Wu, Y.-L.; Ferencik, M.; Lee, M.-S.; Redberg, R.F.; A Kawatkar, A.; Musigdilok, V.V.; Sharp, A.L. Automated interpretation of stress echocardiography reports using natural language processing. Eur. Heart J. Digit. Health 2022, 3, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, E.; Elbattah, M.; Gignon, M.; Dequen, G. Learning Embeddings from Free-text Triage Notes using Pretrained Transformer Models. In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2022), Workshop on Scaling-Up Health-IT, Vienna, Austria, 9–11 February 2022; pp. 835–841. [Google Scholar]
- Rietberg, M.T.; Nguyen, V.B.; Geerdink, J.; Vijlbrief, O.; Seifert, C. Accurate and Reliable Classification of Unstructured Reports on Their Diagnostic Goal Using BERT Models. Diagnostics 2023, 13, 1251. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Riloff, E.; Meystre, S.M. Exploiting Unlabeled Texts with Clustering-based Instance Selection for Medical Relation Classification. AMIA Annu. Symp. Proc. 2018, 2017, 1060–1069. [Google Scholar] [PubMed]
- Robinson, S.; Rana, B.; Oxborough, D.; Steeds, R.; Monaghan, M.; Stout, M.; Pearce, K.; Harkness, A.; Ring, L.; Paton, M.; et al. A practical guideline for performing a comprehensive transthoracic echocardiogram in adults: The British Society of Echocardiography minimum dataset. Echo Res. Pract. 2020, 7, G59–G93. [Google Scholar] [CrossRef] [PubMed]
- Andrade, J.C.B.; Jaramillo, C.M.Z. Gate-Based Rules for Extracting Attribute Values. Comput. Y Sist. 2021, 25, 851–862. [Google Scholar] [CrossRef]
- Cunningham, H.; Tablan, V.; Roberts, A.; Bontcheva, K. Getting More Out of Biomedical Documents with GATE’s Full Lifecycle Open Source Text Analytics. PLoS Comput. Biol. 2013, 9, e1002854. [Google Scholar] [CrossRef] [PubMed]
- Yeung, A.; Iaboni, A.; Rochon, E.; Lavoie, M.; Santiago, C.; Yancheva, M.; Novikova, J.; Xu, M.; Robin, J.; Kaufman, L.D.; et al. Correlating natural language processing and automated speech analysis with clinician assessment to quantify speech-language changes in mild cognitive impairment and Alzheimer’s dementia. Alzheimer’s Res. Ther. 2021, 13, 109. [Google Scholar] [CrossRef]
- Rahman, M.; Nowakowski, S.; Agrawal, R.; Naik, A.; Sharafkhaneh, A.; Razjouyan, J. Validation of a Natural Language Processing Algorithm for the Extraction of the Sleep Parameters from the Polysomnography Reports. Healthcare 2022, 10, 1837. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.S.; Rodriguez, Z.; Warren, K.K.; Cowan, T.; Masucci, M.D.; Granrud, O.E.; Holmlund, T.B.; Chandler, C.; Foltz, P.W.; Strauss, G.P. Natural Language Processing and Psychosis: On the Need for Comprehensive Psychometric Evaluation. Schizophr. Bull. 2022, 48, 939–948. [Google Scholar] [CrossRef]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef]
- Khalifa, A.; Meystre, S. Adapting existing natural language processing resources for cardiovascular risk factors identification in clinical notes. J. Biomed. Inform. 2015, 58, S128–S132. [Google Scholar] [CrossRef]
- Yang, H.; Spasic, I.; Keane, J.A.; Nenadic, G. A Text Mining Approach to the Prediction of Disease Status from Clinical Discharge Summaries. J. Am. Med. Inform. Assoc. 2009, 16, 596–600. [Google Scholar] [CrossRef] [PubMed]
- Cunliffe, D.; Vlachidis, A.; Williams, D.; Tudhope, D. Natural language processing for under-resourced languages: Developing a Welsh natural language toolkit. Comput. Speech Lang. 2021, 72, 101311. [Google Scholar] [CrossRef]
