Prediction of Sleep Apnea Events Using a CNN–Transformer Network and Contactless Breathing Vibration Signals
Abstract
1. Introduction
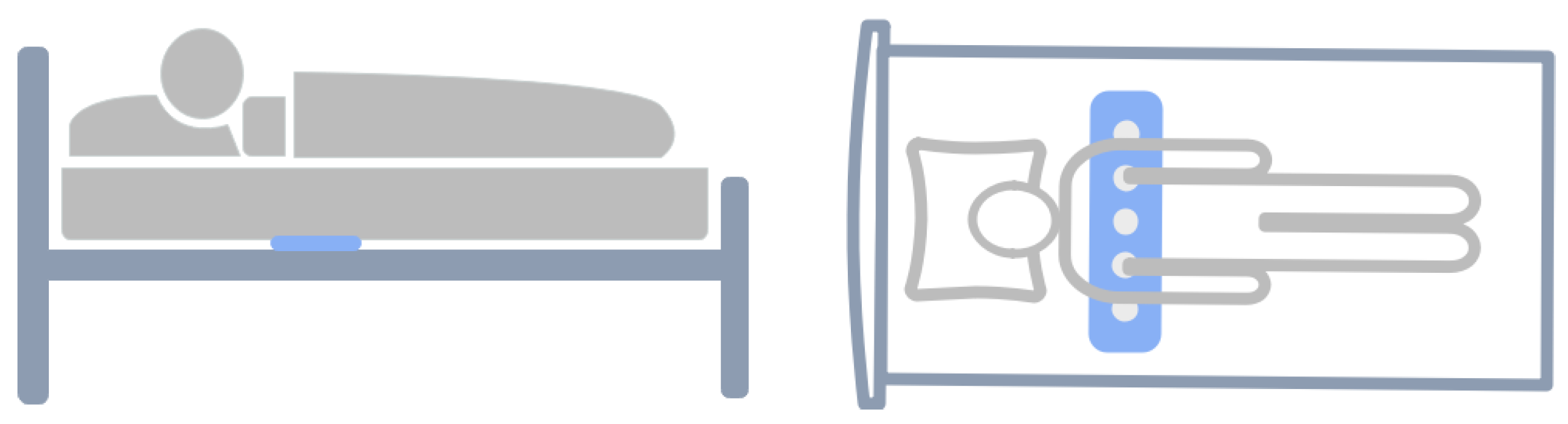
- A novel contactless scheme based on deep learning and breathing vibration signals is developed for sleep apnea event prediction. Our method can effectively predict respiratory events without disturbing the sleep of the subjects.
- A novel CNN–transformer network is proposed for prediction. It leverages the advantages of both CNN and transformer architectures, to effectively capture both local and global features present in the respiratory signals for prediction.
- The proposed method is validated on a dataset of 105 subjects from a public hospital and obtained a prediction accuracy of 85.9%. The method outperformed classical time series classification methods in terms of accuracy, sensitivity, and F1 score, demonstrating its effectiveness for the prediction of sleep apnea events. Two types of cross-validation were performed to demonstrate the generalization of our model.
2. Materials and Methods
2.1. Data Collection and Preparation
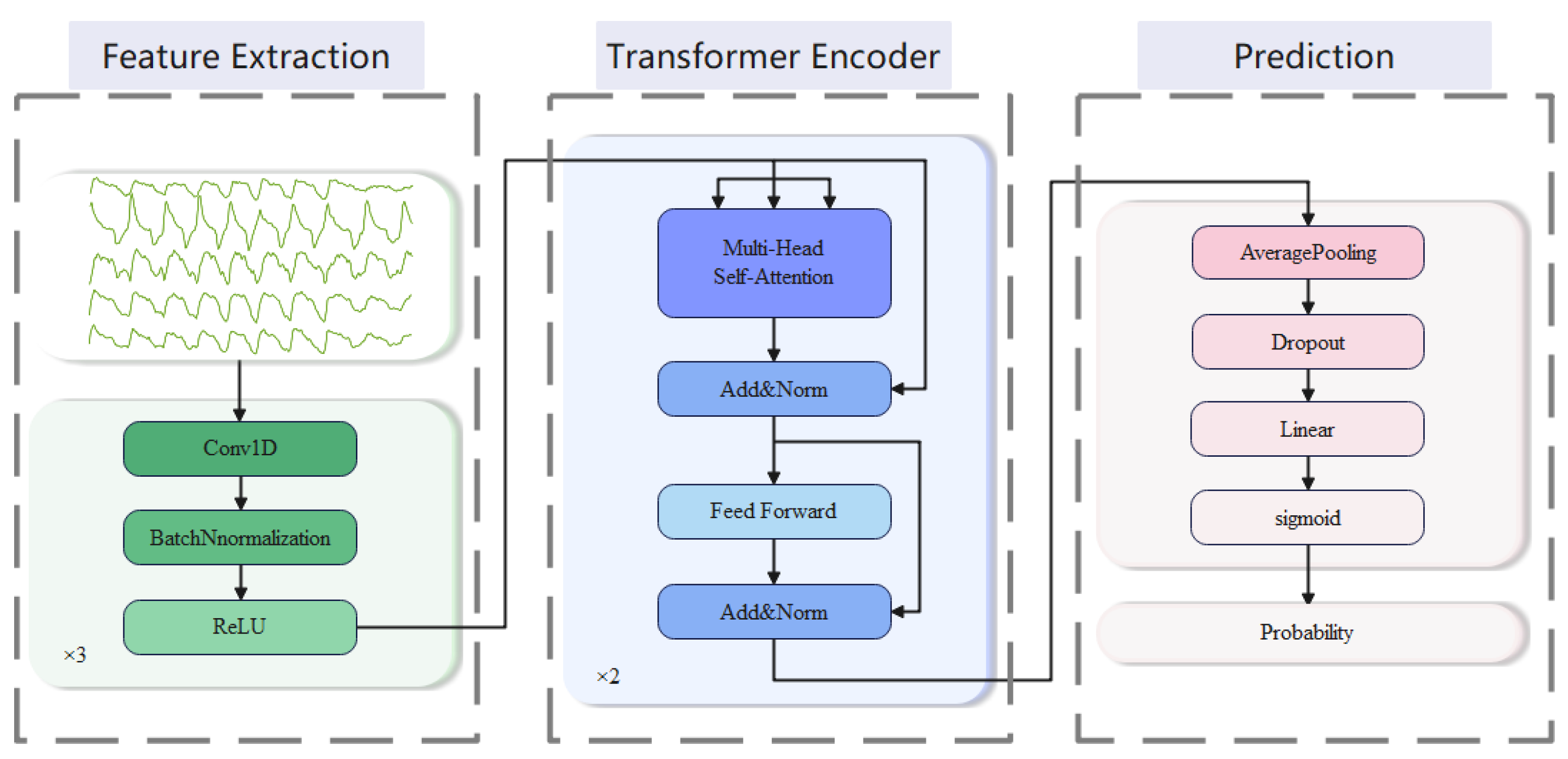
2.2. Analysis Model
- Feature extraction: We employ three 1D convolutional blocks to extract features. Each of the blocks consists of three sub-layers, which perform in turn: 1D-CNN layer, batch normalization (BN) layer, and ReLU activation layer. The first block has a convolutional kernel size of 3, while the next two blocks have convolutional kernel sizes of 29. The number of output channels is set to 64, and the padding is 1 in the first block and 14 in the last two blocks. The smaller kernel size captures local features with a smaller receptive field, while the larger kernel size captures global features with a larger receptive field. By combining them, the model can capture both local and global features, leading to a more comprehensive representation of the input signals.
- Transformer encoder: We employ a stack of 2 transformer encoders to encode the high-dimensional features output. These encoded representations can effectively capture long-range dependencies in the sequence, providing strong support for subsequent classification tasks. Each transformer encoder consists of a multi-head self-attention layer and a position-wise feed-forward layer (FFN) [28].
- 3.
- Prediction: After the output of a transformer encoder, an average pooling layer and a dropout layer are typically applied. We use the average pooling layer to reduce the dimensionality of the output. A dropout layer is used to prevent overfitting with a parameter set to 0.5. Then, the result is mapped to the target output dimension through a linear layer and finally mapped to between 0 and 1 through a sigmoid layer to obtain the output probability.
