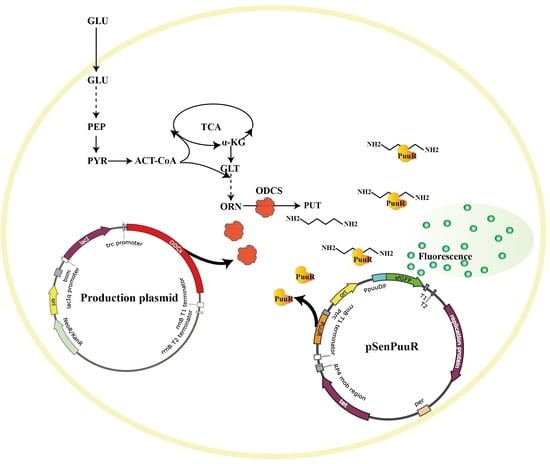
Development of a Transcriptional Factor PuuR-Based Putrescine-Specific Biosensor in Corynebacterium glutamicum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Cultivation Conditions
2.2. Construction of Recombinant Plasmids
2.3. Fluorescence Detection
2.4. Construction of Putrescine-Producing Strains
2.5. Cell Fragmentation and Putrescine Detection
2.6. Screening of ODC
3. Results
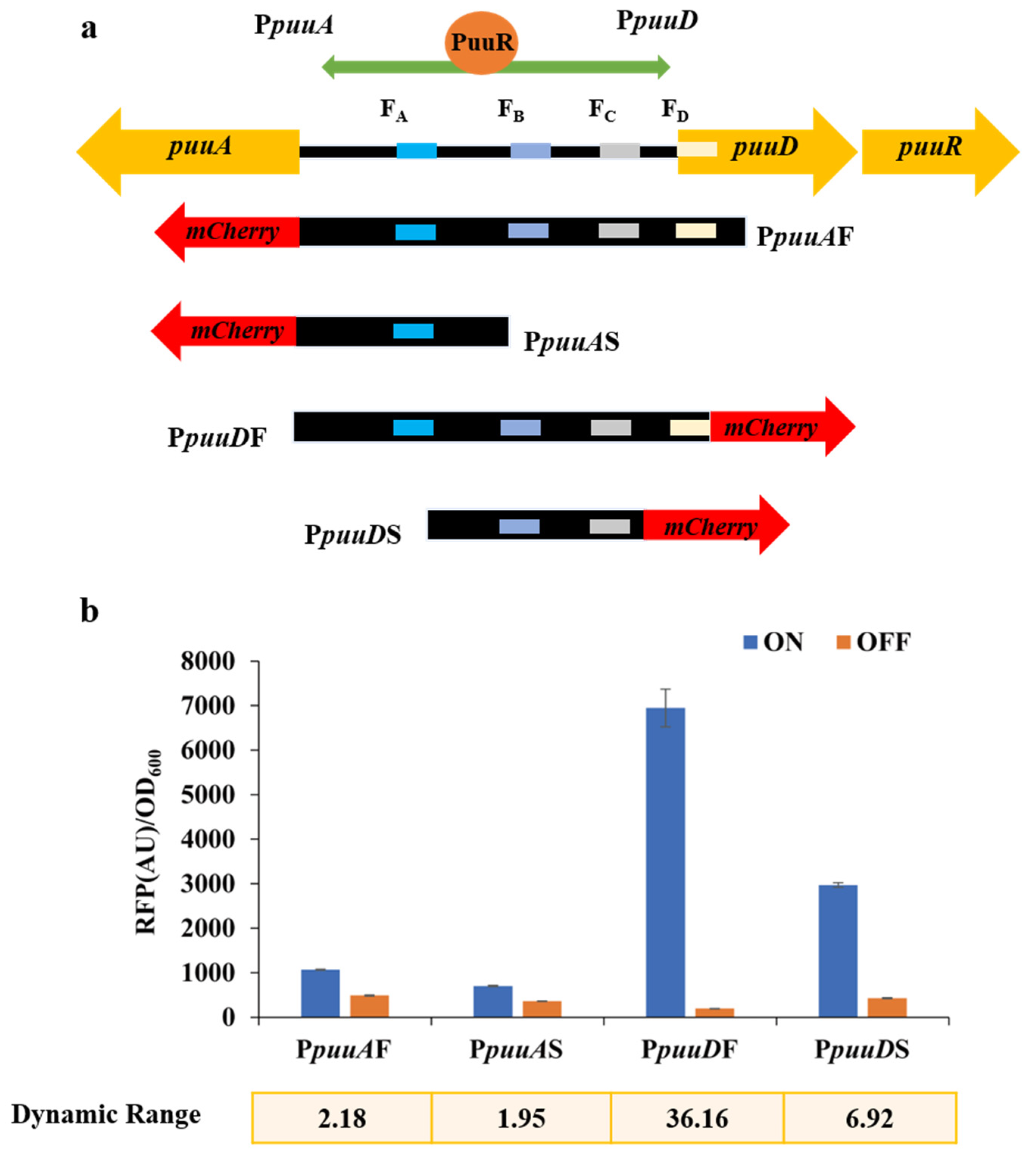
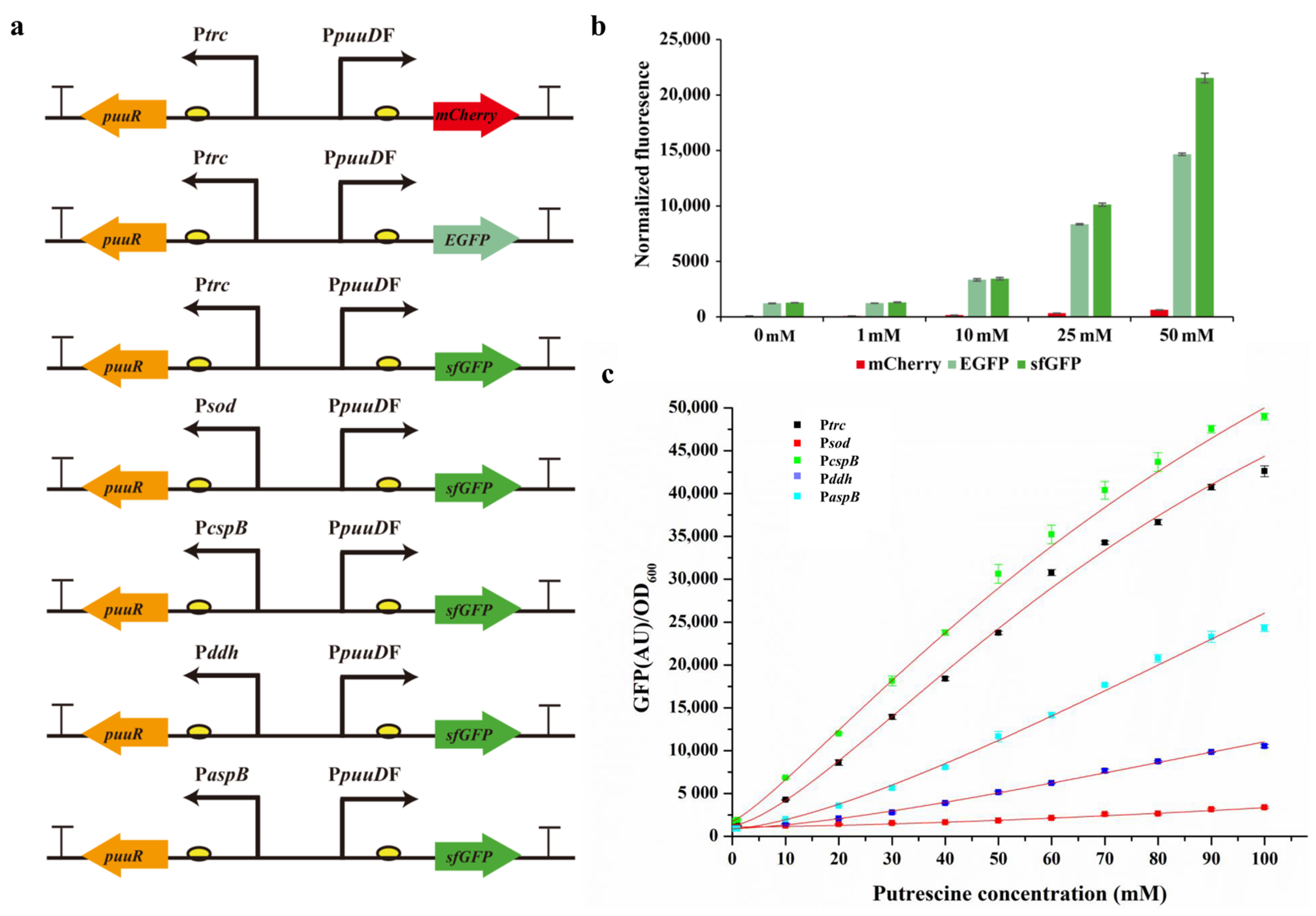
3.1. Development of the TF PuuR-Based Putrescine Biosensor
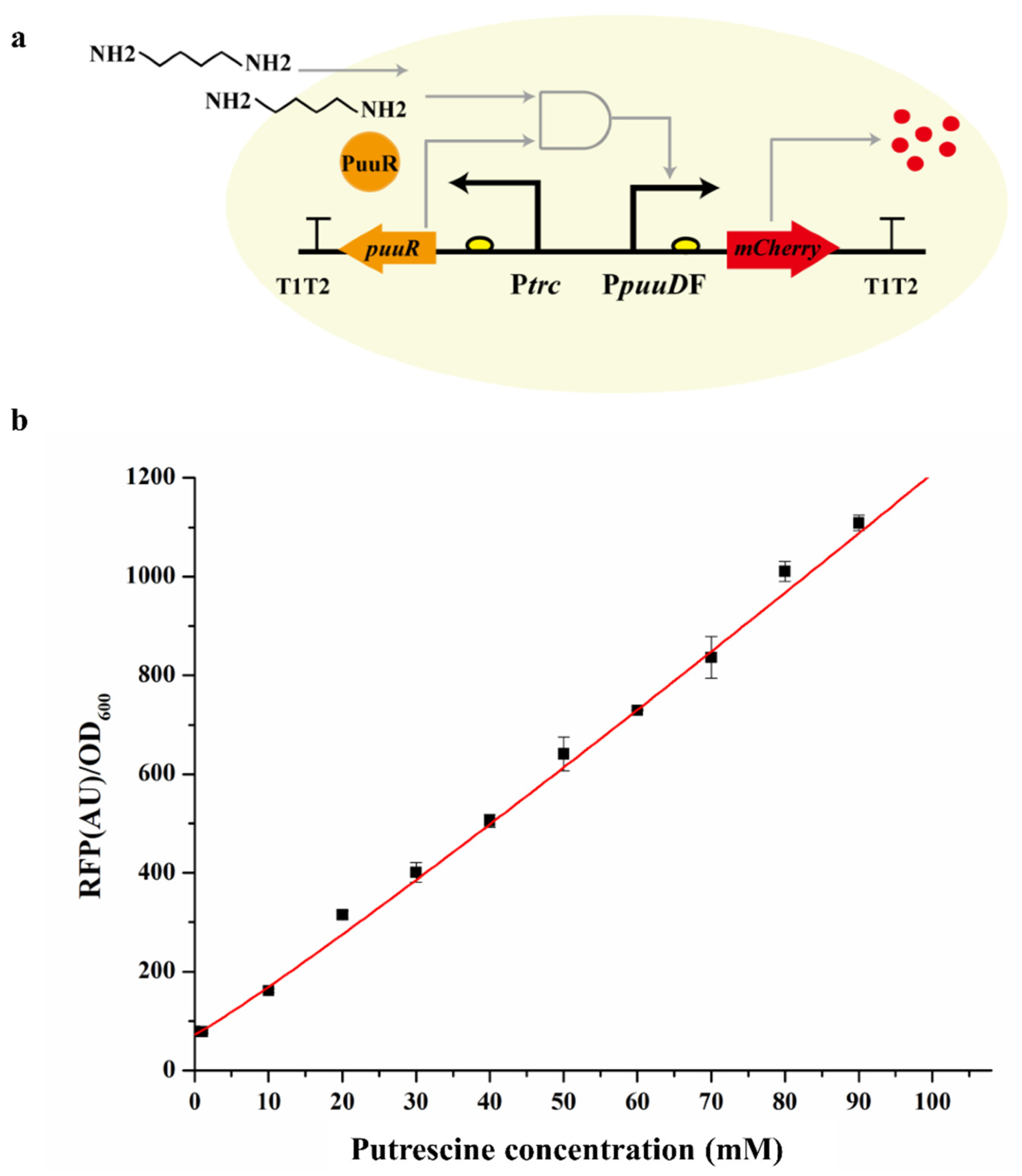
3.2. Dose Response of the Sensor to Extracellular Putrescine Concentration
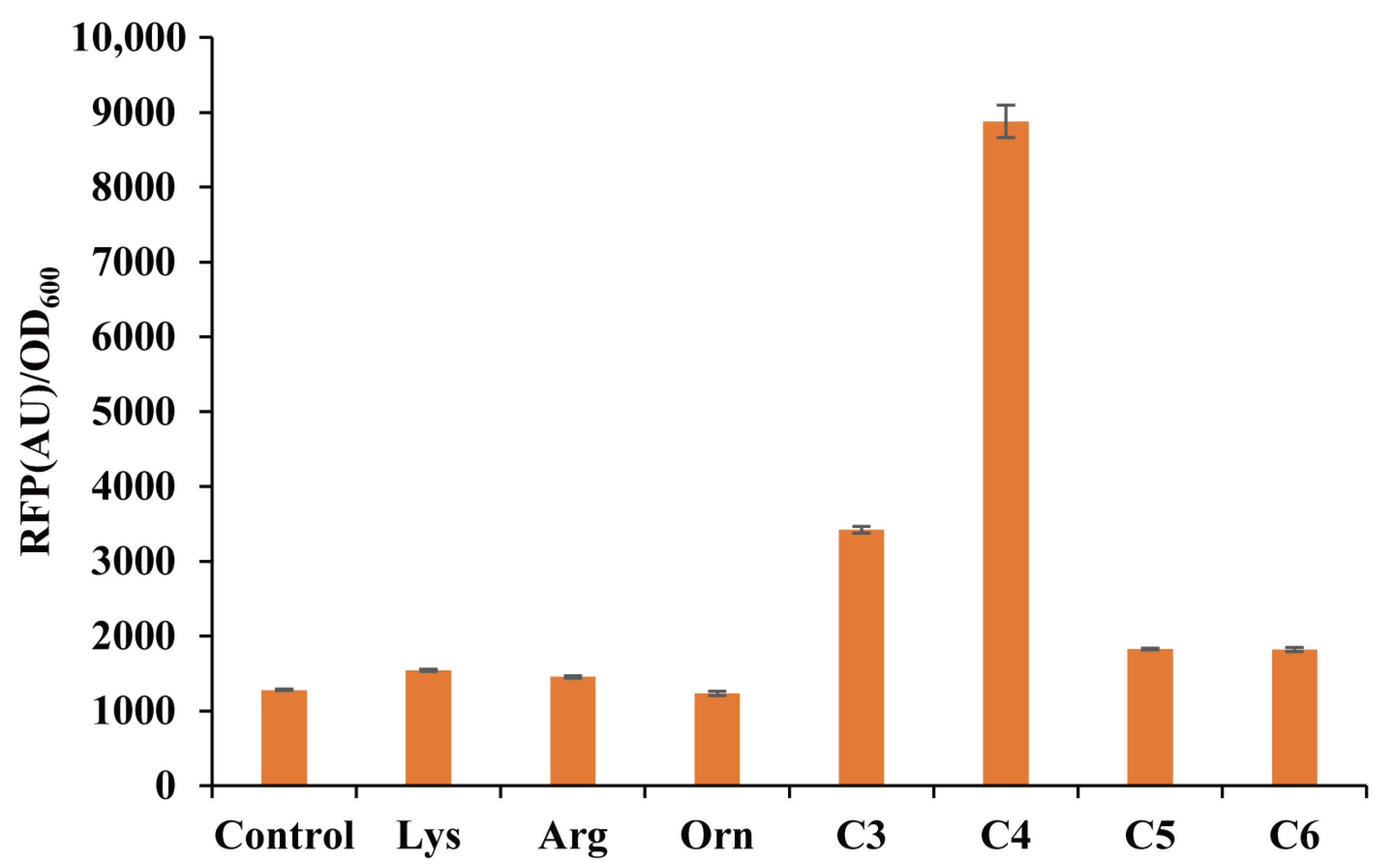
3.3. Ligand Specificity of the pSenPuuR Sensor
3.4. Optimization and Characterization of pSenPuuR
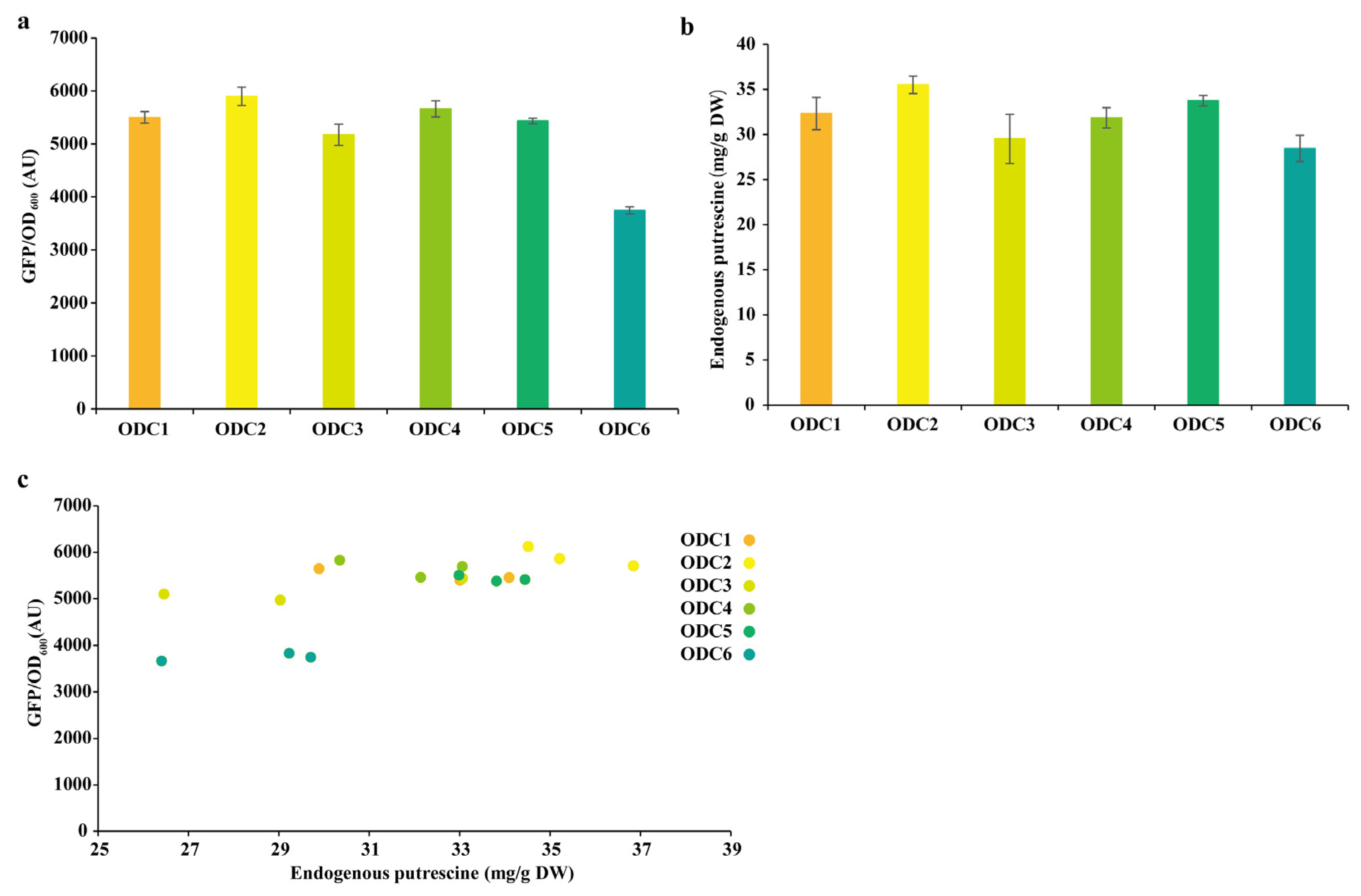
3.5. Construction of Putrescine-Producing Strains
3.6. Database Mining and Phylogenetic Tree Construction of ODCs
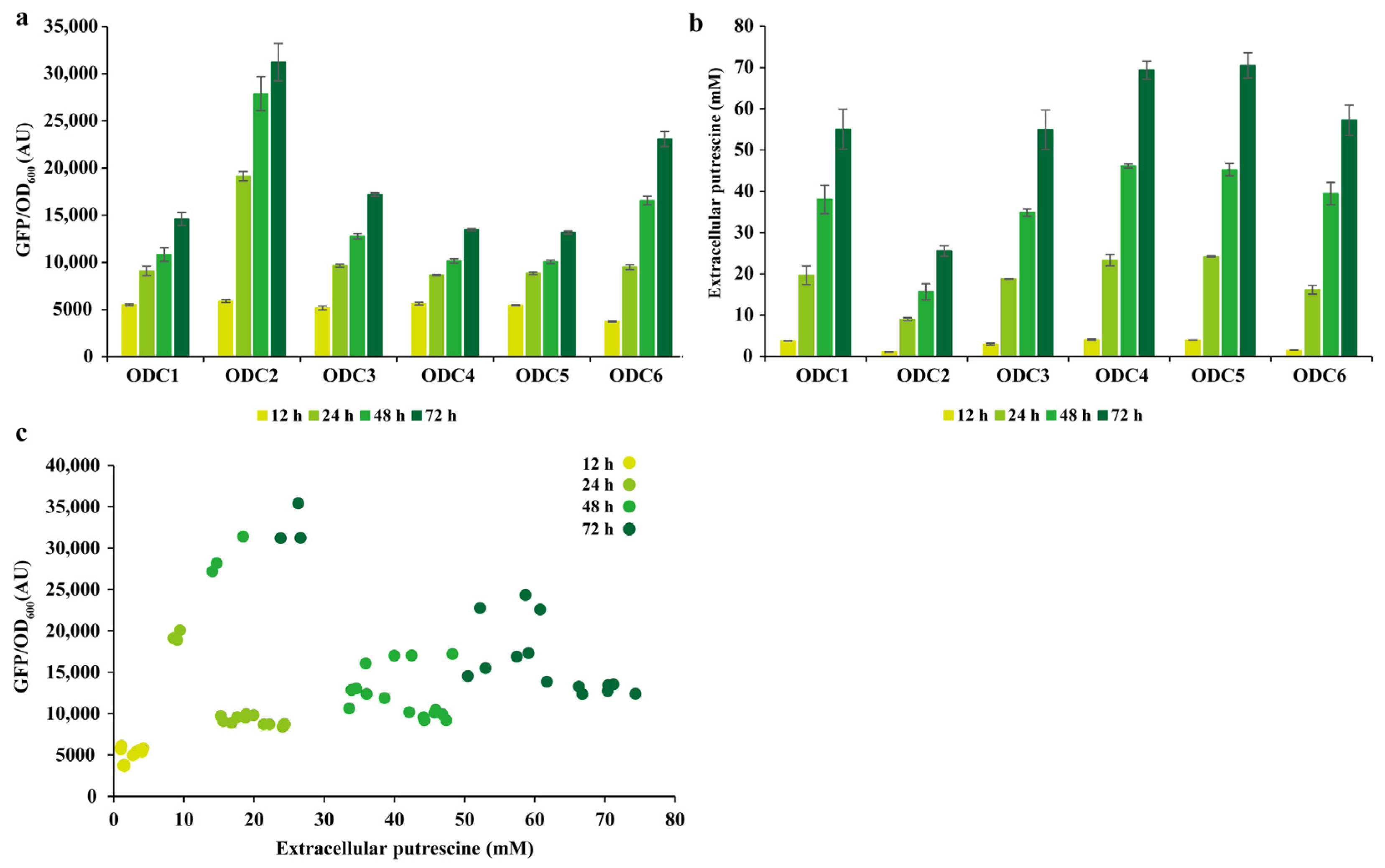
3.7. pSenPuuR for the Detection of Putrescine Production in C. glutamicum
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sanders, J.; Scott, E.; Weusthuis, R.; Mooibroek, H. Bio-refinery as the bio-inspired process to bulk chemicals. Macromol. Biosci. 2007, 7, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.; Peter, F.; Sanders, J. Biomass in the manufacture of industrial products—The use of proteins and amino acids. Appl. Microbiol. Biotechnol. 