- Digan, W.; Névéol, A.; Neuraz, A.; Wack, M.; Baudoin, D.; Burgun, A.; Rance, B. Can reproducibility be improved in clinical natural language processing? A study of 7 clinical NLP suites. J. Am. Med. Inform. Assoc. 2020, 28, 504–515. [Google Scholar] [CrossRef] [PubMed]
- Amato, F.; Cozzolino, G.; Moscato, V. Analyse digital forensic evidences through a semantic-based methodology and NLP techniques. Futur. Gener. Comput. Syst. 2019, 98, 297–307. [Google Scholar] [CrossRef]
- Drousiotis, E.; Pentaliotis, P.; Shi, L.; Cristea, A.I. Balancing Fined-Tuned Machine Learning Models Between Continuous and Discrete Variables—A Comprehensive Analysis Using Educational Data. In Proceedings of the International Conference on Artificial Intelligence in Education, Durham, UK, 27–31 July 2022; pp. 256–268. [Google Scholar]
- Belz, A.; Kow, E. Discrete vs. continuous rating scales for language evaluation in NLP. In Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies: Short Papers—Volume 2, Portland, OR, USA, 19–24 June 2011; Association for Computational Linguistics: Stroudsburg, PA, USA, 2011; pp. 230–235. [Google Scholar]
- Cartuyvels, R.; Spinks, G.; Moens, M.-F. Discrete and continuous representations and processing in deep learning: Looking forward. AI Open 2021, 2, 143–159. [Google Scholar] [CrossRef]
- Sutton, R.S.; McAllester, D.; Singh, S.; Mansour, Y. Policy Gradient Methods for Reinforcement Learning with Function Approximation. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2023; Available online: https://papers.nips.cc/paper_files/paper/1999/hash/464d828b85b0bed98e80ade0a5c43b0f-Abstract.html (accessed on 7 August 2023).
- Hu, R.; Andreas, J.; Rohrbach, M.; Darrell, T.; Saenko, K. Learning to Reason: End-to-End Module Networks for Visual Question Answering. arXiv 2017, arXiv:1704.05526. [Google Scholar]
- Maddison, C.J.; Mnih, A.; Teh, Y.W. The Concrete Distribution: A Continuous Relaxation of Discrete Random Variables. In Proceedings of the 5th International Conference on Learning Representations, ICLR 2017, Toulon, France, 24–26 April 2017; Conference Track Proceedings, OpenReview.net. Available online: https://openreview.net/forum?id=S1jE5L5gl (accessed on 7 August 2023).
- Bengio, Y.; Ducharme, R.; Vincent, P.; Jauvin, C. A Neural Probabilistic Language Model. In Advances in Neural Information Processing Systems 13 (NIPS 2000); MIT Press: Denver, CO, USA, 2001. [Google Scholar]
- Johnson, J.; Hariharan, B.; Van Der Maaten, L.; Hoffman, J.; Fei-Fei, L.; Zitnick, C.L.; Girshick, R. Inferring and Executing Programs for Visual Reasoning. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 3008–3017. [Google Scholar]
- Andreas, J.; Rohrbach, M.; Darrell, T.; Klein, D. Learning to Compose Neural Networks for Question Answering. In Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, San Diego, CA, USA, 12–17 June 2016. [Google Scholar]
- Hu, R.; Andreas, J.; Darrell, T.; Saenko, K. Explainable neural computation via stack neural module networks. Appl. AI Lett. 2021, 2, e39. [Google Scholar] [CrossRef]
- Mascharka, D.; Tran, P.; Soklaski, R.; Majumdar, A. Transparency by Design: Closing the Gap Between Performance and Interpretability in Visual Reasoning. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 4942–4950. [Google Scholar]
- Yi, K.; Wu, J.; Gan, C.; Torralba, A.; Kohli, P.; Tenenbaum, J. Neural-Symbolic VQA: Disentangling Reasoning from Vision and Language Understanding. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2023; Available online: https://proceedings.neurips.cc/paper_files/paper/2018/hash/5e388103a391daabe3de1d76a6739ccd-Abstract.html (accessed on 8 August 2023).
