Multimodal Classification Framework Based on Hypergraph Latent Relation for End-Stage Renal Disease Associated with Mild Cognitive Impairment
Abstract
1. Introduction
2. Data and Methods
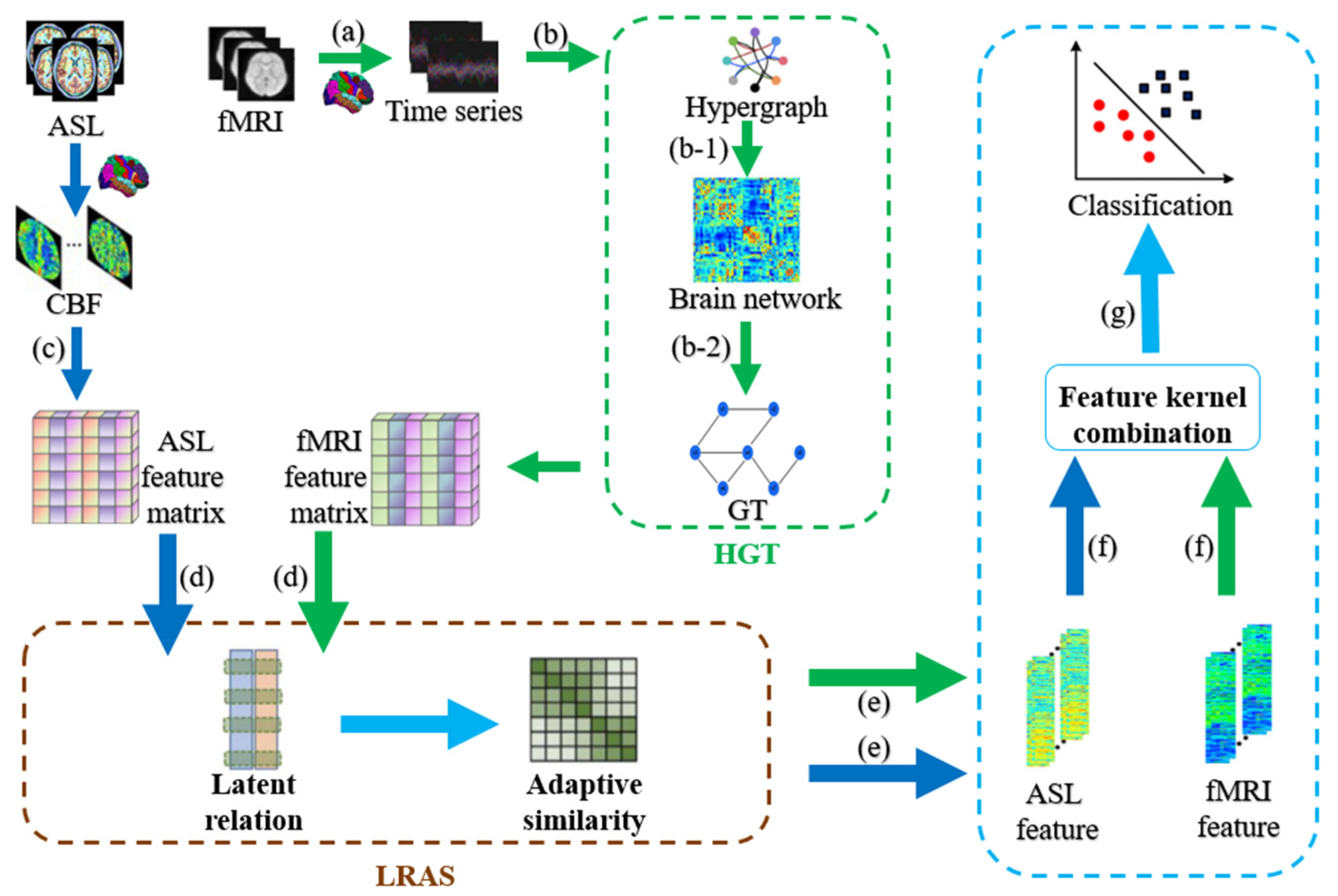
2.1. HLR-Based Multimodal Classification Framework
2.2. Data Preprocessing
2.3. Hypergraph Feature Matrix
2.4. Adaptive Similarity Learning
2.5. Multimodal Feature Selection Based on Adaptive Similarity Learning
2.6. Multimodal Feature Selection Based on Latent Relation
- (a)
- Update :
- (b)
- Update :
- (c)
- Update :
- (d)
- Update W:
- (e)
- Update S:
| Algorithm 1 Objective function optimization algorithm |
| Input: //The feature matrix of the m-th modality; |
| //The label corresponding to the m-th modality subjects; |
| K//The adaptive similarity neighbors; |
| //The group sparsity regularization parameter; |
| β//The regularization parameter for adaptive similarity learning. |
| Output: //The weight matrix of features. |
| Initialize S//Constructed by Equation (4); |
| While not converges Fix other variables Update U by Equation (7) with the constraint Then Fix other variables Compute SVD of Q Update V by Equation (10) Then Fix other variables Compute P Update E by Equation (11) Then Fix other variables Define D Calculated derivative Update W by Equation (13) Then Fix other variables KKT conditions Update S by Equation (15) End while |
3. Experiment and Analysis
3.1. Parameters Selection
3.2. Contrast Experiment
3.3. Discriminative Brain Regions
3.4. Data Visualization and Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Deyn, P.P.; Saxena, V.K.; Abts, H.; Borggreve, F.; D’Hooge, R.; Marescau, B.; Crols, R. Clinical and pathophysiological aspects of neurological complications in renal failure. Acta Neurol. Belg. 1992, 92, 191–206. [Google Scholar]
- Saji, N.; Sato, T.; Sakuta, K.; Aoki, J.; Kobayashi, K.; Matsumoto, N.; Uemura, J.; Shibazaki, K.; Kimura, K. Chronic Kidney Disease Is an Independent Predictor of Adverse Clinical Outcomes in Patients with Recent Small Subcortical Infarcts. Cerebrovasc. Dis. Extra 2014, 4, 174–181. [Google Scholar] [CrossRef]
- Karunaratne, K.; Taube, D.; Khalil, N.; Perry, R.; A Malhotra, P. Neurological complications of renal dialysis and transplantation. Pract. Neurol. 2018, 18, 115–125. [Google Scholar] [CrossRef]
- Raphael, K.L.; Wei, G.; Greene, T.; Baird, B.C.; Beddhu, S. Cognitive Function and the Risk of Death in Chronic Kidney Disease. Am. J. Nephrol. 2012, 35, 49–57. [Google Scholar] [CrossRef]
- Zhu, W.; Sun, L.; Huang, J.; Han, L.; Zhang, D. Dual Attention Multi-Instance Deep Learning for Alzheimer’s Disease Diagnosis with Structural MRI. IEEE Trans. Med. Imaging 2021, 40, 2354–2366. [Google Scholar] [CrossRef]