2007, 75, 751–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chae, T.U.; Ahn, J.H.; Ko, Y.S.; Kim, J.W.; Lee, J.A.; Lee, E.H.; Lee, S.Y. Metabolic engineering for the production of dicarboxylic acids and diamines. Metab. Eng. 2020, 58, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.G.; Xia, X.X.; Lee, S.Y. Metabolic engineering of Escherichia coli for the production of putrescine: A four carbon diamine. Biotechnol. Bioeng. 2009, 104, 651–662. [Google Scholar] [CrossRef]
- Noh, M.; Yoo, S.M.; Kim, W.J.; Lee, S.Y. Gene Expression Knockdown by Modulating Synthetic Small RNA Expression in Escherichia coli. Cell Syst. 2017, 5, 418–426. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, A.Q.; Schneider, J.; Reddy, G.K.; Wendisch, V.F. Fermentative production of the diamine putrescine: System metabolic engineering of Corynebacterium glutamicum. Metabolites 2015, 5, 211–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasikumar, K.; Hannibal, S.; Wendisch, V.F.; Nampoothiri, K.M. Production of Biopolyamide Precursors 5-Amino Valeric Acid and Putrescine from Rice Straw Hydrolysate by Engineered Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 2021, 9, 635509. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.; Eberhardt, D.; Wendisch, V.F. Improving putrescine production by Corynebacterium glutamicum by fine-tuning ornithine transcarbamoylase activity using a plasmid addiction system. Appl. Microbiol. Biotechnol. 2012, 95, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Shen, Y.P.; Jiang, X.L.; Feng, L.S.; Liu, J.Z. Metabolic evolution and a comparative omics analysis of Corynebacterium glutamicum for putrescine production. J. Ind. Microbiol. Biotechnol. 2018, 45, 123–139. [Google Scholar] [CrossRef]
