Human Patient-Derived Brain Tumor Models to Recapitulate Ependymoma Tumor Vasculature
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Brain Tumors
2.2. Brain Tumor Tissue Dissociation
2.3. 3D Silk Protein-Based Scaffolds and Extracellular Matrix (ECM) Gel Preparation
2.4. Cell Seeding in 2D and 3D
2.5. Endothelial Cell 3D Co-Culture
2.6. Tissue Viability Assay
2.7. Flow Cytometry Cell Counting
2.8. Immunofluorescence Staining and Imaging
2.9. RNA-Seq
2.10. Differential Gene Expression
2.11. Statistical Analysis
3. Results
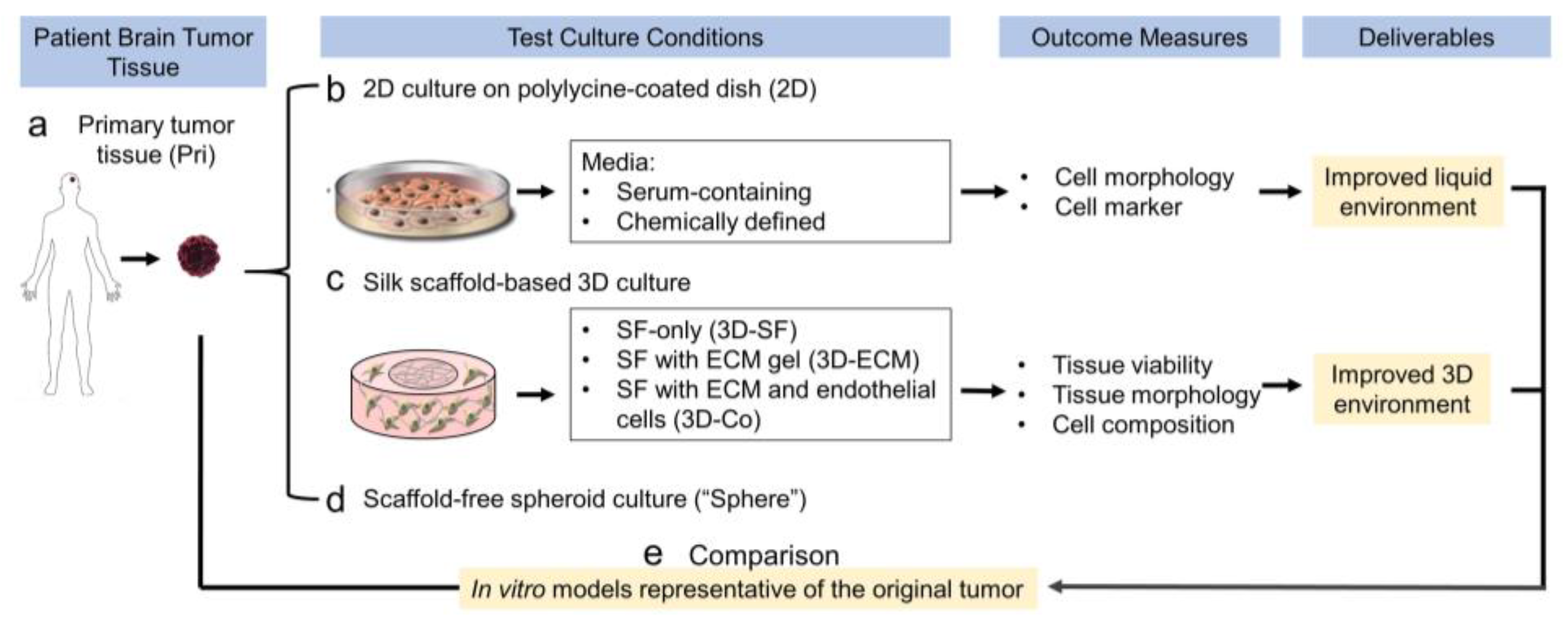
3.1. Study Design of the Optimization of Cell Culture Conditions for Intracranial Ependymoma
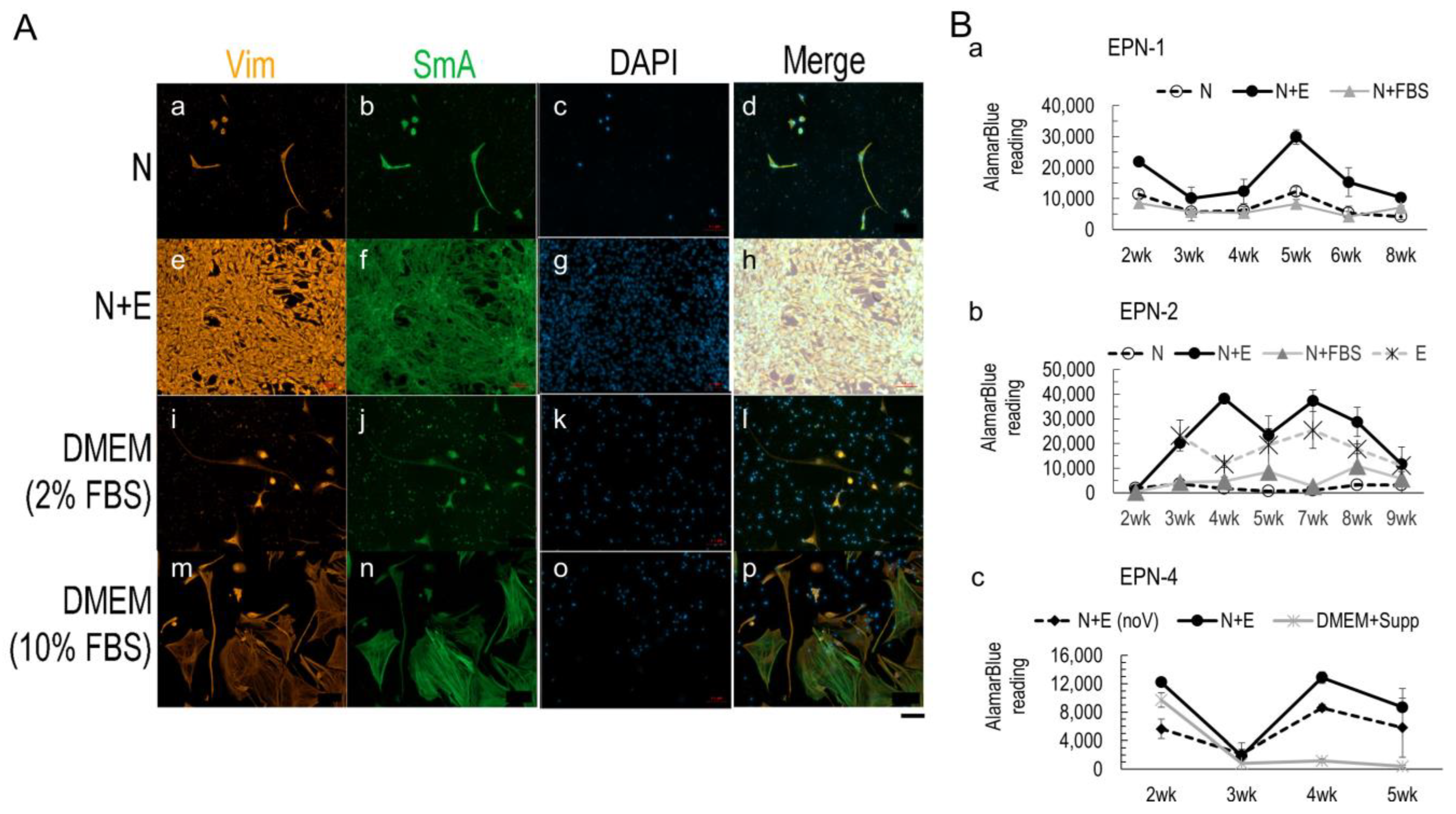
3.2. Media Conditions Affect Ependymoma Cell Growth and 3D Model Viability
3.3. Nestin+ Cell Enrichment in 2D Ependymoma Cultures by the Improved N+E Media
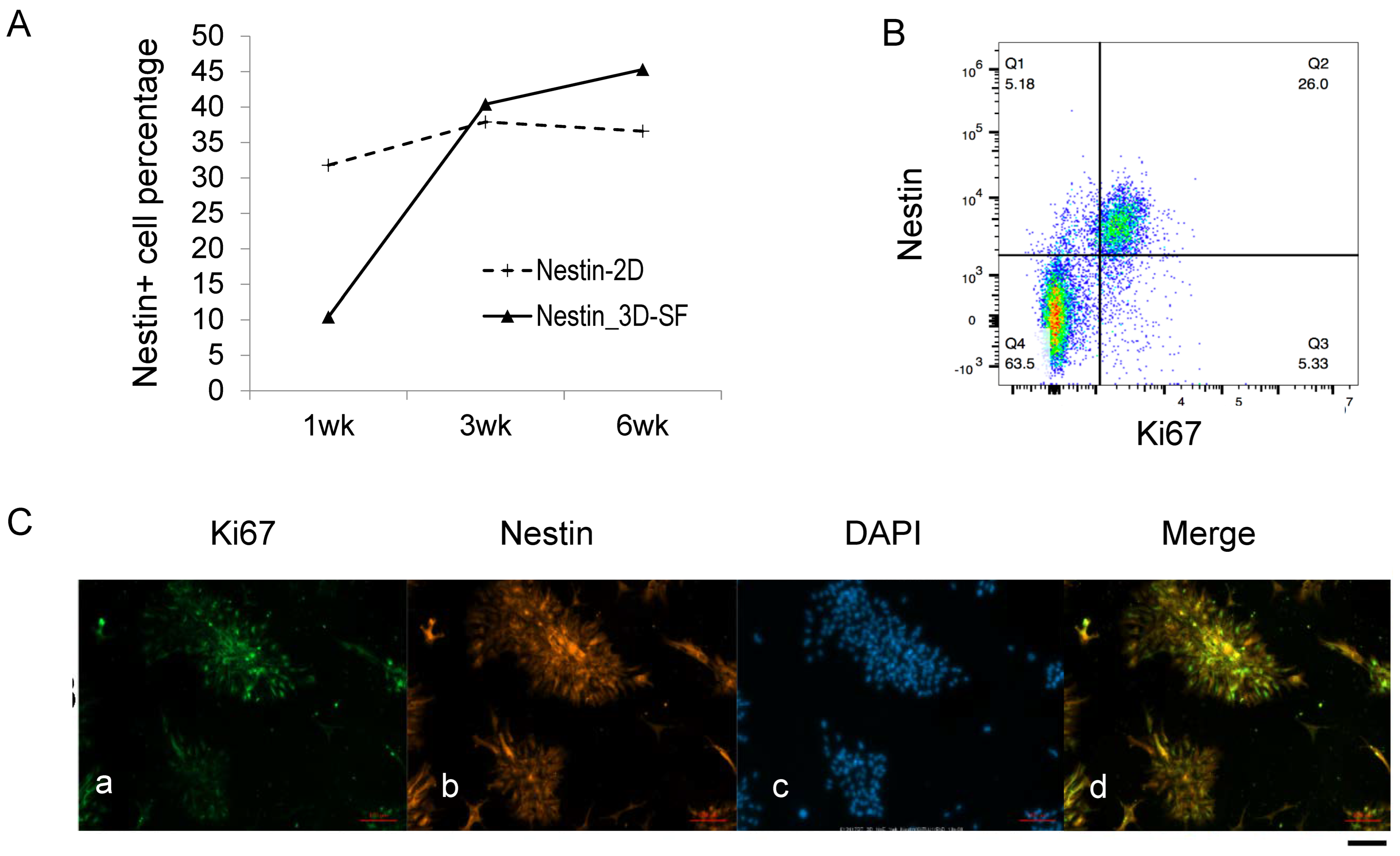
3.4. Nestin+ Cells Are the Predominant Proliferating Cells during Ependymoma In Vitro Growth
3.5. Nestin+/Ki67+ Cells form Tumor Stem Cell Niche during Ependymoma In Vitro Growth
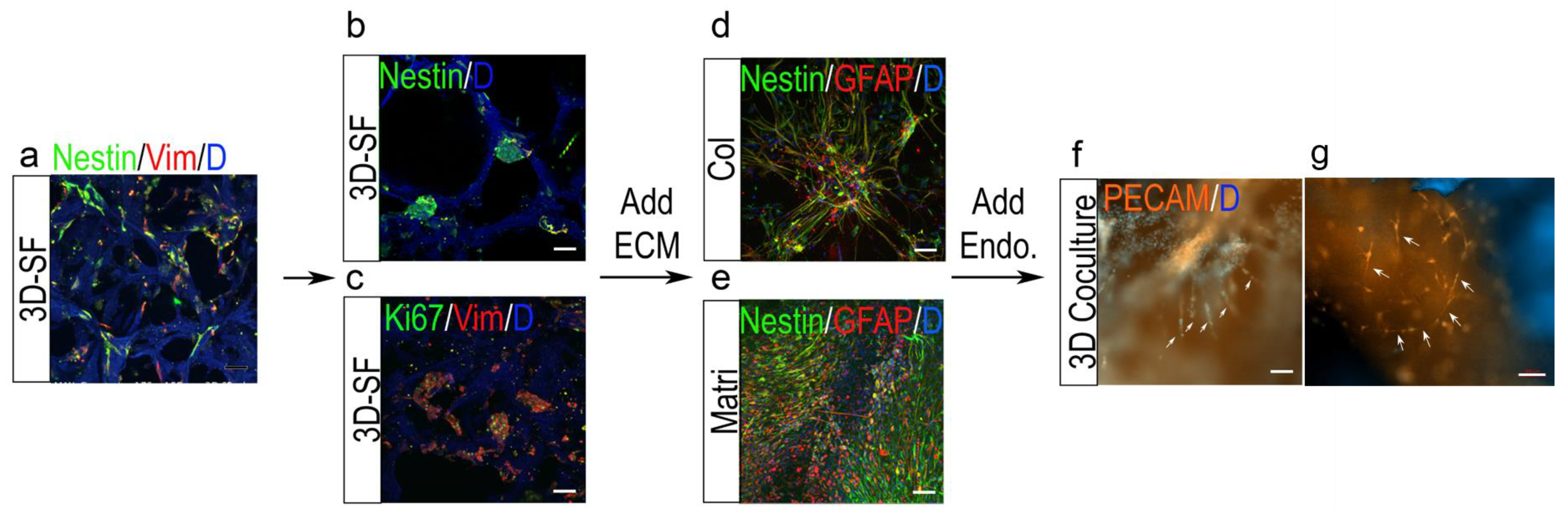
3.6. Nestin+ Cell Enrichment by ECM in 3D Ependymoma Models
3.7. 3D Tumor-Endothelial Co-Culture Model for Ependymoma
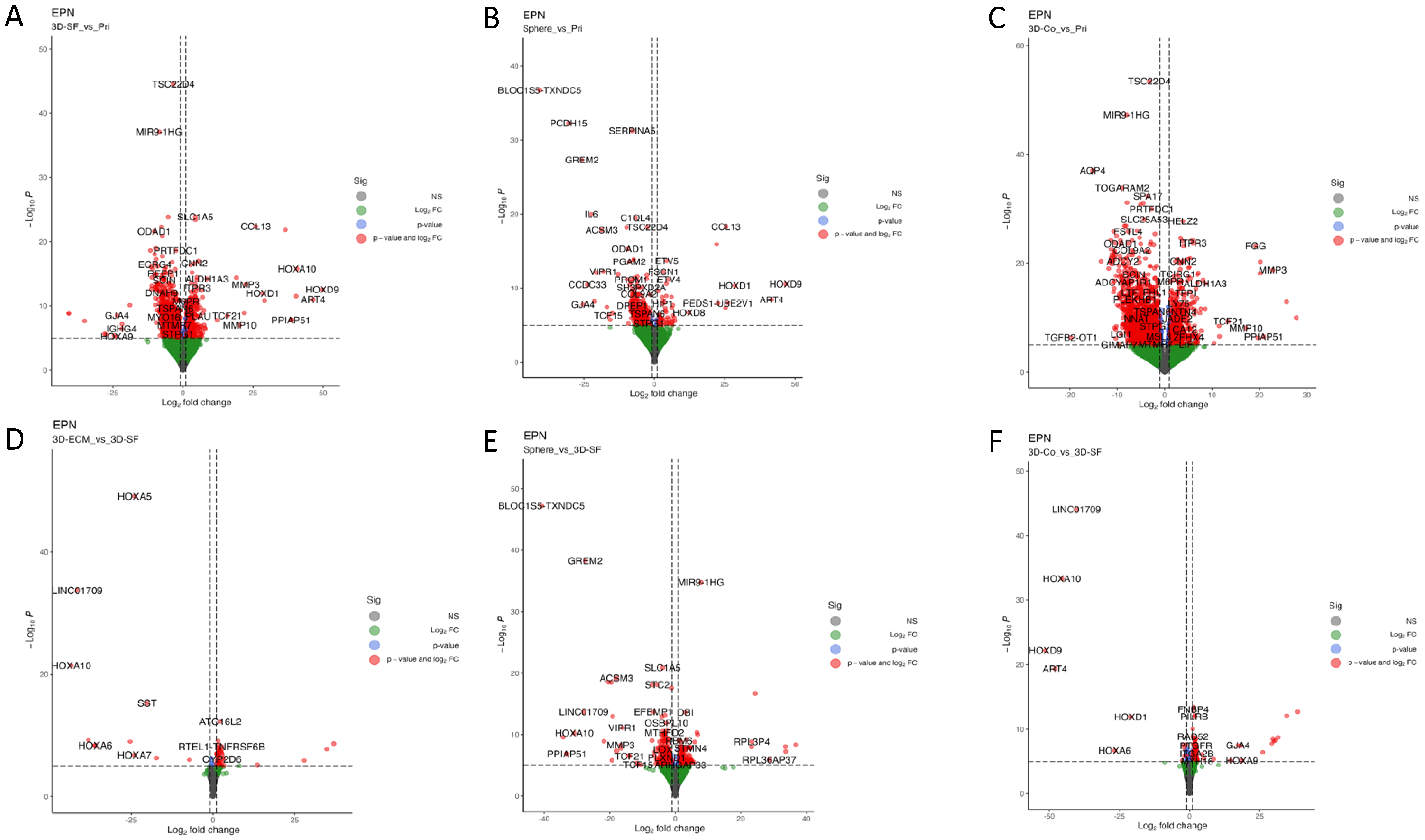
3.8. Transcriptomic Profiles of Ependymoma Cultures in Comparison with the Original Tumors
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Merchant, T.E.; Pollack, I.F.; Loeffler, J.S. Brain tumors across the age spectrum: Biology, therapy, and late effects. Semin. Radiat. Oncol. 2010, 20, 58–66. [Google Scholar] [CrossRef]
- Witt, H.; Mack, S.C.; Ryzhova, M.; Bender, S.; Sill, M.; Isserlin, R.; Benner, A.; Hielscher, T.; Milde, T.; Remke, M.; et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell 2011, 20, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Kilday, J.; Bouffet, E. Chemotherapy in Childhood Brain Tumors. Curr. Pediatr. Rep. 2014, 2, 49. [Google Scholar] [CrossRef][Green Version]
- Pajtler, K.W.; Witt, H.; Sill, M.; Jones, D.T.; Hovestadt, V.; Kratochwil, F.; Wani, K.; Tatevossian, R.; Punchihewa, C.; Johann, P.; et al. Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell 2015, 27, 728–743. [Google Scholar] [CrossRef]