2.3. Model Evaluation
3. Experiments and Results
3.1. Experiment Details
3.2. Ablation Study
3.3. Performance Comparison
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Somers, V.K.; White, D.P.; Amin, R.; Abraham, W.T.; Costa, F.; Culebras, A.; Daniels, S.; Floras, J.S.; Hunt, C.E.; Olson, L.J.; et al. Sleep apnea and cardiovascular disease: An American Heart Association/American College of Cardiology Foundation Scientific Statement from the American Heart Association Council for High Blood Pressure Research Professional Education Committee, Council on Clinical Cardiology, Stroke Council, and Council on Cardiovascular Nursing. J. Am. Coll. Cardiol. 2008, 52, 686–717. [Google Scholar] [CrossRef]
- Shamsuzzaman, A.S.M.; Gersh, B.J.; Somers, V.K. Obstructive Sleep Apnea Implications for Cardiac and Vascular Disease. JAMA 2003, 290, 1906–1914. [Google Scholar] [CrossRef]
- Marin, J.M.; Carrizo, S.J.; Vicente, E.; Agusti, A.G.N. Long-term cardiovascular outcomes in men with obstructive sleep apnoea-hypopnoea with or without treatment with continuous positive airway pressure: An observational study. Lancet 2005, 365, 1046–1053. [Google Scholar] [CrossRef] [PubMed]
- Berry, R.B.; Brooks, R.; Gamaldo, C.E.; Harding, S.M.; Lloyd, R.M.; Marcus, C.L.; Vaughn, B.V. The AASM manual for the scoring of sleep and associated events. Rules Terminol. Tech. Specif. Darien Ill. Am. Acad. Sleep Med. 2012, 176, 2012. [Google Scholar]
- Senaratna, C.V.; Perret, J.L.; Lodge, C.J.; Lowe, A.J.; Campbell, B.E.; Matheson, M.C.; Hamilton, G.S.; Dharmage, S.C. Prevalence of obstructive sleep apnea in the general population: A systematic review. Sleep Med. Rev. 2017, 34, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ren, R.; Lei, F.; Zhou, J.; Zhang, J.; Wing, Y.K.; Sanford, L.D.; Tang, X. Worldwide and regional prevalence rates of co-occurrence of insomnia and insomnia symptoms with obstructive sleep apnea: A systematic review and meta-analysis. Sleep Med. Rev. 2019, 45, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.G. APAP, BPAP, CPAP, and New Modes of Positive Airway Pressure Therapy. In Advances in the Diagnosis and Treatment of Sleep Apnea; Springer: Cham, Switzerland, 2022. [Google Scholar]
- Heinzer, R.; Petitpierre, N.J.; Marti-Soler, H.; Haba-Rubio, J. Prevalence and characteristics of positional sleep apnea in the HypnoLaus population-based cohort. Sleep Med. 2018, 48, 157–162. [Google Scholar] [CrossRef]
- Jeon, Y.J.; Park, S.H.; Kang, S.J. Self-x based closed loop wearable IoT for real-time detection and resolution of sleep apnea. Internet Things 2023, 22, 100767. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Q.; Wang, Y.P.; Qiu, C. A Real-time Auto-Adjustable Smart Pillow System for Sleep Apnea Detection and Treatment. In Proceedings of the 2013 Acm/Ieee International Conference on Information Processing in Sensor Networks (Ipsn), Philadelphia, PA, USA, 8–11 April 2013; pp. 179–190. [Google Scholar]
- Pant, H.; Kumar Dhanda, H.; Taran, S. Sleep Apnea Detection Using Electrocardiogram Signal Input to FAWT and Optimize Ensemble Classifier. Measurement 2021, 189, 110485. [Google Scholar] [CrossRef]
- Nassi, T.E.; Ganglberger, W.; Sun, H.; Bucklin, A.A.; Biswal, S.; Van Putten, M.; Thomas, R.; Westover, B. Automated Scoring of Respiratory Events in Sleep with a Single Effort Belt and Deep Neural Networks. IEEE Trans. Biomed. Eng. 2021, 69, 2094–2104. [Google Scholar] [CrossRef]
- Chen, X.; Chen, Y.; Ma, W.; Fan, X.; Li, Y. Toward sleep apnea detection with lightweight multi-scaled fusion network. Knowl. -Based Syst. 2022, 247, 108783. [Google Scholar] [CrossRef]
