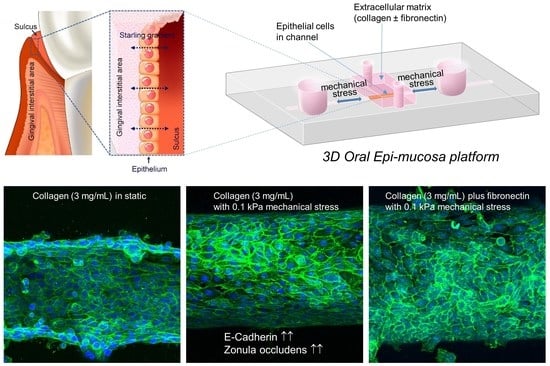
Mechanical Regulation of Oral Epithelial Barrier Function
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
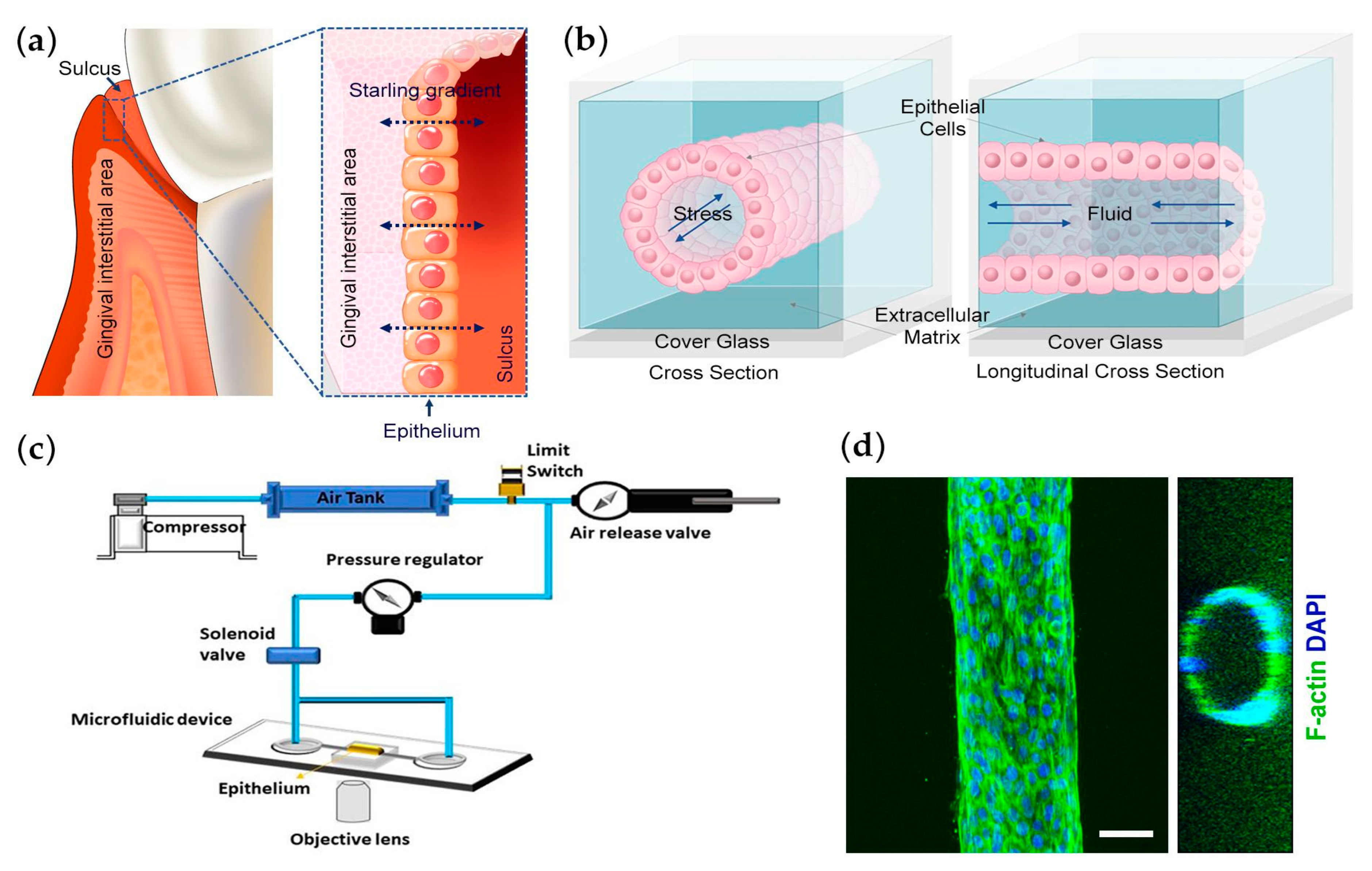
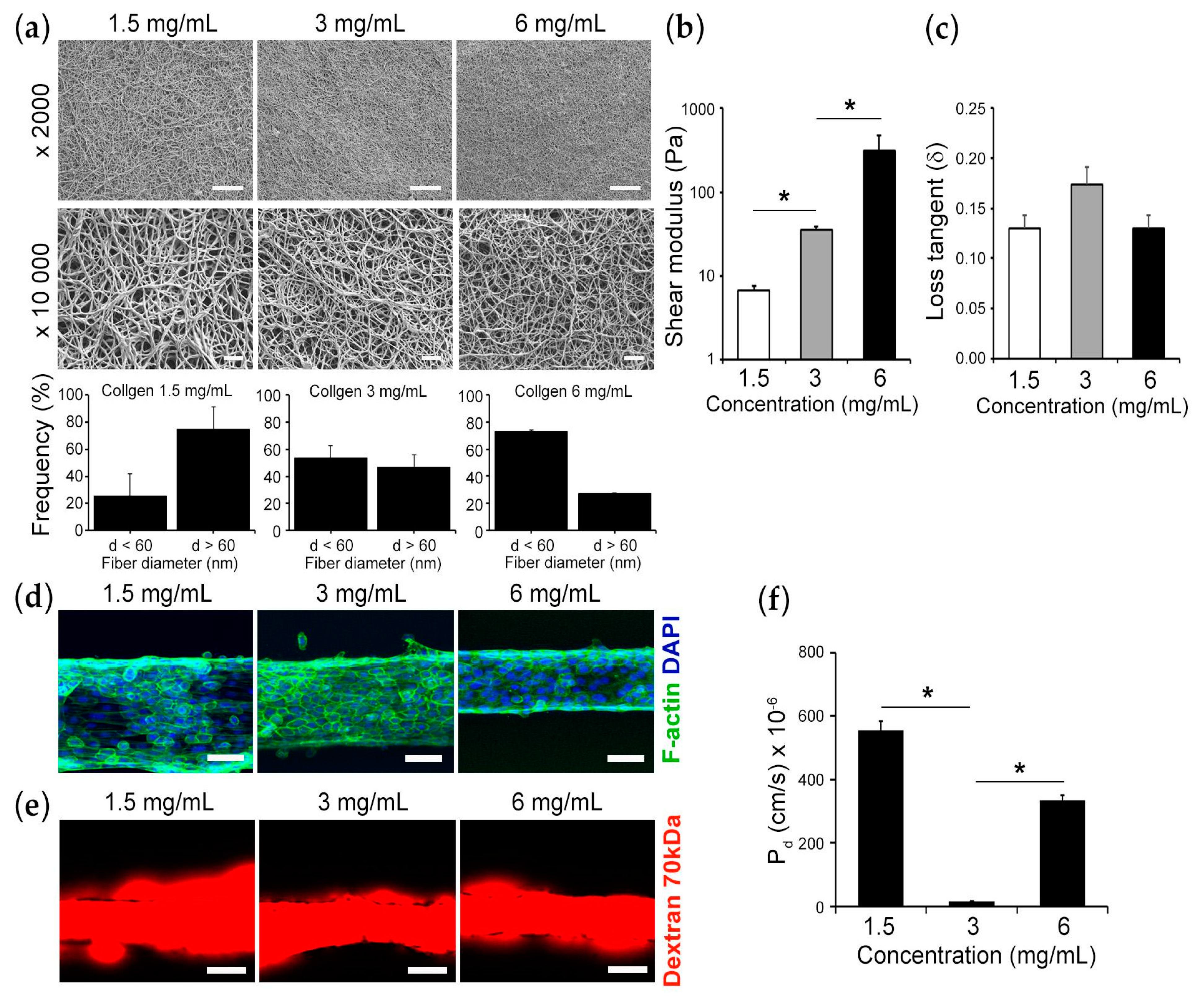
2.2. Formation of Collagen Gels
2.3. Fabrication of Microfluidic Platform
2.4. Scanning Electron Microscopy
2.5. Mechanical Testing
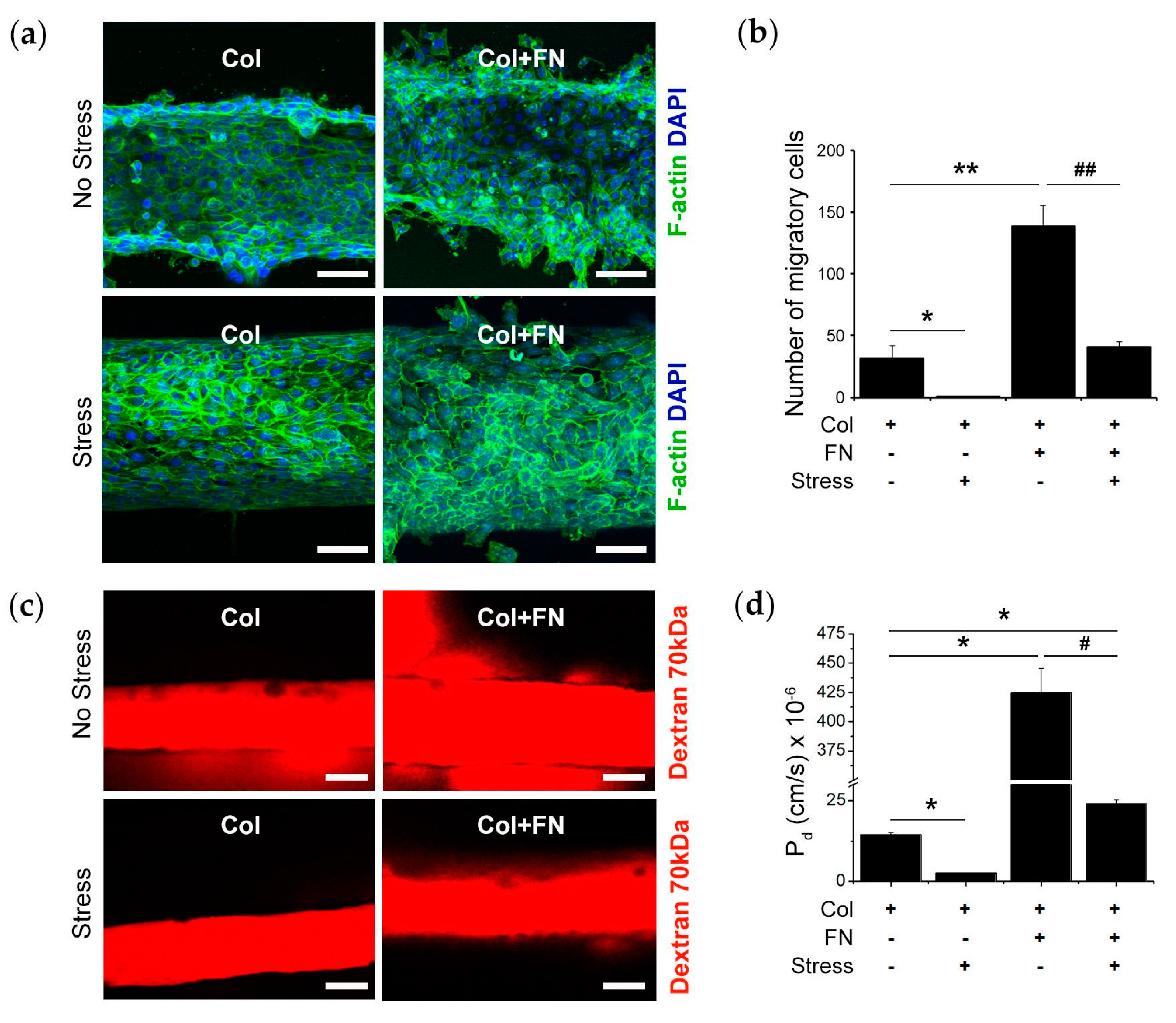
2.6. Epithelial Permeability Measurement
2.7. Immunofluorescence Staining
2.8. RT-PCR and RT-qPCR Analysis
2.9. Statistical Analysis
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brune, K.; Frank, J.; Schwingshackl, A.; Finigan, J.; Sidhaye, V.K. Pulmonary epithelial barrier function: Some new players and mechanisms. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 308, L731–L745. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ahmad, R.; Li, W.; Swain, M.; Li, Q. Biomechanics of oral mucosa. J. R. Soc. Interface 2015, 12, 325. [Google Scholar] [CrossRef]
- Nguyen, H.T.T.; Ono, M.; Hara, E.S.; Komori, T.; Edamatsu, M.; Yonezawa, T.; Kimura-Ono, A.; Maekawa, K.; Kuboki, T.; Oohashi, T. Type XVIII Collagen Modulates Keratohyalin Granule Formation and Keratinization in Oral Mucosa. Int. J. Mol. Sci. 2019, 20, 4739. [Google Scholar] [CrossRef]
- Bierbaumer, L.; Schwarze, U.Y.; Gruber, R.; Neuhaus, W. Cell culture models of oral mucosal barriers: A review with a focus on applications, culture conditions and barrier properties. Tissue Barriers 2018, 6, 1479568. [Google Scholar] [CrossRef]
- Groeger, S.; Meyle, J. Oral Mucosal Epithelial Cells. Front. Immunol. 2019, 10, 208. [Google Scholar] [CrossRef]
- Brill, N. The gingival pocket fluid. Studies of its occurrence, composition and effect. Acta Odontol. Scand. 1962, 20 (Suppl. 32), 1–115. [Google Scholar]
- Dutzan, N.; Abusleme, L.; Bridgeman, H.; Greenwell-Wild, T.; Zangerle-Murray, T.; Fife, M.E.; Bouladoux, N.; Linley, H.; Brenchley, L.; Wemyss, K.; et al. On-going Mechanical Damage from Mastication Drives Homeostatic Th17 Cell Responses at the Oral Barrier. Immunity 2017, 46, 133–147. [Google Scholar] [CrossRef]
- Lesch, C.A.; Squier, C.A.; Cruchley, A.; Williams, D.M.; Speight, P. The permeability of human oral mucosa and skin to water. J. Dent. Res. 1989, 68, 1345–1349. [Google Scholar] [CrossRef]
