1. Introduction
Good sleep helps the body to eliminate fatigue and maintain normal brain functioning [
1]. In contrast, a lack of sleep can lead to depression, obesity, coronary heart disease, and other diseases [
2,
3,
4,
5]. However, sleep disorders are becoming an alarmingly common health problem, affecting the health status of thousands of people [
6,
7]. To assess sleep quality and diagnose sleep disorders, signals such as electroencephalography (EEG), electrooculography (EOG), and electromyography (EMG) collected through polysomnography (PSG) are usually used to stage sleep. An entire night’s sleep can be divided into wake (W), non-rapid eye movement (NREM, S1, S2, S3, and S4), and rapid eye movement(REM) stages, according to the Rechtschaffen and Kales (R&K) standard [
8] or wake (W), non-rapid eye movement (NREM, N1, N2, and N3), and rapid eye movement(REM) stages, according to the American Academy of Sleep Medicine (AASM) standard [
9]. In the clinic, sleep staging is performed manually by experienced experts. The procedure is time-consuming and labor-intensive. Meanwhile, there is a subjective element in the judgment of experts and different experts do not fully agree on the classification of sleep stages [
10,
11]. To relieve the burden on physicians and save medical resources, many studies have focused on automatic sleep staging using biosignals through machine learning approaches [
12,
13,
14,
15].
Automatic sleep staging methods can be divided into traditional machine learning-based methods and deep learning-based methods. Traditional machine learning-based methods usually consist of handcrafted feature extraction and traditional classification methods. Handcrafted feature extraction extracts the features of signals from the time domain, frequency domain, etc., based on medical knowledge. These extracted features are then fed into traditional classifiers, such as support vector machines (SVM) [
16,
17,
18], random forests (RF) [
19,
20], etc., for automatic sleep staging. Instead of requiring medical knowledge as a prerequisite, the deep learning-based method uses networks to automatically extract features. Thus, it has been widely explored in recent research [
21]. Some convolutional neural networks (CNN) [
22,
23,
24] or recurrent neural networks (RNN) [
25] models have achieved good results in automatic sleep staging. Furthermore, some studies have combined different network architectures, to incorporate their advantages, such as the combination of CNN and RNN [
26,
27,
28], the combination of RNN and RNN [
29,
30], and the combination of CNN and Transformer architectures [
31,
32]. With the development of machine learning methods, performance in sleep staging has been greatly improved, with an excellent performance on certain public datasets [
33,
34,
35,
36]. However, both the traditional machine learning-based methods and deep learning-based methods apply an EEG signal as the main or only input signal. The process of acquiring EEG signals is very tedious and uncomfortable for the subject.