- Waxman, J.A.; Graupe, D.; Carley, D.W. Automated prediction of apnea and hypopnea, using a LAMSTAR artificial neural network. Am. J. Resp. Crit. Care 2010, 181, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Taghizadegan, Y.; Jafarnia Dabanloo, N.; Maghooli, K.; Sheikhani, A. Obstructive sleep apnea event prediction using recurrence plots and convolutional neural networks (RP-CNNs) from polysomnographic signals. Biomed. Signal Process. Control 2021, 69, 102928. [Google Scholar] [CrossRef]
- Zhang, Z.; Conroy, T.B.; Krieger, A.C.; Kan, E.C. Detection and Prediction of Sleep Disorders by Covert Bed-Integrated RF Sensors. IEEE Trans. Biomed. Eng. 2022, 70, 1208–1218. [Google Scholar] [CrossRef]
- Dubatovka, A.; Buhmann, J.M. Automatic Detection of Atrial Fibrillation from Single-Lead ECG Using Deep Learning of the Cardiac Cycle. BME Front. 2022, 2022, 9813062. [Google Scholar] [CrossRef]
- Meng, L.; Tan, W.; Ma, J.; Wang, R.; Yin, X.; Zhang, Y. Enhancing dynamic ECG heartbeat classification with lightweight transformer model. Artif. Intell. Med. 2022, 124, 102236. [Google Scholar] [CrossRef]
- Wang, M.; Rahardja, S.; Fränti, P.; Rahardja, S. Single-lead ECG recordings modeling for end-to-end recognition of atrial fibrillation with dual-path RNN. Biomed. Signal Process. Control 2023, 79, 104067. [Google Scholar] [CrossRef]
- Faust, O.; Barika, R.; Shenfield, A.; Ciaccio, E.J.; Acharya, U.R. Accurate detection of sleep apnea with long short-term memory network based on RR interval signals. Knowl. Based Syst. 2020, 212, 106591. [Google Scholar] [CrossRef]
- Van Steenkiste, T.; Groenendaal, W.; Deschrijver, D.; Dhaene, T. Automated Sleep Apnea Detection in Raw Respiratory Signals Using Long Short-Term Memory Neural Networks. IEEE J. Biomed. Health Inform. 2019, 23, 2354–2364. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Yoon, H.; Kim, H.S.; Kim, H.B.; Kwon, H.B.; Oh, S.M.; Lee, Y.J.; Park, K.S. Real-time apnea-hypopnea event detection during sleep by convolutional neural networks. Comput. Biol. Med. 2018, 100, 123–131. [Google Scholar] [CrossRef]
- Leino, A.; Nikkonen, S.; Kainulainen, S.; Korkalainen, H.; Toyras, J.; Myllymaa, S.; Leppanen, T.; Yla-Herttuala, S.; Westeren-Punnonen, S.; Muraja-Murro, A.; et al. Neural network analysis of nocturnal SpO2 signal enables easy screening of sleep apnea in patients with acute cerebrovascular disease. Sleep Med. 2021, 79, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Cui, S.; Zhao, X.; Cong, F. Detection of obstructive sleep apnea from single-channel ECG signals using a CNN-transformer architecture. Biomed. Signal Process. Control 2023, 82, 104581. [Google Scholar] [CrossRef]
- Zarei, A.; Beheshti, H.; Asl, B.M. Detection of sleep apnea using deep neural networks and single-lead ECG signals. Biomed. Signal Process. Control 2022, 71, 103125. [Google Scholar] [CrossRef]
- Almutairi, H.; Hassan, G.M.; Datta, A. Classification of Obstructive Sleep Apnoea from single-lead ECG signals using convolutional neural and Long Short Term Memory networks. Biomed. Signal Process. Control 2021, 69, 102906. [Google Scholar] [CrossRef]
- Philips. Alice PDx. Available online: https://www.philips.co.in/healthcare/product/HC1043844/alice-pdx-portable-sleep-diagnostic-system (accessed on 6 June 2023).
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.; Kaiser, L.; Polosukhin, I. Attention is all you need. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. Adv. Neural Inf. Process. Syst. 2019, 32, 8024–8035. [Google Scholar]
- Kingma, D.P. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]



| Participants (#) (male) | 105 (87) |
| Age (years) | 51.0 ± 13.1 |
| BMI (kg/m2) | 28.7 ± 4.7 |
| AHI (events/h) | 21.9 ± 18.8 |
| Normal/mild/moderate/severe OSA cases (#) | 17/35/20/33 |