- Xi, Z.; Liu, T.; Shi, H.; Jiao, Z. Hypergraph representation of multimodal brain networks for patients with end-stage renal disease associated with mild cognitive impairment. Math. Biosci. Eng. 2023, 20, 1882–1902. [Google Scholar] [CrossRef]
- Jiao, Z.; Chen, S.; Shi, H.; Xu, J. Multi-Modal Feature Selection with Feature Correlation and Feature Structure Fusion for MCI and AD Classification. Brain Sci. 2022, 12, 80. [Google Scholar] [CrossRef]
- Li, Y.; Liu, J.; Gao, X.; Jie, B.; Kim, M.; Yap, P.-T.; Wee, C.-Y.; Shen, D. Multimodal hyper-connectivity of functional networks using functionally-weighted LASSO for MCI classification. Med. Image Anal. 2019, 52, 80–96. [Google Scholar] [CrossRef]
- Jann, K.; Gee, D.G.; Kilroy, E.; Schwab, S.; Smith, R.X.; Cannon, T.D.; Wang, D.J. Functional connectivity in BOLD and CBF data: Similarity and reliability of resting brain networks. Neuroimage 2015, 106, 111–122. [Google Scholar] [CrossRef]
- Liu, G.; Lin, Z.; Yan, S.; Sun, J.; Yu, Y.; Ma, Y. Robust Recovery of Subspace Structures by Low-Rank Representation. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 35, 171–184. [Google Scholar] [CrossRef]
- Rehman, M.U.; Akhtar, S.; Zakwan, M.; Mahmood, M.H. Novel architecture with selected feature vector for effective classification of mitotic and non-mitotic cells in breast cancer histology images. Biomed. Signal Process. Control 2022, 71, 103212. [Google Scholar] [CrossRef]
- Bommert, A.; Sun, X.; Bischl, B.; Rahnenführer, J.; Lang, M. Benchmark for filter methods for feature selection in high-dimensional classification data. Comput. Stat. Data Anal. 2020, 143, 106839. [Google Scholar] [CrossRef]
- Lei, B.Y.; Cheng, N.N.; Alejandro, F.F.; Tan, E.L.; Cao, J.W.; Yang, P.; Ahmed, E.; Du, J.; Xu, Y.W.; Wang, T.F. Self-calibrated brain network estimation and joint non-convex multi-task learning for identification of early Alzheimer’s disease. Med. Image Anal. 2020, 61, 101652. [Google Scholar] [CrossRef]
- Hao, X.; Bao, Y.; Guo, Y.; Yu, M.; Zhang, D.; Risacher, S.L.; Saykin, A.J.; Yao, X.; Shen, L. Multimodal neuroimaging feature selection with consistent metric constraint for diagnosis of Alzheimer’s disease. Med. Image Anal. 2020, 60, 101625. [Google Scholar] [CrossRef]
- Jie, B.; Zhang, D.Q.; Cheng, B.; Shen, D.G. Manifold regularized multi-task feature selection for multimodal classification in Alzheimer’s disease. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Nagoya, Japan, 22–26 September 2013; pp. 275–283. [Google Scholar]