- Li, Z.; Liu, J.Z. Transcriptomic Changes in Response to Putrescine Production in Metabolically Engineered Corynebacterium glutamicum. Front. Microbiol. 2017, 8, 1987. [Google Scholar] [CrossRef]
- Wang, L.; Li, G.; Deng, Y. Diamine Biosynthesis: Research Progress and Application Prospects. Appl. Environ. Microbiol. 2020, 86, e01972-20. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Tague, N.; Whelan, S.A.; Dunlop, M.J. Programmable gene regulation for metabolic engineering using decoy transcription factor binding sites. Nucleic Acids Res. 2021, 49, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Park, S.Y.; Park, Y.S.; Eun, H.; Lee, S.Y. Metabolic Engineering of Escherichia coli for Natural Product Biosynthesis. Trends Biotechnol. 2020, 38, 745–765. [Google Scholar] [CrossRef] [PubMed]
- Chai, M.; Deng, C.; Chen, Q.; Lu, W.; Liu, Y.F.; Li, J.H.; Du, G.C.; Lv, X.Q.; Liu, L. Synthetic Biology Toolkits and Metabolic Engineering Applied in Corynebacterium glutamicum for Biomanufacturing. ACS Synth. Biol. 2021, 10, 3237–3250. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Keasling, J. Biosensors and their applications in microbial metabolic engineering. Trends Microbiol. 2011, 19, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Mahr, R.; Frunzke, J. Transcription factor-based biosensors in biotechnology: Current state and future prospects. Appl. Microbiol. Biotechnol. 