- Amirian, E.S.; Armstrong, T.S.; Aldape, K.D.; Gilbert, M.R.; Scheurer, M.E. Predictors of survival among pediatric and adult ependymoma cases: A study using Surveillance, Epidemiology, and End Results data from 1973 to 2007. Neuroepidemiology 2012, 39, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Safaee, M.; Oh, M.C.; Kim, J.M.; Aranda, D.; Tarapore, P.E.; Cage, T.A.; Gupta, N.; Parsa, A.T. Histologic grade and extent of resection are associated with survival in pediatric spinal cord ependymomas. Childs Nerv. Syst. 2013, 29, 2057–2064. [Google Scholar] [CrossRef]
- Grill, J.; Pascal, C.; Chantal, K. Childhood ependymoma: A systematic review of treatment options and strategies. Paediatr. Drugs 2003, 5, 533–543. [Google Scholar] [CrossRef]
- Timmermann, B.; Kortmann, R.D.; Kühl, J.; Meisner, C.; Slavc, I.; Pietsch, T.; Bamberg, M. Combined postoperative irradiation and chemotherapy for anaplastic ependymomas in childhood: Results of the German prospective trials HIT 88/89 and HIT 91. Int. J. Radiat. Oncol. Biol. Phys. 2000, 46, 287–295. [Google Scholar] [CrossRef]
- Coradini, D.; Casarsa, C.; Oriana, S. Epithelial cell polarity and tumorigenesis: New perspectives for cancer detection and treatment. Acta Pharmacol. Sin. 2011, 32, 552–564. [Google Scholar]
- Lourdusamy, A.; Luo, L.Z.; Storer, L.C.; Cohen, K.J.; Resar, L.; Grundy, R.G. Transcriptomic analysis in pediatric spinal ependymoma reveals distinct molecular signatures. Oncotarget 2017, 8, 115570–115581. [Google Scholar] [CrossRef]
- Fukuoka, K.; Kanemura, Y.; Shofuda, T.; Fukushima, S.; Yamashita, S.; Narushima, D.; Kato, M.; Honda-Kitahara, M.; Ichikawa, H.; Kohno, T.; et al. Significance of molecular classification of ependymomas: C11orf95-RELA fusion-negative supratentorial ependymomas are a heterogeneous group of tumors. Acta Neuropathol. Commun. 2018, 6, 134. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.L.; Hsieh, T.H.; Liu, Y.R.; Chen, Y.W.; Lee, Y.Y.; Chang, F.C.; Lin, S.C.; Huang, M.C.; Ming-Tak, H.D.; Wong, T.T.; et al. Significance of cyclin D1 overexpression in progression and radio-resistance of pediatric ependymomas. Oncotarget 2017, 9, 2527–2542. [Google Scholar] [CrossRef] [PubMed]
- Wani, K.; Armstrong, T.S.; Vera-Bolanos, E.; Raghunathan, A.; Ellison, D.; Gilbertson, R.; Vaillant, B.; Goldman, S.; Packer, R.J.; Fouladi, M.; et al. A prognostic gene expression signature in infratentorial ependymoma. Acta Neuropathol. 2012, 123, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Dai, J.X.; Pan, Y.B.; Ma, Y.B.; Chu, S.H. Identification of biomarkers and construction of a microRNA-mRNA regulatory network for ependymoma using integrated bioinformatics analysis. Oncol. Lett. 2019, 18, 6079–6089. [Google Scholar] [CrossRef] [PubMed]