- Shao, W.; Peng, Y.; Zu, C.; Wang, M.; Zhang, D. Hypergraph based multi-task feature selection for multimodal classification of Alzheimer’s disease. Comput. Med. Imaging Graph. 2020, 80, 101663. [Google Scholar] [CrossRef]
- Shi, Y.; Zu, C.; Hong, M.; Zhou, L.; Wang, L.; Wu, X.; Wang, Y. ASMFS: Adaptive-similarity-based multimodal feature selection for classification of Alzheimer’s disease. Pattern Recognit. 2022, 126, 108566. [Google Scholar] [CrossRef]
- Song, C.; Liu, T.; Wang, H.; Shi, H.; Jiao, Z. Multi-modal feature selection with self-expression topological manifold for end-stage renal disease associated with mild cognitive impairment. Math. Biosci. Eng. 2023, 20, 14827–14845. [Google Scholar] [CrossRef]
- Zhang, Y.; Sheng, Q.; Fu, X.; Shi, H.; Jiao, Z. Integrated Prediction Framework for Clinical Scores of Cognitive Functions in ESRD Patients. Comput. Intell. Neurosci. 2022, 2022, 8124053. [Google Scholar] [CrossRef]
- Xu, C.Y.; Chen, C.C.; Guo, Q.W.; Lin, Y.W.; Meng, X.Y. Comparison of moCA-B and MES scales for the recognition of amnestic mild cognitive impairment. J. Alzheimer’s Dis. Relat. Disord. 2021, 4, 33–36. [Google Scholar] [CrossRef]
- Rubinov, M.; Sporns, O. Complex network measures of brain connectivity: Uses and interpretations. Neuroimage 2010, 52, 1059–1069. [Google Scholar] [CrossRef]
- Nathalie, T.M.; Brigitte, L.; Dimitri, P.; Fabrice, C.; Olivier, E.; Nicolas, D.; Bernard, M.; Marc, J. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002, 15, 273–289. [Google Scholar] [CrossRef]
- Wang, Z.; Aguirre, G.K.; Rao, H.; Wang, J.; Fernández-Seara, M.A.; Childress, A.R.; Detre, J.A. Empirical optimization of ASL data analysis using an ASL data processing toolbox: ASLtbx. Magn. Reson. Imaging 2008, 26, 261–269. [Google Scholar] [CrossRef]
- Ji, Y.; Zhang, Y.; Shi, H.; Jiao, Z.; Wang, S.-H.; Wang, C. Constructing Dynamic Brain Functional Networks via Hyper-Graph Manifold Regularization for Mild Cognitive Impairment Classification. Front. Neurosci. 2021, 15, 669345. [Google Scholar] [CrossRef]
- Sheng, Q.; Zhang, Y.; Shi, H.; Jiao, Z. Global iterative optimization framework for predicting cognitive function statuses of patients with end-stage renal disease. Int. J. Imaging Syst. Technol. 2023, 33, 837–852. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, J.; Wu, Q.; Kuang, W.; Huang, X.; He, Y.; Gong, Q. Disrupted Brain Connectivity Networks in Drug-Naive, First-Episode Major Depressive Disorder. Biol. Psychiatry 2011, 70, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Tsang, I.W.; Xu, Z.; Lv, J. Latent Representation Guided Multi-view Clustering. IEEE Trans. Knowl. Data Eng. 2022. [Google Scholar] [CrossRef]