- Peng, B.; Alcaide, E.; Anthony, Q.; Albalak, A.; Arcadinho, S.; Cao, H.; Cheng, X.; Chung, M.; Grella, M.; Kiran, K.; et al. RWKV: Reinventing RNNs for the Transformer Era. arXiv 2023, arXiv:2305.13048. [Google Scholar]
- Karypis, G. CLUTO—A Clustering Toolkit. Report, Apr. Available online: http://conservancy.umn.edu/handle/11299/215521 (accessed on 2 August 2023).

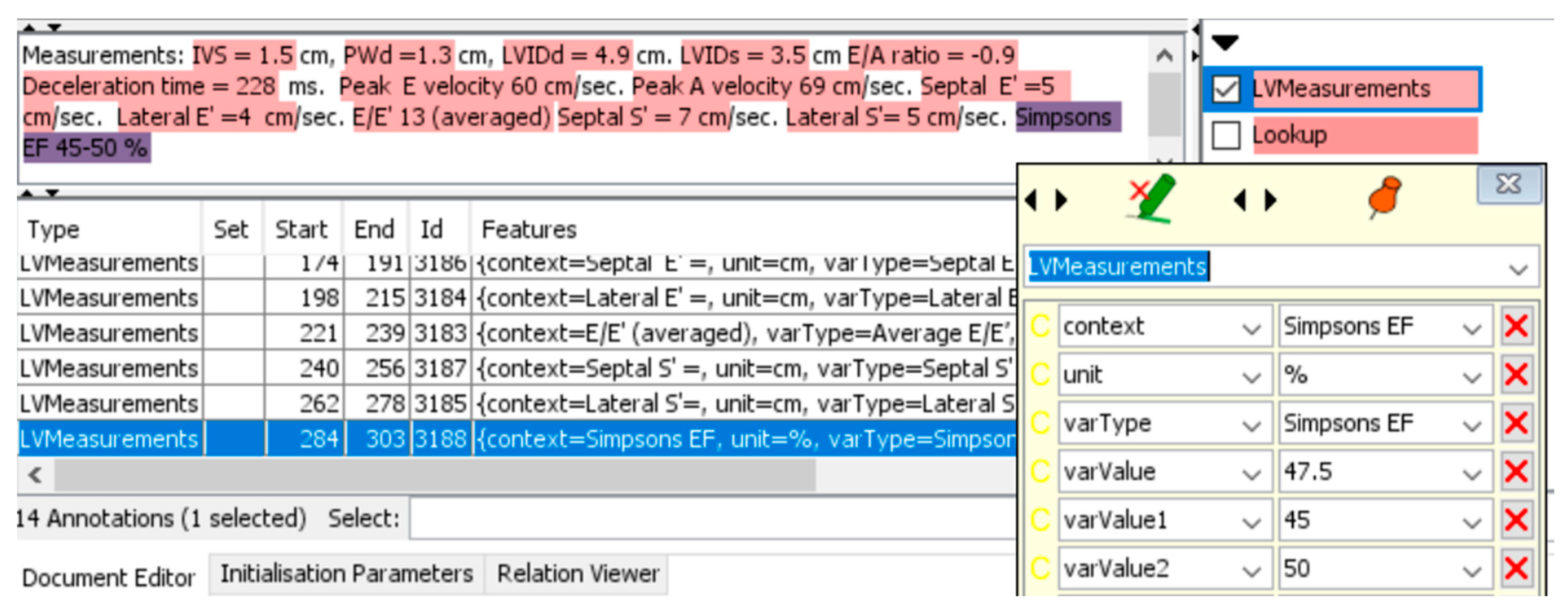

| Abbreviation Clinician | Abbreviation NLP | Definition | Category | Data Type |
|---|---|---|---|---|
| LVEF | EF | Left Ventricular Ejection Fraction | Left ventricle | Continuous |
| LVIDd | LVIDd | Left Ventricular Internal Diameter in Diastole | Continuous | |
| LVIDs | LVIDs | Left Ventricular Internal Diameter in Systole | Continuous | |
| LVSF | LV Systolic Function | Left Ventricular Shortening Fraction | Discrete | |
| IVSd | IVS | Interventricular Septum Thickness in Diastole | Continuous | |
| PWd | PWd | Posterior Wall Thickness in Diastole | Continuous | |
| LVOTd | LVOT Diam | Left Ventricular Outflow Tract Diameter | Continuous | |
| LVOTvti | LVOT VTI | Left Ventricular Outflow Tract Velocity-Time Integral | Continuous | |
| LVOTpv | LVOT Vmax | Left Ventricular Outflow Tract Peak Velocity | Continuous | |
| RV_TDI_S | RV S’ | Right Ventricle Tissue Doppler Imaging S Wave | Right ventricle | Continuous |
| RVD1 | RVD1 | Right Ventricle Diameter at Basal Level | Continuous | |
| TAPSE | RV TAPSE | Tricuspid Annular Plane Systolic Excursion | Continuous | |
| LA_area | LA Area | Left Atrial Area | Left atrium | Continuous |