- Squier, C.A.; Kremer, M.J. Biology of oral mucosa and esophagus. J. Natl. Cancer Inst. Monogr. 2001, 2001, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Berg, A.; Kirkebo, A.; Heyeraas, K.J. Micropuncture measurements of interstitial fluid pressure in rat nasal mucosa during early inflammatory reactions. J. Appl. Physiol. 1998, 85, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Fjaertoft, M.; Johannessen, A.C.; Heyeraas, K.J. Micropuncture measurements of interstitial fluid pressure in normal and inflamed gingiva in rats. J. Periodontal Res. 1992, 27, 534–538. [Google Scholar] [CrossRef]
- Del Fabbro, M.; Galardi, E.; Weinstein, R.; Bulfamante, G.; Miserocchi, G. Fluid dynamics of gingival tissues. J. Periodontal Res. 1998, 33, 328–334. [Google Scholar] [CrossRef]
- Chen, J.; Suenaga, H.; Hogg, M.; Li, W.; Swain, M.; Li, Q. Determination of oral mucosal Poisson’s ratio and coefficient of friction from in-vivo contact pressure measurements. Comput. Methods Biomech. Biomed. Eng. 2016, 19, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Barros, S.P.; Williams, R.; Offenbacher, S.; Morelli, T. Gingival crevicular fluid as a source of biomarkers for periodontitis. Periodontol. 2000 2016, 70, 53–64. [Google Scholar] [CrossRef]
- Griffiths, G.S. Formation, collection and significance of gingival crevice fluid. Periodontol. 2000 2003, 31, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Lagos, M.L.; Sant’ana, A.C.; Greghi, S.L.; Passanezi, E. Keratinized Gingiva Determines a Homeostatic Behavior of Gingival Sulcus through Transudation of Gingival Crevice Fluid. Int. J. Dent. 2011, 2011, 953135. [Google Scholar] [CrossRef]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef]
- Ingber, D.E. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat. Rev. Genet. 2022, 23, 467–491. [Google Scholar] [CrossRef]
- Jeong, S.Y.; Lee, S.; Choi, W.H.; Jee, J.H.; Kim, H.R.; Yoo, J. Fabrication of Dentin-Pulp-Like Organoids Using Dental-Pulp Stem Cells. Cells 2020, 9, 642. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Y. Engineered organoids in oral and maxillofacial regeneration. iScience 2023, 26, 105757. [Google Scholar] [CrossRef]
- Arumugasaamy, N.; Navarro, J.; Leach, J.K.; Kim, P.C.W.; Fisher, J.P. In Vitro Models for Studying Transport Across Epithelial Tissue Barriers. Ann. Biomed. Eng. 2019, 47, 1–21. [Google Scholar] [CrossRef]