- Ding, C.; Li, T.; Peng, W.; Park, H. Orthogonal nonnegative matrix t-factorizations for clustering. In Proceedings of the 12th ACM SIGKDD international conference on Knowledge discovery and data mining, Philadelphia, PA, USA, 20–23 August 2006; pp. 126–135. [Google Scholar] [CrossRef]
- Bazaraa, M.S.; Sherali, H.D.; Shetty, C.M. Nonlinear Programming: Theory and Algorithms; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Wall, M.E.; Rechtsteiner, A.; Rocha, L.M. Singular Value Decomposition and Principal Component Analysis. In A Practical Approach to Microarray Data Analysis; Springer: Boston, MA, USA, 2003; pp. 91–109. [Google Scholar] [CrossRef]
- Nie, F.; Huang, H.; Cai, X.; Ding, C. Efficient and robust feature selection via joint -norms minimization. Adv. Neural Inf. Process. Syst. 2021, 2, 1813–1821. [Google Scholar]
- Kabgani, A.; Soleimani-Damaneh, M.; Zamani, M. Optimality conditions in optimization problems with convex feasible set using convexificators. Math. Methods Oper. Res. 2017, 86, 103–121. [Google Scholar] [CrossRef]
- Bach, F.R.; Lanckriet, G.R.; Jordan, M.I. Multiple kernel learning, conic duality, and the SMO algorithm. In Proceedings of the Twenty-First International Conference on Machine Learning, Banff, AB, Canada, 4–8 July 2004. [Google Scholar] [CrossRef]
- Xu, X.; Li, W.; Mei, J.; Tao, M.; Wang, X.; Zhao, Q.; Liang, X.; Wu, W.; Ding, D.; Wang, P. Feature Selection and Combination of Information in the Functional Brain Connectome for Discrimination of Mild Cognitive Impairment and Analyses of Altered Brain Patterns. Front. Aging Neurosci. 2020, 12, 28. [Google Scholar] [CrossRef]
- Huang, S.; Li, J.; Ye, J.; Wu, T.; Chen, K. Fleisher and E. Reiman. Identifying Alzheimer’s disease-related brain regions from multimodal neuroimaging data using sparse composite linear discrimination analysis. Adv. Neural Inf. Process. Syst. 2011, 24, 1431–1439. [Google Scholar]
- Zhang, Y.; Wang, S.; Sui, Y.; Yang, M.; Liu, B.; Cheng, H.; Sun, J.; Jia, W.; Phillips, P.; Gorriz, J.M. Multivariate Approach for Alzheimer’s Disease Detection Using Stationary Wavelet Entropy and Predator-Prey Particle Swarm Optimization. J. Alzheimer’s Dis. 2018, 65, 855–869. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, S.; Dong, Z. Classification of alzheimer disease based on structural magnetic resonance imaging by kernel support vector machine decision tree. Prog. Electromagn. Res. 2014, 144, 171–184. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, S. Detection of Alzheimer’s disease by displacement field and machine learning. PeerJ 2015, 3, e1251. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, Y.; Liu, G.; Phillips, P.; Yuan, T.-F. Detection of Alzheimer’s Disease by Three-Dimensional Displacement Field Estimation in Structural Magnetic Resonance Imaging. J. Alzheimer’s Dis. 2016, 50, 233–248. [Google Scholar] [CrossRef]