- Peyre, M.; Commo, F.; Dantas-Barbosa, C.; Andreiuolo, F.; Puget, S.; Lacroix, L.; Drusch, F.; Scott, V.; Varlet, P.; Mauguen, A.; et al. Portrait of ependymoma recurrence in children: Biomarkers of tumor progression identified by dual-color microarray-based gene expression analysis. PLoS ONE 2010, 5, e12932. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, J.M.; Shelat, A.A.; Carcaboso, A.M.; Kranenburg, T.A.; Arnold, L.A.; Boulos, N.; Wright, K.; Johnson, R.A.; Poppleton, H.; Mohankumar, K.M.; et al. An integrated in vitro and in vivo high-throughput screen identifies treatment leads for ependymoma. Cancer Cell 2011, 20, 384–399. [Google Scholar] [CrossRef]
- Parker, M.; Mohankumar, K.M.; Punchihewa, C.; Weinlich, R.; Dalton, J.D.; Li, Y.; Lee, R.; Tatevossian, R.G.; Phoenix, T.N.; Thiruvenkatam, R.; et al. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature 2014, 506, 451–455. [Google Scholar] [CrossRef]
- Mack, S.C.; Witt, H.; Piro, R.M.; Gu, L.; Zuyderduyn, S.; Stütz, A.M.; Wang, X.; Gallo, M.; Garzia, L.; Zayne, K.; et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 2014, 506, 445–450. [Google Scholar] [CrossRef]
- Amani, V.; Donson, A.M.; Lummus, S.C.; Prince, E.W.; Griesinger, A.M.; Witt, D.A.; Hankinson, T.C.; Handler, M.H.; Dorris, K.; Vibhakar, R.; et al. Characterization of 2 novel ependymoma cell lines with chromosome 1q gain derived from posterior fossa tumors of childhood. J. Neuropathol. Exp. Neurol. 2017, 76, 595–604. [Google Scholar] [CrossRef]
- Pierce, A.M.; Witt, D.A.; Donson, A.M.; Gilani, A.; Sanford, B.; Sill, M.; Van Court, B.; Oweida, A.; Prince, E.W.; Steiner, J.; et al. Establishment of patient-derived orthotopic xenograft model of 1q+ posterior fossa group A ependymoma. Neuro Oncol. 2019, 21, 1540–1551. [Google Scholar] [CrossRef]
- Huszthy, P.C.; Daphu, I.; Niclou, S.P.; Stieber, D.; Nigro, J.M.; Sakariassen, P.Ø.; Miletic, H.; Thorsen, F.; Bjerkvig, R. In vivo models of primary brain tumors: Pitfalls and perspectives. Neuro Oncol. 2012, 14, 979–993. [Google Scholar] [CrossRef] [PubMed]