2016, 100, 79–90. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Zhang, B.; Liu, Y.M.; Yang, K.Q.; Liu, S.J. New Intracellular Shikimic Acid Biosensor for Monitoring Shikimate Synthesis in Corynebacterium glutamicum. ACS Synth. Biol. 2018, 7, 591–601. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Z.; Su, T.; Sun, H.; Zhu, Y.; Qi, Q.; Wang, Q. Tuning the binding affinity of heme-responsive biosensor for precise and dynamic pathway regulation. iScience 2020, 23, 101067. [Google Scholar] [CrossRef]
- Lim, H.G.; Jang, S.; Jang, S.; Seo, S.W.; Jung, G.Y. Design and optimization of genetically encoded biosensors for high-throughput screening of chemicals. Curr. Opin. Biotechnol. 2018, 54, 18–25. [Google Scholar] [CrossRef]
- Ding, N.; Zhou, S.; Deng, Y. Transcription-Factor-based Biosensor Engineering for Applications in Synthetic Biology. ACS Synth. Biol. 2021, 10, 911–922. [Google Scholar] [CrossRef]
- Pang, Q.X.; Han, H.; Liu, X.Q.; Wang, Z.G.; Liang, Q.F.; Hou, J.; Qi, Q.S.; Wang, Q. In vivo evolutionary engineering of riboswitch with high-threshold for N-acetylneuraminic acid production. Metab. Eng. 2020, 59, 36–43. [Google Scholar] [CrossRef]
- Jang, S.; Jang, S.; Im, D.K.; Kang, T.J.; Oh, M.K.; Jung, G.Y. Artificial Caprolactam-Specific Riboswitch as an Intracellular Metabolite Sensor. ACS Synth. Biol. 2019, 8, 1276–1283. [Google Scholar] [CrossRef] [PubMed]