| LA_vol | LA Volume | Left Atrial Volume | Continuous | |
| RA_area | RA Area | Right Atrial Area | Right atrium | Continuous |
| AS_sev | AV Stenosis | Aortic Stenosis Severity | Aortic valve | Discrete |
| AR_sev | AR level | Aortic Regurgitation Severity | Discrete | |
| AVpv | AV Vmax | Aortic Valve Peak Velocity | Continuous | |
| AVpg | AV max PG | Aortic Valve Peak Gradient | Continuous | |
| AVmg | AV MPG | Aortic Valve Mean Gradient | Continuous | |
| AVpht | AR PHT | Aortic Valve Pressure Half-Time | Continuous | |
| AVvti | AV VTI | Aortic Valve Velocity-Time Integral | Continuous | |
| MS_sev | MV Stenosis | Mitral Stenosis Severity | Mitral valve | Discrete |
| MR_sev | MV Regurgitation Level | Mitral Regurgitation Severity | Discrete | |
| MAPSE | Lateral MAPSE | Lateral Mitral Annular Plane Systolic Excursion | Continuous | |
| MV_E_vel | Peak E Velocity | Mitral Valve E Wave Velocity | Continuous | |
| MV_A_vel | Peak A Velocity | Mitral Valve A Wave Velocity | Continuous | |
| MV_decT | DcT | Mitral Valve Deceleration Time | Continuous | |
| MV_Earatio | E/A ratio | Mitral Valve E/A Ratio | Continuous | |
| MV_EE_avg | Average E/E’ | Mitral Valve E/E’ Average | Continuous | |
| TDI_lat_S | Lateral S’ Velocity | Tissue Doppler Imaging Lateral S Wave | Continuous | |
| TDI_lat_E | Lateral E’ Velocity | Tissue Doppler Imaging Lateral E Wave | Continuous | |
| TDI_sep_S | Septal S’ Velocity | Tissue Doppler Imaging Septal S Wave | Continuous | |
| TDI_sep_E | Septal E’ Velocity | Tissue Doppler Imaging Septal E Wave | Continuous | |