- Gao, X.; Wu, Y.; Liao, L.; Tian, W. Oral Organoids: Progress and Challenges. J. Dent. Res. 2021, 100, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Alimperti, S.; Mirabella, T.; Bajaj, V.; Polacheck, W.; Pirone, D.M.; Duffield, J.; Eyckmans, J.; Assoian, R.K.; Chen, C.S. Three-dimensional biomimetic vascular model reveals a RhoA, Rac1, and N-cadherin balance in mural cell-endothelial cell-regulated barrier function. Proc. Natl. Acad. Sci. USA 2017, 114, 8758–8763. [Google Scholar] [CrossRef]
- Salipante, P.F.; Hudson, S.D.; Alimperti, S. Blood vessel-on-a-chip examines the biomechanics of microvasculature. Soft Matter 2021, 18, 117–125. [Google Scholar] [CrossRef]
- Au, B.; Boulton, M.R.; Narini, P.P.; McCulloch, C.A.; Hay, J.B. Lymph and interstitial fluid dynamics in labial gingival tissues of sheep. J. Periodontal Res. 1996, 31, 570–578. [Google Scholar] [CrossRef]
- Del Fabbro, M.; Francetti, L.; Bulfamante, G.; Cribiu, M.; Miserocchi, G.; Weinstein, R.L. Fluid dynamics of gingival tissues in transition from physiological condition to inflammation. J. Periodontol. 2001, 72, 65–73. [Google Scholar] [CrossRef]
- Glim, J.E.; Everts, V.; Niessen, F.B.; Ulrich, M.M.; Beelen, R.H. Extracellular matrix components of oral mucosa differ from skin and resemble that of foetal skin. Arch. Oral. Biol. 2014, 59, 1048–1055. [Google Scholar] [CrossRef] [PubMed]
- Goktas, S.; Dmytryk, J.J.; McFetridge, P.S. Biomechanical behavior of oral soft tissues. J. Periodontol. 2011, 82, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Arikawa, H.; Fujii, K.; Shinohara, N.; Kawahata, N. Viscoelastic properties of oral soft tissue. 1. A method of determining elastic modulus of oral soft tissue. Dent. Mater. J. 1985, 4, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Kydd, W.L.; Mandley, J. The stiffness of palatal mucoperiosteum. J. Prosthet. Dent. 1967, 18, 116–121. [Google Scholar] [CrossRef]
- Tomlin, H.R.; Wilson, H.J. The measurement of thickness and hardness of oral soft tissues. Br. Dent. J. 1968, 124, 22–27. [Google Scholar] [PubMed]
- Nikoloudaki, G.; Creber, K.; Hamilton, D.W. Wound healing and fibrosis: A contrasting role for periostin in skin and the oral mucosa. Am. J. Physiol. Cell Physiol. 2020, 318, C1065–C1077. [Google Scholar] [CrossRef] [PubMed]
- Hassell, T.M. Tissues and cells of the periodontium. Periodontol. 2000 1993, 3, 9–38. [Google Scholar] [CrossRef]
- Stanton, G.; Levy, M.; Stahl, S.S. Collagen restoration in healing human gingiva. J. Dent. Res. 1969, 48, 27–31. [Google Scholar] [CrossRef]