- Whitehouse, J.P.; Hii, H.; Mayoh, C.; Wong, M.; Ajuyah, P.; Barahona, P.; Cui, L.; Dholaria, H.; White, C.L.; Buntine, M.K.; et al. In vivo loss of tumorigenicity in a patient-derived orthotopic xenograft mouse model of ependymoma. Front. Oncol. 2023, 13, 1123492. [Google Scholar] [CrossRef] [PubMed]
- Pertuiset, B.; Dougherty, D.; Cromeyer, C.; Hoshino, T.; Berger, M.; Rosenblum, M.L. Stem cell studies of human malignant brain tumors. part 2: Proliferation kinetics of brain-tumor cells in vitro in early-passage cultures. J. Neurosurg. 1985, 63, 426–432. [Google Scholar] [CrossRef]
- Engebraaten, O.; Bjerkvig, R.; Lund-Johansen, M.; Wester, K.; Pedersen, P.H.; Mork, S.; Backlund, E.O.; Laerum, O.D. Interaction between human brain tumour biopsies and fetal rat brain tissue in vitro. Acta Neuropathol. 1990, 81, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Jennings, M.T.; Kaariainen, I.T.; Gold, L.; Maciunas, R.J.; Commers, P.A. TGF beta 1 and TGF beta 2 are potential growth regulators for medulloblastomas, primitive neuroectodermal tumors, and ependymomas: Evidence in support of an autocrine hypothesis. Hum. Pathol. 1994, 25, 464–475. [Google Scholar] [CrossRef]
- Ishiwata, I.; Ishiwata, C.; Iguchi, M.; Soma, M.; Sato, Y.; Sonobe, M.; Kiguchi, K.; Tachibana, T.; Ishikawa, H. Biological characteristics of cultured cells derived from various types of human brain tumors. Hum. Cell 2004, 17, 117–124. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Hirakawa, K.; Ueda, K.; Okada, T.S. Establishment of a human ependymoma cell line. No Shinkei = Brain Nerve 1983, 35, 283–288. [Google Scholar]
- Hussain, D.; Punjaruk, W.; Storer, L.C.; Shaw, L.; Ottoman, R.; Peet, A.; Miller, S.; Bandopadhyay, G.; Heath, R.; Kumari, R.; et al. Pediatric brain tumor cancer stem cells: Cell cycle dynamics, DNA repair, and etoposide extrusion. Neuro Oncol. 2011, 13, 70–83. [Google Scholar] [CrossRef]
- Sanden, E.; Eberstal, S.; Visse, E.; Siesjo, P.; Darabi, A. A standardized and reproducible protocol for serum-free monolayer culturing of primary paediatric brain tumours to be utilized for therapeutic assays. Sci. Rep. 2015, 5, 12218. [Google Scholar] [CrossRef]
- Pavon, L.F.; Sibov, T.T.; de Toledo, S.R.C.; de Oliveira, D.M.; Cabral, F.R.; de Souza, J.G.; Boufleur, P.; Marti, L.C.; Malheiros, J.M.; da Cruz, E.F.; et al. Establishment of primary cell culture and an intracranial xenograft model of pediatric ependymoma: A prospect for therapy development and understanding of tumor biology. Oncotarget 2018, 9, 21731–21743. [Google Scholar] [CrossRef][Green Version]