| Module | Layer | Output Size | Parameters |
|---|---|---|---|
| Feature Extraction | Convolutional block | 32 × 200 | Kernel size: 3, stride: 1, padding: 1 |
| Convolutional block | 32 × 200 | Kernel size: 29, stride: 1, padding: 14 | |
| Convolutional block | 64 × 200 | Kernel size: 29, stride: 1, padding: 14 | |
| Transformer Encoder | Transformer | 200 × 64 | d_model: 64, nhead: 8 dim_feedforward: 128 dropout: 0.3, num_layers: 2 |
| Prediction | Average Pooling | 64 | Kernel size: 200 |
| Dropout | 64 | p: 0.5 | |
| Linear | 1 |
| Fold 1 | Fold 2 | Fold 3 | Fold 4 | Fold 5 | Total | |
|---|---|---|---|---|---|---|
| Normal cases | 3 | 3 | 4 | 4 | 3 | 17 |
| Mild cases | 7 | 7 | 7 | 7 | 7 | 35 |
| Moderate cases | 4 | 4 | 4 | 4 | 4 | 20 |
| Severe cases | 7 | 7 | 6 | 6 | 7 | 33 |
| Total | 21 | 21 | 21 | 21 | 21 | 105 |
| Five-fold CV | LOO CV | |||||
|---|---|---|---|---|---|---|
| Accuracy | Sensitivity | F1 | Accuracy | Sensitivity | F1 | |
| CNN | 0.798 | 0.685 | 0.770 | 0.715 | 0.600 | 0.678 |
| Transformer | 0.803 | 0.812 | 0.805 | 0.718 | 0.698 | 0.711 |
| Proposed | 0.859 | 0.847 | 0.858 | 0.741 | 0.726 | 0.737 |
| Five-Fold CV | LOO CV | |||||
|---|---|---|---|---|---|---|
| Accuracy | Sensitivity | F1 | Accuracy | Sensitivity | F1 | |
| GRU | 0.734 | 0.692 | 0.724 | 0.569 | 0.583 | 0.575 |
| LSTM | 0.728 | 0.597 | 0.685 | 0.572 | 0.586 | 0.578 |
| BiLSTM | 0.809 | 0.746 | 0.797 | 0.622 | 0.600 | 0.614 |
| CNN-GRU | 0.801 | 0.714 | 0.783 | 0.723 | 0.605 | 0.686 |
| CNN-LSTM | 0.801 | 0.754 | 0.791 | 0.720 | 0.633 | 0.694 |
| CNN-BiLSTM | 0.824 | 0.734 | 0.805 | 0.719 | 0.652 | 0.699 |
| Proposed (95%CI 1) | 0.859 (0.856 0.860) | 0.847 (0.843 0.867) | 0.858 (0.856 0.859) | 0.741 (0.736 0.743) | 0.726 (0.718 0.737) | 0.737 (0.735 0.738) |
| Accuracy | Sensitivity | F1-Score | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Severe | Moderate | Mild | Severe | Moderate | Mild | Severe | Moderate | Mild | |
| CNN | 0.677 | 0.678 | 0.734 | 0.660 | 0.562 | 0.510 | 0.722 | 0.655 | 0.544 |
| Transformer | 0.707 | 0.708 | 0.723 | 0.727 | 0.692 | 0.646 | 0.763 | 0.719 | 0.598 |
| CNN-BiLSTM | 0.700 | 0.695 | 0.730 | 0.700 | 0.629 | 0.577 | 0.756 | 0.696 | 0.575 |
| Proposed | 0.738 | 0.716 | 0.743 | 0.761 | 0.694 | 0.692 | 0.794 | 0.730 | 0.636 |
| Study | No. of Subjects | Sensor Type | Method | Per-Segment | Per-Subject | ||
|---|---|---|---|---|---|---|---|
| Sen | Acc | Sen | Acc | ||||
| [15] | 16 | Respiratory belts | ShuffleNet | 0.803 | 0.808 | 0.766 | 0.767 |
| [16] | 27 | Radio-frequency sensors | Random Forest | 0.746 | 0.819 | 0.727 | 0.817 |
| Proposed | 105 | Piezoelectric sensors. | CNN–Transformer | 0.847 | 0.859 | 0.726 | 0.741 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Yang, S.; Li, H.; Wang, L.; Wang, B. Prediction of Sleep Apnea Events Using a CNN–Transformer Network and Contactless Breathing Vibration Signals. Bioengineering 2023, 10, 746. https://doi.org/10.3390/bioengineering10070746
Chen Y, Yang S, Li H, Wang L, Wang B. Prediction of Sleep Apnea Events Using a CNN–Transformer Network and Contactless Breathing Vibration Signals. Bioengineering. 2023; 10(7):746. https://doi.org/10.3390/bioengineering10070746
Chicago/Turabian StyleChen, Yuhang, Shuchen Yang, Huan Li, Lirong Wang, and Bidou Wang. 2023. "Prediction of Sleep Apnea Events Using a CNN–Transformer Network and Contactless Breathing Vibration Signals" Bioengineering 10, no. 7: 746. https://doi.org/10.3390/bioengineering10070746
APA StyleChen, Y., Yang, S., Li, H., Wang, L., & Wang, B. (2023). Prediction of Sleep Apnea Events Using a CNN–Transformer Network and Contactless Breathing Vibration Signals. Bioengineering, 10(7), 746. https://doi.org/10.3390/bioengineering10070746