Taking into account the comfort of physiological signal acquisition, some studies have tried to use certain easy-to-collect signals for sleep staging, such as cardiopulmonary signals [
37,
38,
39], acoustic signals [
40,
41], and EOG signals [
42,
43,
44]. These signals are relatively easy and comfortable to acquire compared to EEG signals, but the sleep staging performance with cardiopulmonary and acoustic signals was not satisfactory for clinical application. Noteworthy, the accuracy of sleep staging using a single-channel EOG signal was similar to that of a single-channel EEG signal in some studies [
28,
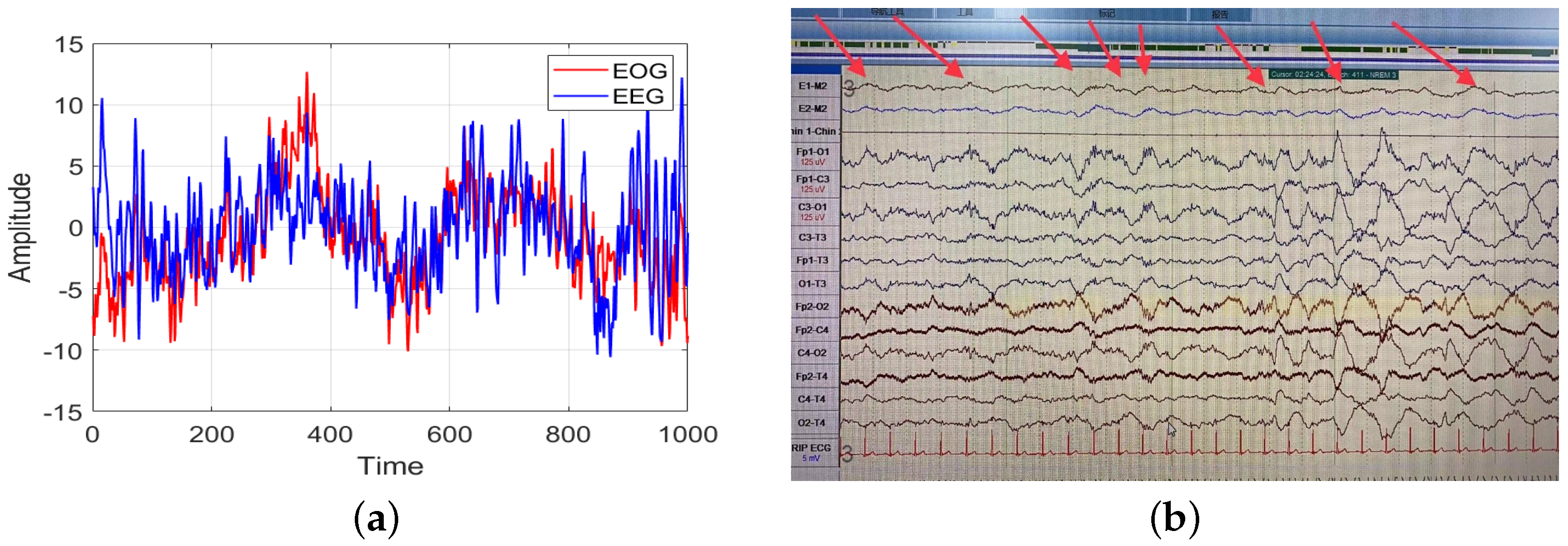
42]. This suggested that an EOG signal could also be used for sleep staging with good performance, allowing comfortable sleep monitoring. Despite the good results yielded by EOG signals in automatic sleep staging, the positions of the acquisition electrodes for the EOG signal and the prefrontal EEG signal are close to each other, which means that part of the EEG signal may be coupled in the EOG signal. Comparison of an EEG signal and an EOG signal in the N3 stage revealed slow wave signals with similar frequencies to those in the Fp1-O1 channel and the E1-M2 and E2-M2 channels (
Figure 1). The slow wave signal, as the main characteristic wave of N3 sleep stage, appears in an EOG signal. Therefore, it is not clear whether the sleep staging ability of an EOG signal comes from the coupled EEG signal, and how this coupled EEG could affect the sleep staging results.
To explore the above issue, we conducted experiments applying data from two public datasets and one clinical dataset. First, we processed the EEG signal with a blind source separation algorithm named second-order blind identification (SOBI) [
45] to obtain an EEG signal without EOG signals. Second, the raw EOG signal and the clean EEG signal were coupled to obtain a clean EOG signal and EOG signal coupled with different contents of the EEG signal. Third, the coupled EOG signals with different EEG signal contents were fed into a hierarchical neural network named two-step hierarchical neural network (THNN), which consists of a multi-scale CNN and a bidirectional gating unit (Bi-GRU), for automatic sleep staging. We also performed automatic sleep staging using the EEG signal with the THNN, to explore the difference in performance between the EOG signal and the EEG signal. Finally, we considered the impact of EEG signal coupling in EOG signals on sleep staging.
2. Materials and Methods
In this section, we introduce the subjects selected for this exploration, the blind source separation method used, as well as the specific structure, details, and training strategy of the THNN.
2.1. Subjects and PSG Recordings
In this work, we applied two widely used public datasets and one clinical dataset to conduct the experiment. The details of the three datasets are shown in
Table 1.