- Mohsin, M.; Abdin, M.Z.; Nischal, L.; Kardam, H.; Ahmad, A. Genetically encoded FRET-based nanosensor for in vivo measurement of leucine. Biosens. Bioelectron. 2013, 50, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Li, Q.G.; Wang, Y.; Zhou, W.J.; Guo, Y.M.; Chen, J.Z.; Zheng, P.; Sun, J.B.; Ma, Y.H. Isoleucyl-tRNA synthetase mutant based whole-cell biosensor for high-throughput selection of isoleucine overproducers. Biosens. Bioelectron. 2021, 172, 112783. [Google Scholar] [CrossRef]
- Younger, A.K.; Dalvie, N.C.; Rottinghaus, A.G.; Leonard, J.N. Engineering Modular Biosensors to Confer Metabolite-Responsive Regulation of Transcription. ACS Synth. Biol. 2017, 6, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.K.; Church, G.M. Genetically encoded sensors enable real-time observation of metabolite production. Proc. Natl. Acad. Sci. USA 2016, 113, 2388–2393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binder, S.; Schendzielorz, G.; Stäbler, N.; Krumbach, K.; Hoffmann, K.; Bott, M.; Eggeling, L. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol. 2012, 13, R40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeom, S.J.; Kim, M.; Kwon, K.K.; Fu, Y.Y.; Park, S.H.; Lee, H.; Kim, H.; Lee, D.H.; Kim, D.M.; Lee, S.G. A synthetic microbial biosensor for high-throughput screening of lactam biocatalysts. Nat. Commun. 2018, 9, 5053. [Google Scholar] [CrossRef] [Green Version]
- Doong, S.J.; Gupta, A.; Prather, K.L.J. Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli. Proc. Natl. Acad. Sci. USA 2018, 115, 2964–2969. [Google Scholar] [CrossRef] [Green Version]