| TS_sev | TV Stenosis | Tricuspid Stenosis Severity | Tricuspid valve | Discrete |
| TR_sev | TR level | Tricuspid Regurgitation Severity | Discrete | |
| TR_pv | TR Vmax | Tricuspid Regurgitation Peak Velocity | Continuous | |
| TR_pg | TR max PG | Tricuspid Regurgitation Peak Gradient | Continuous | |
| PS_sev | PV Stenosis | Pulmonary Stenosis Severity | Pulmonary valve | Discrete |
| PV_Vmax | PV Vmax | Pulmonary Valve Maximum Velocity | Continuous | |
| AO_SOV | Sinuses of Valsalva | Sinus of Valsalva | Aorta | Continuous |
| AO_STJ | Sinotubular Junction | Aortic Outflow Sinotubular Junction | Continuous | |
| AO_ASC | Ascending Aorta | Aortic Outflow Ascending Aorta | Continuous |
| Aortic Valve (AV) Velocity Time Integral (VTI) | AV Regurgitation Level |
|---|---|
| AV Vmax 4.2 m/s MPD 46 mmHg VTI 103 cm | Aortic Valve (biological AVR): AVR in situ, well seated. … No significant regurgitation. … Aorta: |
| AV Vmax 4 m/s MPD 42 mmHg VTI 87.9 cm | Aortic Valve (unclear imaging of AVR): … AVR seen in situ with a mild paraprosthetic regurgitation. … Aorta: |
| Ao VTI 36 cm; AVA (VTI) 1.8 cm2 | Aortic Valve: … with valve type/size in situ No aortic regurgitation Right Ventricle: |
| Ao VTI 36 cm; | Aortic Valve: … No aortic stenosis. Trivial aortic regurgitation. AV Vmax: 1.6 m/s. Aorta: |
| Aortic Valve: Appears Trileaflet. Thickening of LCC/NCC with reduced mobility of these cusps. V max 2.8 m/s, PPD: 32 mmHg, MPD: 19 mmHg, VTI: 62.3 cm. | Aortic Valve (TAVI): … No aortic stenosis/obstruction indicated. Trivial–mild paravalvular aortic regurgitation. … Aorta: |