- Yuan, M.; White, D.; Resar, L.; Bar, E.; Groves, M.; Cohen, A.; Jackson, E.; Bynum, J.; Rubens, J.; Mumm, J.; et al. Conditional reprogramming culture conditions facilitate growth of lower-grade glioma models. Neuro Oncol. 2021, 23, 770–782. [Google Scholar] [CrossRef] [PubMed]
- Yamada, A.; Tsuchida, T.; Kato, T.; Kawamoto, K. Ultrastructural analysis of brain tumors using collagen gel culture. Brain Tumor Pathol. 2002, 19, 11–14. [Google Scholar] [CrossRef] [PubMed]
- Brisson, C.; Lelong-Rebel, I.; Mottolese, C.; Jouvet, A.; Fevre-Montange, M.; Saint Pierre, G.; Rebel, G.; Belin, M.F. Establishment of human tumoral ependymal cell lines and coculture with tubular-like human endothelial cells. Int. J. Oncol. 2002, 21, 775–785. [Google Scholar] [CrossRef] [PubMed]
- Thirant, C.; Bessette, B.; Varlet, P.; Puget, S.; Cadusseau, J.; dos Reis Tavares, S.; Studler, J.M.; Silvestre, D.C.; Susini, A.; Villa, C.; et al. Clinical relevance of tumor cells with stem-like properties in pediatric brain tumors. PLoS ONE 2011, 6, e16375. [Google Scholar] [CrossRef]
- Sood, D.; Tang-Schomer, M.; Pouli, D.; Mizzoni, C.; Raia, N.; Tai, A.; Arkun, K.; Wu, J.; Black, L.D., III; Scheffler, B.; et al. 3D extracellular matrix microenvironment in bioengineered tissue models of primary pediatric and adult brain tumors. Nat. Commun. 2019, 10, 4529. [Google Scholar] [CrossRef]
- Johnson, R.A.; Wright, K.D.; Poppleton, H.; Mohankumar, K.M.; Finkelstein, D.; Pounds, S.B.; Rand, V.; Leary, S.E.; White, E.; Eden, C.; et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature 2010, 466, 632–636. [Google Scholar] [CrossRef]
- Yu, L.; Baxter, P.A.; Voicu, H.; Gurusiddappa, S.; Zhao, Y.; Adesina, A.; Man, T.K.; Shu, Q.; Zhang, Y.J.; Zhao, X.M.; et al. A clinically relevant orthotopic xenograft model of ependymoma that maintains the genomic signature of the primary tumor and preserves cancer stem cells in vivo. Neuro Oncol. 2010, 12, 580–594. [Google Scholar] [CrossRef]
- Tang-Schomer, M.D.; White, J.D.; Tien, L.W.; Schmitt, L.I.; Valentin, T.M.; Graziano, D.J.; Hopkins, A.M.; Omenetto, F.G.; Haydon, P.G.; Kaplan, D.L. Bioengineered functional brain-like cortical tissue. Proc. Natl. Acad. Sci. USA 2014, 111, 13811–13816. [Google Scholar] [CrossRef]