- Mahr, R.; Gätgens, C.; Gätgens, J.; Polen, T.; Kalinowski, J.; Frunzke, J. Biosensor-driven adaptive laboratory evolution of L-valine production in Corynebacterium glutamicum. Metab. Eng. 2015, 32, 184–194. [Google Scholar] [CrossRef]
- Zhao, N.; Song, J.; Zhang, H.; Lin, Y.; Han, S.; Huang, Y.; Zheng, S. Development of a Transcription Factor-Based Diamine Biosensor in Corynebacterium glutamicum. ACS Synth. Biol. 2021, 10, 3074–3083. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, N.; Kurihara, S.; Kitahara, Y.; Asada, K.; Kato, K.; Suzuki, H. Mechanism for regulation of the putrescine utilization pathway by the transcription factor PuuR in Escherichia coli K-12. J. Bacteriol. 2012, 194, 3437–3447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.F.; Xia, X.X.; Lee, S.Y.; Qian, Z.G. Engineering tunable biosensors for monitoring putrescine in Escherichia coli. Biotechnol. Bioeng. 2018, 115, 1014–1027. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, L.; Xie, S.; Zhao, N.; Han, S.; Lin, Y.; Zheng, S. Recombineering using RecET in Corynebacterium glutamicum ATCC14067 via a self-excisable cassette. Sci. Rep. 2017, 7, 7916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, N.N.; Li, L.; Luo, G.J.; Xie, S.; Lin, Y.; Han, S.Y.; Huang, Y.Y.; Zheng, S.P. Multiplex gene editing and large DNA fragment deletion by the CRISPR/Cpf1-RecE/T system in Corynebacterium glutamicum. J. Ind. Microbiol. Biotechnol. 2020, 47, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Yamanobe, T.; Kurihara, Y.; Uehara, H.; Komoto, T. Structure and characterization of nylon 46. J. Mol. Struct. 2007, 829, 80–87. [Google Scholar] [CrossRef]