| AV Vmax 4 m/s MPD 42 mmHg VTI 87.9 cm LVOT 2.7 cm Peak V = 0.7 m/s, MPD 1.2 mmHg, VTI 16.2 cm | Aortic Valve: AVR in situ. …No significant obstruction/stenosis indicated. No obvious aortic regurgitation. … Aorta: |
| AV Vmax: 4.8 m/s, PPD: 91 mmHg, MPD: 57 mmHg, VTI: 107 cm | Aortic Valve: … Mild eccentric paravalvular aortic regurgitation seen. Aorta: |
| AV mean PG: 54 mmHg. AV VTI: 78 cm. AVA VTI: 0.88 cm2. AVAi VTI: 0.45 cm/m2 | Aortic Valve: …. Mild transvalvular aortic regurgitation. … |
| AV VTI 61.3 cm | Aorta: Aortic Valve: … Aortic regurgitation present …Overall assessment is of mild aortic regurgitation. AV Vmax: 1.5 m/s. Aorta: |
| Ao VTI 106 cm; | Aortic Valve: … ? BAV. Moderate AS. Trivial AR …. Aorta: |
| AV VTI: 78 cm. | Aortic Valve: …. Overall assessment is of severe aortic regurgitation. … Aorta: |
| Baseline Patient Total = 78,536 | |
|---|---|
| Age (years), mean (SD) | 59.4 (33.0) |
| Female gender, n (%) | 36,959 (47.1%) |
| Mortalities, n (%) | 25,048 (31.9%) |
| Number of repeat echocardiograms | |
| 1 | 53,508 (68.1%) |
| 2 | 13,031 (16.6%) |
| 3 | 5064 (6.4%) |
| 4 | 2675 (3.4%) |
| 5 | 1632 (2.1%) |
| 6 | 933 (1.2%) |
| 7 | 531 (0.7%) |
| 8 | 356 (0.5%) |
| 9 | 233 (0.3%) |
| >10 | 573 (0.7%) |
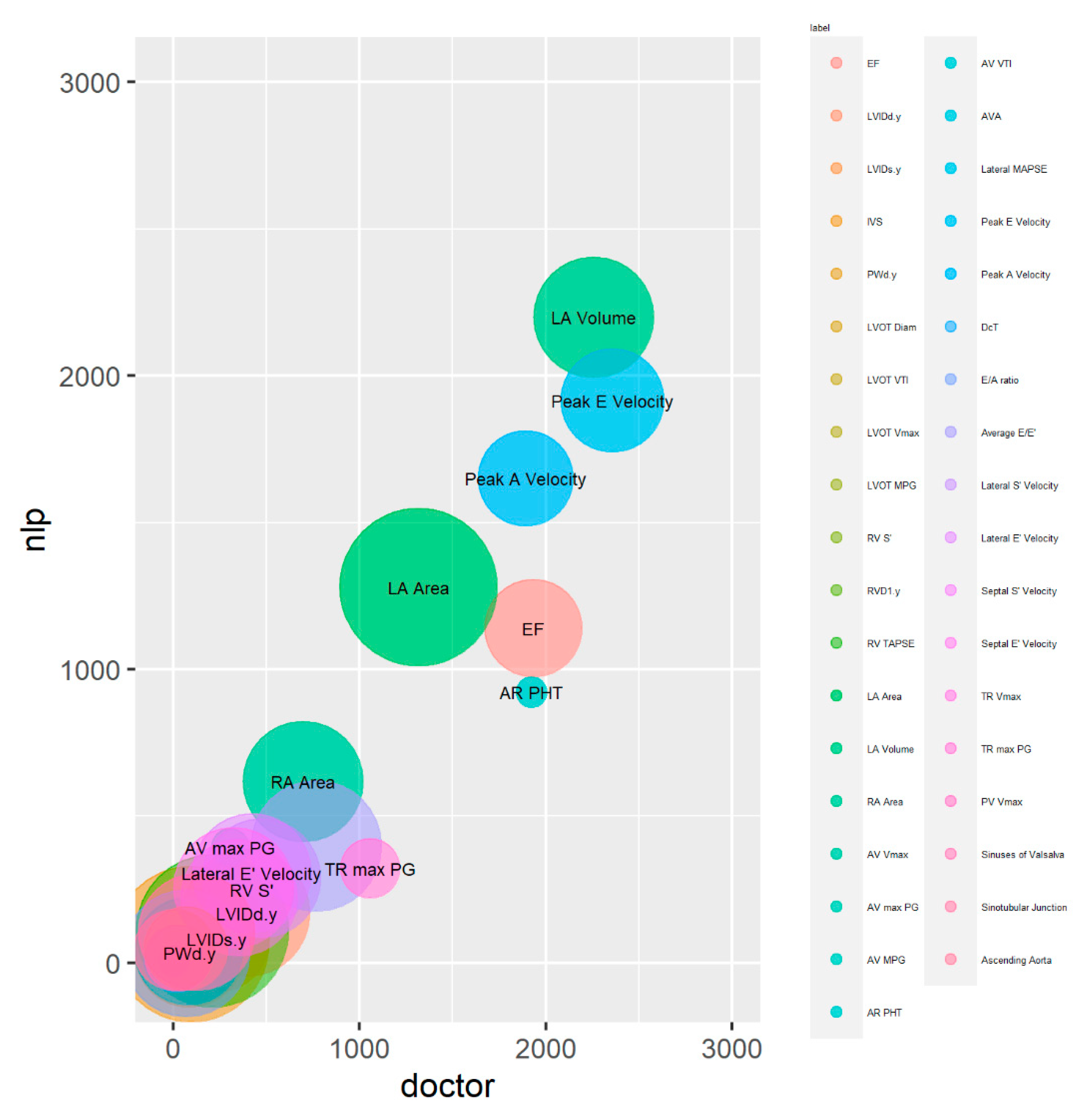
| Variable Name | R Squared | ICC | ICC p-Value |
|---|---|---|---|
| EF | 0.47 | 0.64 | 0.00 |
| LVIDd | 0.10 | 0.18 | 0.07 |
| LVIDs | 0.14 | 0.25 | 0.04 |
| IVS | 0.31 | 0.47 | 0.00 |