- Chandran, A.; Bhandary, R.; Shenoy, N.; Shetty, U.A. Analysis of collagen fibers in human gingival tissues using picrosirius red stain under polarized microscope. J. Indian Soc. Periodontol. 2021, 25, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, S.; Leiva-Sabadini, C.; Schuh, C.; Aguayo, S. Bacterial adhesion to collagens: Implications for biofilm formation and disease progression in the oral cavity. Crit. Rev. Microbiol. 2022, 48, 83–95. [Google Scholar] [CrossRef]
- Bjornfot Holmstrom, S.; Clark, R.; Zwicker, S.; Bureik, D.; Kvedaraite, E.; Bernasconi, E.; Nguyen Hoang, A.T.; Johannsen, G.; Marsland, B.J.; Bostrom, E.A.; et al. Gingival Tissue Inflammation Promotes Increased Matrix Metalloproteinase-12 Production by CD200R(low) Monocyte-Derived Cells in Periodontitis. J. Immunol. 2017, 199, 4023–4035. [Google Scholar] [CrossRef] [PubMed]
- Oda, D.; Bigler, L.; Lee, P.; Blanton, R. HPV immortalization of human oral epithelial cells: A model for carcinogenesis. Exp. Cell Res. 1996, 226, 164–169. [Google Scholar] [CrossRef]
- Handfield, M.; Mans, J.J.; Zheng, G.; Lopez, M.C.; Mao, S.; Progulske-Fox, A.; Narasimhan, G.; Baker, H.V.; Lamont, R.J. Distinct transcriptional profiles characterize oral epithelium-microbiota interactions. Cell. Microbiol. 2005, 7, 811–823. [Google Scholar] [CrossRef]
- Jeon, H.H.; Yu, Q.; Lu, Y.; Spencer, E.; Lu, C.; Milovanova, T.; Yang, Y.; Zhang, C.; Stepanchenko, O.; Vafa, R.P.; et al. FOXO1 regulates VEGFA expression and promotes angiogenesis in healing wounds. J. Pathol. 2018, 245, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ideguchi, H.; Kajikawa, T.; Mastellos, D.C.; Lambris, J.D.; Hajishengallis, G. Complement Is Required for Microbe-Driven Induction of Th17 and Periodontitis. J. Immunol. 2022, 209, 1370–1378. [Google Scholar] [CrossRef] [PubMed]
- Raub, C.B.; Suresh, V.; Krasieva, T.; Lyubovitsky, J.; Mih, J.D.; Putnam, A.J.; Tromberg, B.J.; George, S.C. Noninvasive assessment of collagen gel microstructure and mechanics using multiphoton microscopy. Biophys. J. 2007, 92, 2212–2222. [Google Scholar] [CrossRef]
- Jin, L.; Kou, N.; An, F.; Gao, Z.; Tian, T.; Hui, J.; Chen, C.; Ma, G.; Mao, H.; Liu, H. Analyzing Human Periodontal Soft Tissue Inflammation and Drug Responses In Vitro Using Epithelium-Capillary Interface On-a-Chip. Biosensors 2022, 12, 345. [Google Scholar] [CrossRef] [PubMed]
- Tolo, K.; Jonsen, J. In vitro penetration of tritiated dextrans through rabbit oral mucosa. Arch. Oral. Biol. 1975, 20, 419–422. [Google Scholar] [CrossRef]