- Hui, H.; Bai, Y.; Fan, T.P.; Zheng, X. Biosynthesis of Putrescine from L-arginine Using Engineered Escherichia coli Whole Cells. Catalysts 2020, 10, 947. [Google Scholar] [CrossRef]
- Choi, H.; Kyeong, H.H.; Choi, J.M.; Kim, H.S. Rational design of ornithine decarboxylase with high catalytic activity for the production of putrescine. Appl. Microbiol. Biotechnol. 2014, 98, 7483–7490. [Google Scholar] [CrossRef]
- Thongbhubate, K.; Irie, K.; Sakai, Y.; Itoh, A.; Suzuki, H. Improvement of putrescine production through the arginine decarboxylase pathway in Escherichia coli K-12. AMB Express 2021, 11, 168. [Google Scholar] [CrossRef]
- Della Corte, D.; van Beek, H.L.; Syberg, F.; Schallmey, M.; Tobola, F.; Cormann, K.U.; Schlicker, C.; Baumann, P.T.; Krumbach, K.; Sokolowsky, S.; et al. Engineering and application of a biosensor with focused ligand specificity. Nat. Commun. 2020, 11, 4851. [Google Scholar] [CrossRef]
- Mannan, A.A.; Liu, D.; Zhang, F.; Oyarzún, D.A. Fundamental Design Principles for Transcription-Factor-Based Metabolite Biosensors. ACS Synth. Biol. 2017, 6, 1851–1859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Ho, J.M.L.; Shis, D.L.; Gupta, C.; Long, J.; Wagner, D.S.; Ott, W.; Josić, K.; Bennett, M.R. Tuning the dynamic range of bacterial promoters regulated by ligand-inducible transcription factors. Nat. Commun. 