| PWd | 0.39 | 0.43 | 0.04 |
| LVOT Diam | 0.70 | 0.83 a | 0.00 |
| LVOT VTI | 1.00 | 1.00 b | 0.00 |
| LVOT Vmax | 1.00 | 0.02 | 0.42 |
| RV S’ | 0.10 | 0.29 | 0.00 |
| RVD1 | 0.07 | 0.16 | 0.05 |
| RV TAPSE | 0.11 | 0.16 | 0.05 |
| LA Area | 0.95 | 0.97 b | 0.00 |
| LA Volume | 0.98 | 0.99 b | 0.00 |
| RA Area | 0.86 | 0.92 b | 0.00 |
| AV Vmax | 0.13 | 0.29 | 0.00 |
| AV max PG | 0.00 | 0.03 | 0.39 |
| AV MPG | 0.20 | 0.35 | 0.00 |
| AR PHT | 0.46 | 0.63 | 0.00 |
| AV VTI | 1.00 | 1.00 b | 0.00 |
| Lateral MAPSE | 0.60 | 0.76 a | 0.00 |
| Peak E Velocity | 0.59 | 0.76 a | 0.00 |
| Peak A Velocity | 0.81 | 0.90 b | 0.00 |
| DcT | 0.05 | 0.15 | 0.06 |
| E/A ratio | 0.18 | 0.36 | 0.00 |
| Average E/E’ | 0.03 | 0.13 | 0.08 |
| Lateral S’ Velocity | 0.49 | 0.65 | 0.00 |
| Lateral E’ Velocity | 0.62 | 0.77 a | 0.00 |
| Septal S’ Velocity | 0.45 | 0.62 | 0.00 |
| Septal E’ Velocity | 0.49 | 0.69 | 0.00 |
| TR Vmax | 1.00 | 1.00 b | 0.00 |
| TR max PG | 0.30 | 0.45 | 0.00 |
| PV Vmax | 0.62 | 0.77 a | 0.00 |
| Sinuses of Valsalva | 0.73 | 0.85 a | 0.00 |
| Sinotubular Junction | 0.82 | 0.17 | 0.03 |
| Ascending Aorta | 0.64 | 0.78 a | 0.00 |
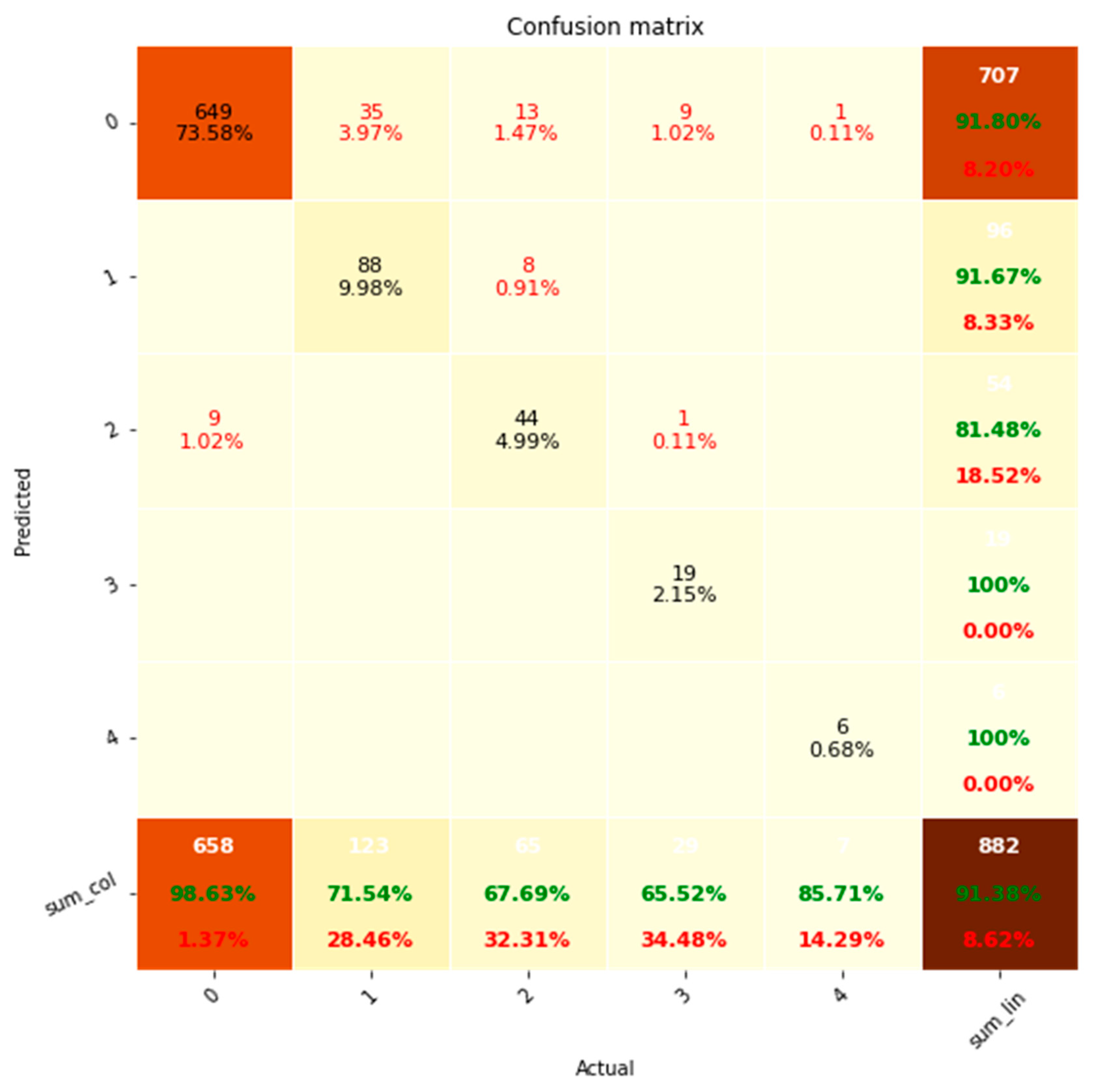
| Outcome | TP | FN | FP | TN | Precision | Recall | F1 Score |
|---|---|---|---|---|---|---|---|
| AR level | 24 | 3 | 4 | 67 | 0.86 | 0.89 | 0.87 |
| LV Systolic Function | 12 | 17 | 3 | 66 | 0.80 | 0.41 | 0.55 |
| MV Regurgitation Level | 59 | 2 | 1 | 36 | 0.98 | 0.97 | 0.98 |
| AV + MV + PV + TV Stenosis | 5 | 4 | 8 | 375 | 0.38 | 0.56 | 0.45 |