- Nasjleti, C.E.; Caffesse, R.G. Dextran penetration through nonkeratinized and keratinized epithelia in monkeys. J. Periodontol. 1984, 55, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- McQuin, C.; Goodman, A.; Chernyshev, V.; Kamentsky, L.; Cimini, B.A.; Karhohs, K.W.; Doan, M.; Ding, L.; Rafelski, S.M.; Thirstrup, D.; et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018, 16, e2005970. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, S.V.; Balzar, M.; Winter, M.J.; Bakker, H.A.; Briaire-de Bruijn, I.H.; Prins, F.; Fleuren, G.J.; Warnaar, S.O. Epithelial cell adhesion molecule (Ep-CAM) modulates cell-cell interactions mediated by classic cadherins. J. Cell Biol. 1997, 139, 1337–1348. [Google Scholar] [CrossRef] [PubMed]
- Sumagin, R.; Parkos, C.A. Epithelial adhesion molecules and the regulation of intestinal homeostasis during neutrophil transepithelial migration. Tissue Barriers 2015, 3, e969100. [Google Scholar] [CrossRef] [PubMed]
- Larjava, H.; Koivisto, L.; Hakkinen, L.; Heino, J. Epithelial integrins with special reference to oral epithelia. J. Dent. Res. 2011, 90, 1367–1376. [Google Scholar] [CrossRef]
- Chang, J.; Hakam, A.E.; McCauley, L.K. Current Understanding of the Pathophysiology of Osteonecrosis of the Jaw. Curr. Osteoporos. Rep. 2018, 16, 584–595. [Google Scholar] [CrossRef]
- Groeger, S.E.; Meyle, J. Epithelial barrier and oral bacterial infection. Periodontol. 2000 2015, 69, 46–67. [Google Scholar] [CrossRef] [PubMed]
- Maymone, M.B.C.; Greer, R.O.; Kesecker, J.; Sahitya, P.C.; Burdine, L.K.; Cheng, A.D.; Maymone, A.C.; Vashi, N.A. Premalignant and malignant oral mucosal lesions: Clinical and pathological findings. J. Am. Acad. Dermatol. 2019, 81, 59–71. [Google Scholar] [CrossRef]
- Huang, C.; Sanaei, F.; Verdurmen, W.P.R.; Yang, F.; Ji, W.; Walboomers, X.F. The Application of Organs-on-a-Chip in Dental, Oral, and Craniofacial Research. J. Dent. Res. 2023, 102, 364–375. [Google Scholar] [CrossRef]
- Makkar, H.; Zhou, Y.; Tan, K.S.; Lim, C.T.; Sriram, G. Modeling Crevicular Fluid Flow and Host-Oral Microbiome Interactions in a Gingival Crevice-on-Chip. Adv. Healthc. Mater. 2023, 12, e2202376. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, C.; Rahimi, B.; Padova, D.; Rooholghodos, S.A.; Bienek, D.R.; Luo, X.; Kaufman, G.; Raub, C.B. Oral mucosa-on-a-chip to assess layer-specific responses to bacteria and dental materials. Biomicrofluidics 2018, 12, 054106. [Google Scholar] [CrossRef] [PubMed]