- Tang-Schomer, M.D. 3D in vitro brain tissue models. In Pre-Clinical and Clinical Methods in Brain Trauma Research, 1st ed.; Srivastava, A.K., Cox, C.S., Eds.; Humana Press: New York, NY, USA, 2018; pp. 15–36. [Google Scholar]
- Tang-Schomer, M.D.; Wu, W.B.; Kaplan, D.L.; Bookland, M.J. In Vitro 3D Regeneration-like Growth of Human Patient Brain Tissue. J. Tissue Eng. Regen Med. 2018, 12, 1247–1260. [Google Scholar] [CrossRef]
- Sood, D.; Chwalek, K.; Stuntz, E.; Pouli, D.; Du, C.; Tang-Schomer, M.; Georgakoudi, I.; Black, L.D., III; Kaplan, D.L. Fetal Brain Extracellular Matrix Boosts Neuronal Network Formation in 3D Bioengineered Model of Cortical Brain Tissue. ACS Biomater. Sci. Eng. 2016, 2, 131. [Google Scholar] [CrossRef]
- Tang-Schomer, M.D.; Chandok, H.; Wu, W.B.; Lau, C.C.; Bookland, M.J.; George, J. 3D patient-derived tumor models to recapitulate pediatric brain tumors In Vitro. Transl. Oncol. 2022, 20, 101407. [Google Scholar] [CrossRef] [PubMed]
- Wagemakers, M.; Sie, M.; Hoving, E.W.; Molema, G.; de Bont, E.S.; den Dunnen, W.F. Tumor vessel biology in pediatric intracranial ependymoma. J. Neurosurg. Pediatr. 2010, 5, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Charles, N.; Holland, E.C. The perivascular niche microenvironment in brain tumor progression. Cell Cycle 2010, 9, 3012–3021. [Google Scholar] [CrossRef]
- Preusser, M.; Wolfsberger, S.; Haberler, C.; Breitschopf, H.; Czech, T.; Slavc, I.; Harris, A.L.; Acker, T.; Budka, H.; Hainfellner, J.A. Vascularization and expression of hypoxia-related tissue factors in intracranial ependymoma and their impact on patient survival. Acta Neuropathol. 2005, 109, 211–216. [Google Scholar] [CrossRef]
- Nambirajan, A.; Sharma, M.C.; Gupta, R.K.; Suri, V.; Singh, M.; Sarkar, C. Study of stem cell marker nestin and its correlation with vascular endothelial growth factor and microvascular density in ependymomas. Neuropathol. Appl. Neurobiol. 2014, 40, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Kinsella, R.J.; Kähäri, A.; Haider, S.; Zamora, J.; Proctor, G.; Spudich, G.; Almeida-King, J.; Staines, D.; Derwent, P.; Kerhornou, A.; et al. Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database 2011, 2011, bar030. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Stephens, M. False discovery rates: A new deal. Biostatistics 2016, 18, 275–294. [Google Scholar] [CrossRef] [PubMed]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. 2018. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 22 June 2023).