- Wu, B.L.; Yue, Z.; Li, X.K.; Li, L.; Zhang, M.; Ren, J.P.; Liu, W.L.; Han, D.M. Brain functional network changes in patients with end-stage renal disease and its correlation with cognitive function. Chin. J. Neuromedicine 2020, 19, 181–187. [Google Scholar] [CrossRef]
- McKhann, G.M.; Knopman, D.S.; Chertkow, H.; Hyman, B.T.; Jack, C.R., Jr.; Kawas, C.H.; Klunk, W.E.; Koroshetz, W.J.; Manly, J.J.; Mayeux, R.; et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. 2011, 7, 263–269. [Google Scholar] [CrossRef]
- Wang, S.-H.; Zhang, Y.; Li, Y.-J.; Jia, W.-J.; Liu, F.-Y.; Yang, M.-M.; Zhang, Y.-D. Single slice based detection for Alzheimer’s disease via wavelet entropy and multilayer perceptron trained by biogeography-based optimization. Multimedia Tools Appl. 2018, 77, 10393–10417. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, S.; Phillips, P.; Dong, Z.; Ji, G.; Yang, J. Detection of Alzheimer’s disease and mild cognitive impairment based on structural volumetric MR images using 3D-DWT and WTA-KSVM trained by PSOTVAC. Biomed. Signal Process. Control 2015, 21, 58–73. [Google Scholar] [CrossRef]
- Jiao, Z.; Ji, Y.; Gao, P.; Wang, S.-H. Extraction and analysis of brain functional statuses for early mild cognitive impairment using variational auto-encoder. J. Ambient. Intell. Humaniz. Comput. 2023, 14, 5439–5450. [Google Scholar] [CrossRef]
- Huang, J.; Zhou, L.; Wang, L.; Zhang, D. Attention-Diffusion-Bilinear Neural Network for Brain Network Analysis. IEEE Trans. Med. Imaging 2020, 39, 2541–2552. [Google Scholar] [CrossRef]
- Xi, Z.; Song, C.; Zheng, J.; Shi, H.; Jiao, Z. Brain Functional Networks with Dynamic Hypergraph Manifold Regularization for Classification of End-Stage Renal Disease Associated with Mild Cognitive Impairment. Comput. Model. Eng. Sci. 2023, 135, 2243–2266. [Google Scholar] [CrossRef]
- Zhao, P.; Wu, H.; Huang, S. Multi-View Graph Clustering by Adaptive Manifold Learning. Mathematics 2022, 10, 1821. [Google Scholar] [CrossRef]
- Ryu, J.; Rehman, M.U.; Nizami, I.F.; Chong, K.T. SegR-Net: A deep learning framework with multi-scale feature fusion for robust retinal vessel segmentation. Comput. Biol. Med. 2023, 163, 107132. [Google Scholar] [CrossRef]
- Mangj, S.M.; Hussan, P.H.; Shakir, W.M.R. Efficient Deep Learning Approach for Detection of Brain Tumor Disease. Int. J. Online Biomed. Eng. 2023, 19, 66–80. [Google Scholar] [CrossRef]