2.1.1. Montreal Archive of Sleep Studies (MASS) Dataset
The MASS dataset was provided by the University of Montreal and the Sacred Heart Hospital in Montreal [
36]. It consists of whole night sleep recordings from 200 subjects aged from 18 years old to 76 years old (97 males and 103 females), divided into five subsets SS1–SS5. The SS1 and SS3 subsets have a length of 30 s for each sleep stage, the other subsets have a sleep stage of 20 s. Each epoch of the recordings in MASS was manually labeled according to the AASM standard or R&K standard by experts. The amplifier system for MASS was the Grass Model 12 or 15 from Grass Technologies. The reference electrodes were CLE or LER. In this experiment, the SS3 subset was used.
2.1.2. Dreams Dataset
The DREAMS dataset was collected during the DREAMS project. It has eight subsets: subject database, patient database, artifact database, sleep spindles database, K-complex database, REM database, PLM database, and apnea database [
35,
46]. These recordings were annotated as microevents or as sleep stages by several experts. In this work, the subject database was applied. The subject database consists of 20 whole-night PSG recordings (16 females and 4 males) derived from healthy subjects, and the sleep stages were categorized into sleep stages according to both the R&K standard and the AASM standard. The data collection instrumentation for DREAMS was a digital 32-channel polygraph (BrainnetTM System of MEDATEC, Brussels, Belgium). The reference electrode was A1.
2.1.3. Huashan Hospital Fudan University (HSFU) Dataset
The HSFU dataset is a non-public database collected in Huashan Hospital, Fudan University, Shanghai, China, during 2019–2020. Twenty-six clinical PSG recordings were collected from people who had sleep disorders. The research was approved by the Ethics Committee of Huashan Hospital (ethical permit No. 2021-811). The PSG recordings were annotated by a qualified sleep expert according to the AASM standard. The specific information of each subject is described in
Table A1. The data collection instrumentation for HSFU was a COMPUMEDICS GREAL HD PSG. The reference electrodes were M1 and M2.
2.2. Blind Source Separation Algorithm
Blind source separation methods are widely used when dealing with coupled signals. Some common blind source separation methods include fast independent component analysis (FastICA), information maximization (Infomax), and second-order blind identification (SOBI); the first two methods require that each channel of the input signal be independent of each other, whereas SOBI has no such requirement for the input signal.Meanwhile, the effectiveness of SOBI for processing mixed signals is not affected by the number of signal channels [
47]. Thus, in this experiment, the SOBI method was applied to remove interference signals, due to its robustness. The SOBI algorithm was proposed by Belouchrani et al. in 1997 [
45]. This algorithm achieves blind source separation by joint approximate diagonalization of the delayed correlation matrix. It is a stable method for blind source separation. SOBI uses second-order statistics, so that it can estimate the components of the source signals with few data points. The pseudo-algorithmic of SOBI is as follows (Algorithm 1): Assuming that the input signal X has M channels and each channel has N samples, i.e.,
. After normalization and whitening of the input signal, joint approximate diagonalization is performed using the covariance of the signal, to obtain the coupling coefficient. Finally, the original signal is obtained using a matrix inverse operation.