- Wikswo, J.P. The relevance and potential roles of microphysiological systems in biology and medicine. Exp. Biol. Med. 2014, 239, 1061–1072. [Google Scholar] [CrossRef]
- Basson, M.D. Paradigms for mechanical signal transduction in the intestinal epithelium. Category: Molecular, cell, and developmental biology. Digestion 2003, 68, 217–225. [Google Scholar] [CrossRef]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef]
- Kilic, A.; Ameli, A.; Park, J.A.; Kho, A.T.; Tantisira, K.; Santolini, M.; Cheng, F.; Mitchel, J.A.; McGill, M.; O’Sullivan, M.J.; et al. Mechanical forces induce an asthma gene signature in healthy airway epithelial cells. Sci. Rep. 2020, 10, 966. [Google Scholar] [CrossRef]
- Kim, H.J.; Li, H.; Collins, J.J.; Ingber, D.E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. USA 2016, 113, E7–E15. [Google Scholar] [CrossRef] [PubMed]
- Vancamelbeke, M.; Vermeire, S. The intestinal barrier: A fundamental role in health and disease. Expert. Rev. Gastroenterol. Hepatol. 2017, 11, 821–834. [Google Scholar] [CrossRef]
- Waters, C.M.; Roan, E.; Navajas, D. Mechanobiology in lung epithelial cells: Measurements, perturbations, and responses. Compr. Physiol. 2012, 2, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Bein, A.; Zilbershtein, A.; Golosovsky, M.; Davidov, D.; Schwartz, B. LPS Induces Hyper-Permeability of Intestinal Epithelial Cells. J. Cell. Physiol. 2017, 232, 381–390. [Google Scholar] [CrossRef]
- Leonardo, T.R.; Shi, J.; Chen, D.; Trivedi, H.M.; Chen, L. Differential Expression and Function of Bicellular Tight Junctions in Skin and Oral Wound Healing. Int. J. Mol. Sci. 2020, 21, 2966. [Google Scholar] [CrossRef] [PubMed]
- Presland, R.B.; Jurevic, R.J. Making sense of the epithelial barrier: What molecular biology and genetics tell us about the functions of oral mucosal and epidermal tissues. J. Dent. Educ. 2002, 66, 564–574. [Google Scholar] [CrossRef]
- Tang, S.; Jiang, X.; Wu, L.; Chen, S.; Chen, L.; Jiang, J.; Yan, P.; Wang, F.; Tu, K.; Wang, D.; et al. Toll-like receptor 4 shRNA attenuates lipopolysaccharide-induced epithelial-mesenchymal transition of intrahepatic biliary epithelial cells in rats. Biomed. Pharmacother. 2018, 107, 1210–1217. [Google Scholar] [CrossRef]
- Epifano, C.; Perez-Moreno, M. Crossroads of integrins and cadherins in epithelia and stroma remodeling. Cell Adhes. Migr. 2012, 6, 261–273. [Google Scholar] [CrossRef]