- Rehman, M.U.; Ryu, J.; Nizami, I.F.; Chong, K.T. RAAGR2-Net: A brain tumor segmentation network using parallel processing of multiple spatial frames. Comput. Biol. Med. 2023, 152, 106426. [Google Scholar] [CrossRef]
- Attallah, O.; Zaghlool, S. AI-Based Pipeline for Classifying Pediatric Medulloblastoma Using Histopathological and Textural Images. Life 2022, 12, 232. [Google Scholar] [CrossRef]




| Gender (Male/Female) | Age | Education Years | MoCA Scores | |
|---|---|---|---|---|
| ESRD group | 24/20 | 49.25 ± 11.15 | 9.58 ± 2.72 | 23.87 ± 4.51 |
| NC group | 13/31 | 46.25 ± 11.39 | 9.65 ± 2.59 | 26.63 ± 3.93 |
| Method | ACC (%) | AUC (%) | SPE (%) | SEN (%) |
|---|---|---|---|---|
| fMRI | 61.04 ± 0.14 | 59.76 ± 0.18 | 56.75 ± 0.23 | 57.35 ± 0.21 |
| ASL | 63.18 ± 0.15 | 67.84 ± 0.18 | 51.35 ± 0.23 | 75.00 ± 0.22 |
| MKSVM [33] | 73.93 ± 0.13 | 63.75 ± 0.24 | 62.07 ± 0.36 | 82.65 ± 0.14 |
| Lasso-MKSVM [35] | 76.17 ± 0.12 | 76.60 ± 0.17 | 67.95 ± 0.23 | 84.55 ± 0.16 |
| M2TFS [15] | 67.90 ± 0.11 | 56.15 ± 0.28 | 59.78 ± 0.35 | 85.40 ± 0.18 |
| HMTFS [16] | 81.14 ± 0.16 | 81.63 ± 0.18 | 77.00 ± 0.22 | 79.00 ± 0.18 |
| ASMFS [17] | 85.08 ± 0.16 | 82.28 ± 0.21 | 78.00 ± 0.28 | 88.50 ± 0.12 |
| SETMFS [18] | 85.83 ± 0.10 | 83.47 ± 0.24 | 86.31 ± 0.19 | 84.97 ± 0.23 |
| OLR | 84.42 ± 0.09 | 80.55 ± 0.13 | 83.50 ± 0.17 | 90.00 ± 0.17 |
| HLR | 88.67 ± 0.08 | 86.20 ± 0.16 | 93.50 ± 0.10 | 86.00 ± 0.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, X.; Song, C.; Zhang, R.; Shi, H.; Jiao, Z. Multimodal Classification Framework Based on Hypergraph Latent Relation for End-Stage Renal Disease Associated with Mild Cognitive Impairment. Bioengineering 2023, 10, 958. https://doi.org/10.3390/bioengineering10080958
Fu X, Song C, Zhang R, Shi H, Jiao Z. Multimodal Classification Framework Based on Hypergraph Latent Relation for End-Stage Renal Disease Associated with Mild Cognitive Impairment. Bioengineering. 2023; 10(8):958. https://doi.org/10.3390/bioengineering10080958
Chicago/Turabian StyleFu, Xidong, Chaofan Song, Rupu Zhang, Haifeng Shi, and Zhuqing Jiao. 2023. "Multimodal Classification Framework Based on Hypergraph Latent Relation for End-Stage Renal Disease Associated with Mild Cognitive Impairment" Bioengineering 10, no. 8: 958. https://doi.org/10.3390/bioengineering10080958
APA StyleFu, X., Song, C., Zhang, R., Shi, H., & Jiao, Z. (2023). Multimodal Classification Framework Based on Hypergraph Latent Relation for End-Stage Renal Disease Associated with Mild Cognitive Impairment. Bioengineering, 10(8), 958. https://doi.org/10.3390/bioengineering10080958