| Algorithm 1 SOBI |
Input: Input: Data, Output: Output: Data, - 1:
Normalization - 2:
Whitening - 3:
Calculate the covariance matrix - 4:
while coefficients not converged or maximum iterations number not reached do - 5:
Joint approximate diagonalization algorithm - 6:
end while - 7:
Calculate the source signal
|
2.3. Two-Step Hierarchical Neural Network
In this work, THNN was applied to conduct automatic sleep staging. The specific structure of THNN is presented in
Figure 2, and the specific parameters are shown in
Table A2. THNN can be divided into two parts: the feature extraction module, and the sequence learning module. The feature extraction module uses a multi-scale convolutional neural network with two scales to extract features from different scales. The sequence learning module uses a Bi-GRU network, which can learn the temporal information in the feature matrix extracted by the feature extraction module. The feature learning module consists of a two-scale CNN network. The two scales of CNN have different sizes of convolutional kernel for extracting large-time-span features and short-time-span features in EEG signals, respectively. Specifically, if the sampling rate of an EEG signal is 128 Hz, and the convolutional kernel length of the small-scale CNN is 64, then each segment of the EEG signal is 0.5 s of the sampling signal, which corresponds to 2 Hz. The large-scale CNN has a convolutional kernel length of 640, thus each segment of the EEG signal is 5 s of the sampling signal, which corresponds to 0.2 Hz. By designing convolutional kernels of different sizes, better feature information can be extracted. Suppose the signal
is the input of THNN, where the
L is the number of epochs and the
P is the length of each epoch. The process of feature learning is represented as follows:
where the
is the small scale branch of CNN,
is the large scale branch of CNN, and
is the concatenation layer. The sequence learning part consists of Bi-GRU, which handles the time-dependent sequence signals well and has a fast operation speed in RNN networks [
48]. GRU has a fast operation speed, but it still takes a long time to train when running serially. Therefore, we added a residual structure to the serial learning module, to speed up the training [
49]. Finally, the probabilities of each sleep stage were output through the softmax layer. The process of the sequence learning part is shown as follows:
where
is the Bi-GRU network,
H is the temporal feature of each sleep stage outputted by the Bi-GRU,
O is the feature after superposition of the residual module, and
Y is the final sleep stage probability of each epoch.
2.4. Data Preprocessing and Experiment Scheme
In this work, we adopted the Fp1 channel EEG signal, Fp2 channel EEG signal, left EOG signal, and right EOG signal from the MASS, DREAMS, and HSFU datasets to conduct the experiments. All the signals used were filtered with a 50 Hz/60 Hz notch filter and a 0.3–35 Hz band-pass filter. and then the signals were resampled to 128 Hz to fit the network, as well as to reduce the complexity of operations. Afterwards, the SOBI method was used to remove the interference signals in the EEG signals and to obtain a clean EEG signal without the EOG signal. Next, the raw EOG signal and the clean EEG signal were processed to obtain a clean EOG signal and the EOG signal coupled with different contents of the EEG signal. The steps are showed in
Figure 3. The specific calculation procedure of the coupled EOG signal is shown in Equation (
7).
where the
is the EEG signal without EOG signals, the
is the EOG signal coupled with the EEG signal, and
a is the superposition factor. The content of EEG signal in the EOG signal was calculated using the correlation coefficient between the coupled EOG signal and the clean EEG signal on the same side.
We performed experiments using EOG signals coupled with different contents of EEG signal, and the correlation coefficients were set as 0.0, 0.1, 0.2, 0.3, and 0.5. We fed each of the five EOG signals into the network for automatic sleep staging. In addition, we used the leave-one-subject-out (LOSO) method for the validation.
5. Conclusions
In this paper, we investigated the effect of the EEG signal coupled in an EOG signal on sleep staging results. Two publicly available datasets and one clinical dataset were used for the experiment. The SOBI method was applied to obtain a clean EEG signal. The clean EEG signal and the raw EOG signal were used to obtain a clean EOG signal and EOG signal coupled with different contents of the EEG signal. Afterwards, a THNN was used to perform automatic sleep staging with coupled EOG signals. The results showed that the sleep staging capability of the EOG signal was not derived from the coupled EEG signal but from its own feature information. Meanwhile, the EOG signal coupled with the EEG signal had better classification performance for the N1, N2, and N3 stages. The coupled EEG signal could complement the feature information lacking in the EOG signal, especially in the N1 and N2 stages. In addition, the amount of coupled EEG signal was similar to the amount of EEG signal contained in the raw EOG signal. Higher or lower levels than this could result in a certain reduction in the accuracy of the sleep staging results. This paper provided an explorative experimental analysis of automatic sleep staging using EOG signals. Moreover, it is excepted to provide an experimental basis for comfortable sleep analysis, home sleep monitoring, etc.