- Yung, W.K.; Luna, M.; Borit, A. Vimentin and glial fibrillary acidic protein in human brain tumors. J. Neurooncol. 1985, 3, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Brewer, G.J.; Torricelli, J.R.; Evege, E.K.; Price, P.J. Optimized Survival of Hippocampal Neurons in B27-Supplemented Neurobasalm, a New Serum-free Medium Combination. J. Neurosci. Methods 1993, 35, 567–576. [Google Scholar]
- Paulus, W.; Grothe, C.; Sensenbrenner, M.; Janet, T.; Baur, I.; Graf, M.; Roggendorf, W. Localization of basic fibroblast growth factor, a mitogen and angiogenic factor, in human brain tumors. Acta Neuropathol. 1990, 79, 418–423. [Google Scholar] [CrossRef]
- Gilbertson, R.J.; Bentley, L.; Hernan, R.; Junttila, T.T.; Frank, A.J.; Haapasalo, H.; Connelly, M.; Wetmore, C.; Curran, T.; Elenius, K.; et al. ERBB receptor signaling promotes ependymoma cell proliferation and represents a potential novel therapeutic target for this disease. Clin. Cancer Res. 2002, 8, 3054–3064. [Google Scholar]
- Friedrich, C.; von Bueren, A.O.; Kolevatova, L.; Bernreuther, C.; Grob, T.; Sepulveda-Falla, D.; van den Boom, L.; Westphal, M.; Simon, R.; Glatzel, M. Epidermal growth factor receptor overexpression is common and not correlated to gene copy number in ependymoma. Childs Nerv. Syst. 2016, 32, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Lehtinen, B.; Raita, A.; Kesseli, J.; Annala, M.; Nordfors, K.; Yli-Harja, O.; Zhang, W.; Visakorpi, T.; Nykter, M.; Haapasalo, H.; et al. Clinical association analysis of ependymomas and pilocytic astrocytomas reveals elevated FGFR3 and FGFR1 expression in aggressive ependymomas. BMC Cancer 2017, 17, 310. [Google Scholar] [CrossRef]
- Taylor, M.D.; Poppleton, H.; Fuller, C.; Su, X.; Liu, Y.; Jensen, P.; Magdaleno, S.; Dalton, J.; Calabrese, C.; Board, J.; et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell 2005, 8, 323–335. [Google Scholar] [CrossRef]
- Lehman, N.L. Central nervous system tumors with ependymal features: A broadened spectrum of primarily ependymal differentiation? J. Neuropathol. Exp. Neurol. 2008, 67, 177–188. [Google Scholar] [CrossRef]
- Vege, K.D.; Giannini, C.; Scheithauer, B.W. The immunophenotype of ependymomas. Appl. Immunohistochem. Mol. Morphol. 2000, 8, 25–31. [Google Scholar] [CrossRef]
- Hemmati, H.D.; Nakano, I.; Lazareff, J.A.; Masterman-Smith, M.; Geschwind, D.H.; Bronner-Fraser, M.; Kornblum, H.I. Cancerous stem cells can arise from pediatric brain tumors. Proc. Natl. Acad. Sci. USA 2003, 100, 15178–15183. [Google Scholar] [CrossRef] [PubMed]




| Reference | Sample Size | Approach | Note | |
|---|---|---|---|---|
| 2D primary culture | Pertuiset et al., 1985 [23] | 2 | Tumor biopsies in 2D culture | The mean tumor cell doubling time was 46 h for two ependymomas. |
| Engebraaten et al., 1990 [24] | 1 | Tumor biopsies cut into fragments of 0.5 mm diameter and placed in agar overlay tissue culture | Co-culture with fetal rat brain tissue to examine tumor invasiveness | |
| Jennings et al., 1994 [25] | 2 | Tumor biopsies within passage 1–4 | TGF beta 1 and TGF beta 2 are potential growth regulators for ependymomas | |
| Ishiwata et al., 2004 [26] | 2 | Tumor biopsies in 2D culture | The ependymoma cells formed a rosette-like cell arrangement. Primary and long-term (>3 mon) cultures were established, but not cell lines. | |
| Cell line development | Nakagawa et al., 1983 [27] | 1 | 2D with Eagle’s MEM plus L-15 medium (6% fetal calf serum) | Grew like a sheet in groups. In soft agar, formed solid tumor differed from the original ependymoma. |
| Hussein et al., 2011 [28] | 2 | Monolayers were grown in tumor medium: DMEM/L-glutamine supplemented with 15% FBS. To generate neurospheres, cells grown as monolayers were washed, dissociated and resuspended into the serum-free stem cell medium: DMEM high glucose and Ham’s F-12 solution (70/30%), 2% B27, 5 ng/mL heparin, supplemented with 20 ng/mL human recombinant epidermal growth factor (hrEGF;), and 20 ng/mL human basic recombinant fibroblast growth factor (bFGF). | Two cell lines were established. Monolayers were passaged for >60 generations. Neurospheres were serially passaged for up to 11 generations. | |
| Sanden et al., 2015 [29] | 1 | UltraCULTURE™ cell culturing medium supplemented with 2 mM L-glutamine,1% Penicillin-Streptomycin, bFGF (40 ng/mL), and EGF (20 ng/mL) every 3–4 days. | Monolayers for >passage 5, but not 3D spheroids | |
| Amani et al., 2017 [19] | 2 | Cells were plated in Optimem media supplemented with 15% fetal bovine serum and cultured in either (1) ultra-low attachment plates to form nonadherent cultures or (2) using standard tissue culture treated plates to generate adherent monolayer cultures. | Two unique cell lines of intracranial, posterior fossa 1q+ ependymoma. The success in establishing these lines potentially stems the fact that the cells were from recurrent and collected from sites of intracranial metastasis rather than primary tumor. In contrast, the laboratory has attempted to establish cell lines from first occurrence ependymoma cases for almost 20 years (∼50 cases), none of which yielded a stable cell line. | |