- Bays, J.L.; Campbell, H.K.; Heidema, C.; Sebbagh, M.; DeMali, K.A. Linking E-cadherin mechanotransduction to cell metabolism through force-mediated activation of AMPK. Nat. Cell Biol. 2017, 19, 724–731. [Google Scholar] [CrossRef]
- Liu, Z.; Tan, J.L.; Cohen, D.M.; Yang, M.T.; Sniadecki, N.J.; Ruiz, S.A.; Nelson, C.M.; Chen, C.S. Mechanical tugging force regulates the size of cell-cell junctions. Proc. Natl. Acad. Sci. USA 2010, 107, 9944–9949. [Google Scholar] [CrossRef] [PubMed]
- Jepsen, K.; Jervoe-Storm, P.M.; Henrichs, I.; Lensing, I.; Muller, A.L.; Cosgarea, R.; Keilig, L.; Bourauel, C.; Jepsen, S. Biomechanical properties of periodontal tissues in non-periodontitis and periodontitis patients assessed with an intraoral computerized electronic measurement device. Clin. Oral Investig. 2023, 27, 797–805. [Google Scholar] [CrossRef]
- Li, Y.; Zhan, Q.; Bao, M.; Yi, J.; Li, Y. Biomechanical and biological responses of periodontium in orthodontic tooth movement: Up-date in a new decade. Int. J. Oral Sci. 2021, 13, 20. [Google Scholar] [CrossRef] [PubMed]
- Chukkapalli, S.S.; Lele, T.P. Periodontal cell mechanotransduction. Open Biol. 2018, 8, 180053. [Google Scholar] [CrossRef]
- Binderman, I.; Bahar, H.; Yaffe, A. Strain relaxation of fibroblasts in the marginal periodontium is the common trigger for alveolar bone resorption: A novel hypothesis. J. Periodontol. 2002, 73, 1210–1215. [Google Scholar] [CrossRef]
- Binderman, I.; Gadban, N.; Yaffe, A. Cytoskeletal disease: A role in the etiology of adult periodontitis. Oral Dis. 2014, 20, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Paszek, M.J.; Zahir, N.; Johnson, K.R.; Lakins, J.N.; Rozenberg, G.I.; Gefen, A.; Reinhart-King, C.A.; Margulies, S.S.; Dembo, M.; Boettiger, D.; et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 2005, 8, 241–254. [Google Scholar] [CrossRef]
- Provenzano, P.P.; Inman, D.R.; Eliceiri, K.W.; Keely, P.J. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene 2009, 28, 4326–4343. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, O.; Koshy, S.T.; Branco da Cunha, C.; Shin, J.W.; Verbeke, C.S.; Allison, K.H.; Mooney, D.J. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat. Mater. 2014, 13, 970–978. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zuidema, A.; Te Molder, L.; Nahidiazar, L.; Hoekman, L.; Schmidt, T.; Coppola, S.; Sonnenberg, A. Hemidesmosomes modulate force generation via focal adhesions. J. Cell Biol. 2020, 219, e201904137. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, K.A.; Wang, C.-C.; Manthiram, A.; Ferreira, P.J. The role of composition in the atomic structure, oxygen loss, and capacity of layered Li–Mn–Ni oxide cathodes. J. Mater. Chem. A 2014, 2, 1353–1362. [Google Scholar] [CrossRef]
- Kim, I.L.; Khetan, S.; Baker, B.M.; Chen, C.S.; Burdick, J.A. Fibrous hyaluronic acid hydrogels that direct MSC chondrogenesis through mechanical and adhesive cues. Biomaterials 2013, 34, 5571–5580. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, S.; Dedden, D.; Nunez, R.V.; Matoba, K.; Takagi, J.; Biertumpfel, C.; Mizuno, N. Structural insights into integrin alpha(5)beta(1) opening by fibronectin ligand. Sci. Adv. 2021, 7, eabe9716. [Google Scholar] [CrossRef]
- Rousselle, P.; Montmasson, M.; Garnier, C. Extracellular matrix contribution to skin wound re-epithelialization. Matrix Biol. 2019, 75–76, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Shafraz, O.; Rübsam, M.; Stahley, S.N.; Caldara, A.L.; Kowalczyk, A.P.; Niessen, C.M.; Sivasankar, S. E-cadherin binds to desmoglein to facilitate desmosome assembly. eLife 2018, 7, e37629. [Google Scholar] [CrossRef] [PubMed]


| Col | Col + FN(5) | Col + FN (50) | |
|---|---|---|---|
| Collagen I (3 mg/mL) | + | + | + |
| FN (5 µg/mL) | − | + | − |
| FN (50 µg/mL) | − | − | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, E.-J.; Kim, Y.; Salipante, P.; Kotula, A.P.; Lipshutz, S.; Graves, D.T.; Alimperti, S. Mechanical Regulation of Oral Epithelial Barrier Function. Bioengineering 2023, 10, 517. https://doi.org/10.3390/bioengineering10050517
Lee E-J, Kim Y, Salipante P, Kotula AP, Lipshutz S, Graves DT, Alimperti S. Mechanical Regulation of Oral Epithelial Barrier Function. Bioengineering. 2023; 10(5):517. https://doi.org/10.3390/bioengineering10050517
Chicago/Turabian StyleLee, Eun-Jin, Yoontae Kim, Paul Salipante, Anthony P. Kotula, Sophie Lipshutz, Dana T. Graves, and Stella Alimperti. 2023. "Mechanical Regulation of Oral Epithelial Barrier Function" Bioengineering 10, no. 5: 517. https://doi.org/10.3390/bioengineering10050517
APA StyleLee, E.-J., Kim, Y., Salipante, P., Kotula, A. P., Lipshutz, S., Graves, D. T., & Alimperti, S. (2023). Mechanical Regulation of Oral Epithelial Barrier Function. Bioengineering, 10(5), 517. https://doi.org/10.3390/bioengineering10050517