2018, 9, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, N.; Yuan, Z.; Zhang, X.; Chen, J.; Zhou, S.; Deng, Y. Programmable cross-ribosome-binding sites to fine-tune the dynamic range of transcription factor-based biosensor. Nucleic Acids Res. 2020, 48, 10602–10613. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, O.; Tauch, A. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J. Biotechnol. 2003, 104, 287–299. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, N.; Wang, J.; Jia, A.; Lin, Y.; Zheng, S. Development of a Transcriptional Factor PuuR-Based Putrescine-Specific Biosensor in Corynebacterium glutamicum. Bioengineering 2023, 10, 157. https://doi.org/10.3390/bioengineering10020157
Zhao N, Wang J, Jia A, Lin Y, Zheng S. Development of a Transcriptional Factor PuuR-Based Putrescine-Specific Biosensor in Corynebacterium glutamicum. Bioengineering. 2023; 10(2):157. https://doi.org/10.3390/bioengineering10020157
Chicago/Turabian StyleZhao, Nannan, Jian Wang, Aiqing Jia, Ying Lin, and Suiping Zheng. 2023. "Development of a Transcriptional Factor PuuR-Based Putrescine-Specific Biosensor in Corynebacterium glutamicum" Bioengineering 10, no. 2: 157. https://doi.org/10.3390/bioengineering10020157
APA StyleZhao, N., Wang, J., Jia, A., Lin, Y., & Zheng, S. (2023). Development of a Transcriptional Factor PuuR-Based Putrescine-Specific Biosensor in Corynebacterium glutamicum. Bioengineering, 10(2), 157. https://doi.org/10.3390/bioengineering10020157