| Pavon et al., 2018 [30] | 5 | The isolated cells were cultured in Dulbecco’s Modified Eagle’s Medium-Low Glucose (DMEM-LG) supplemented with 10% Fetal Bovine Serum and antibiotics. | Primary cell cultures were successfully obtained from five tumor samples. The success rate of isolating EPN cell cultures from all samples was around 70%. GFAP/CD133+CD90+/CD44+ ependymoma cells maintained key histopathological and growth characteristics of the original patient tumor. | |
| Yuan et al., 2021 [31] | 2 | Cells were co-cultured with irradiated 3T3 fibroblasts in the presence of Rho kinase (ROCK) inhibitor Y-27632. | Conditional reprogramming resulted in robust increases in growth for a majority of these tumors, with fibroblast conditioned media and ROCK inhibition both required. Cells were stable for up to 27 passages in terms of their appearance, with a doubling time of approximately 30 h after 8 passages | |
| 3D neurosphere | Yamada et al., 2002 [32] | 2 | The tumors were minced, and small fragments were prepared and embedded in the collagen gel | Ultrastructural observations. A basement membrane was formed surrounding the tumor cell processes facing the collagen gel in two ependymomas. |
| Brisson et al., 2002 [33] | 3D co-culture with endothelial cells in Matrigel | The morphological features (microvilli, cilia, and caveolae) of these cultured cells were similar to those of the tumor in vivo. | ||
| Thirant et al., 2011 [34] | 10 | Cells from fresh or cryo-frozen biopsies were cultured in NSA-H medium with 10 ng/mL FGF, 20 ng/mL EGF, and 1 mg/mL Heparin. The cells were further cultured until appearance of floating cellular spheres. | Limited self-renewal, stopped proliferating within 5 months followed by progressive disappearance. 3 cases no renewal. 3 cases renewal > 7 times, 4 cases < 7 renewal times. | |
| 3D scaffold model | Sood D., et al., 2019 [35] | 1 | 3D silk protein-based scaffold and collagen and Matrigel infusion | The 3D brain ECM-containing microenvironment supports distinctive phenotypes associated with tumor type-specific and ECM-dependent patterns in the tumor cells’ transcriptomic and release profiles. |
| Transgenic mouse model | Johnson et al., 2010 [36] | 1 | A mouse model by selecting neuronal stem cells with a deleted Ink4a/Arf locus that overexpress tyrosine receptor ephrin (EphB2) | Cross species genomics matches driver mutations and cell compartments to model ependymoma. |
| Patient-derived xenograft (PDX) or orthotopic xenograft (PDOX) | Yu et al., 2010 [37] | 1 | Transplanting a fresh surgical EPN tissue from a pediatric patient into the brain of immune deficient mice | A clinically relevant orthotopic xenograft model of ependymoma that maintains the genomic signature of the primary tumor and preserves cancer stem cells in vivo. |
| Pierce et al., 2019 [20] | 2 | Disaggregated tumors from 2 1q+ PFA patients were injected into the flanks of NSG mice. | Establishment of patient-derived orthotopic xenograft model of 1q+ posterior fossa group A ependymoma | |
| Whitehouse et al., 2023 [22] | 1 | The establishment of a patient-derived orthotopic xenograft (PDOX) model of posterior fossa A (PFA) EPN, derived from a metastatic cranial lesion. | Despite the aggressive nature of the tumor in the patient, this PDOX was unable to be maintained past two passages in vivo |
| Case ID | Grade | Age | Sex | Location |
|---|---|---|---|---|
| EPN-1 | II | 15 months | Male | Posterior fossa |
| EPN-2 | II | 11 months | Male | Posterior fossa |
| EPN-3 | III | 2 years | Female | Posterior fossa |
| EPN-4 | III | 7 months | Male | Posterior fossa |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang-Schomer, M.D.; Bookland, M.J.; Sargent, J.E.; N. Jackvony, T. Human Patient-Derived Brain Tumor Models to Recapitulate Ependymoma Tumor Vasculature. Bioengineering 2023, 10, 840. https://doi.org/10.3390/bioengineering10070840
Tang-Schomer MD, Bookland MJ, Sargent JE, N. Jackvony T. Human Patient-Derived Brain Tumor Models to Recapitulate Ependymoma Tumor Vasculature. Bioengineering. 2023; 10(7):840. https://doi.org/10.3390/bioengineering10070840
Chicago/Turabian StyleTang-Schomer, Min D., Markus J. Bookland, Jack E. Sargent, and Taylor N. Jackvony. 2023. "Human Patient-Derived Brain Tumor Models to Recapitulate Ependymoma Tumor Vasculature" Bioengineering 10, no. 7: 840. https://doi.org/10.3390/bioengineering10070840
APA StyleTang-Schomer, M. D., Bookland, M. J., Sargent, J. E., & N. Jackvony, T. (2023). Human Patient-Derived Brain Tumor Models to Recapitulate Ependymoma Tumor Vasculature. Bioengineering, 10(7), 840. https://doi.org/10.3390/bioengineering10070840