| TR Level | 57 | 32 | 2 | 105 | 0.97 | 0.64 | 0.77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, T.; Sunderland, N.; Nightingale, A.; Fudulu, D.P.; Chan, J.; Zhai, B.; Freitas, A.; Caputo, M.; Dimagli, A.; Mires, S.; et al. Development and Evaluation of a Natural Language Processing System for Curating a Trans-Thoracic Echocardiogram (TTE) Database. Bioengineering 2023, 10, 1307. https://doi.org/10.3390/bioengineering10111307
Dong T, Sunderland N, Nightingale A, Fudulu DP, Chan J, Zhai B, Freitas A, Caputo M, Dimagli A, Mires S, et al. Development and Evaluation of a Natural Language Processing System for Curating a Trans-Thoracic Echocardiogram (TTE) Database. Bioengineering. 2023; 10(11):1307. https://doi.org/10.3390/bioengineering10111307
Chicago/Turabian StyleDong, Tim, Nicholas Sunderland, Angus Nightingale, Daniel P. Fudulu, Jeremy Chan, Ben Zhai, Alberto Freitas, Massimo Caputo, Arnaldo Dimagli, Stuart Mires, and et al. 2023. "Development and Evaluation of a Natural Language Processing System for Curating a Trans-Thoracic Echocardiogram (TTE) Database" Bioengineering 10, no. 11: 1307. https://doi.org/10.3390/bioengineering10111307
APA StyleDong, T., Sunderland, N., Nightingale, A., Fudulu, D. P., Chan, J., Zhai, B., Freitas, A., Caputo, M., Dimagli, A., Mires, S., Wyatt, M., Benedetto, U., & Angelini, G. D. (2023). Development and Evaluation of a Natural Language Processing System for Curating a Trans-Thoracic Echocardiogram (TTE) Database. Bioengineering, 10(11), 1307. https://doi.org/10.3390/bioengineering10111307







