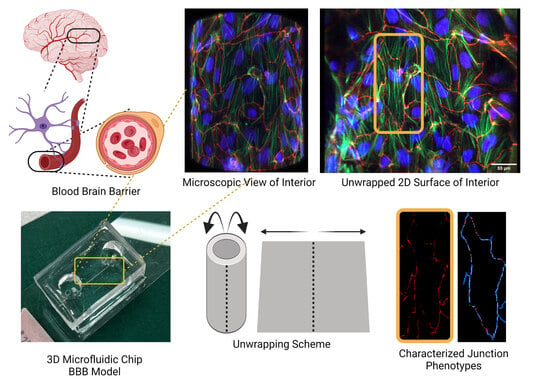
A Rapid-Patterning 3D Vessel-on-Chip for Imaging and Quantitatively Analyzing Cell–Cell Junction Phenotypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
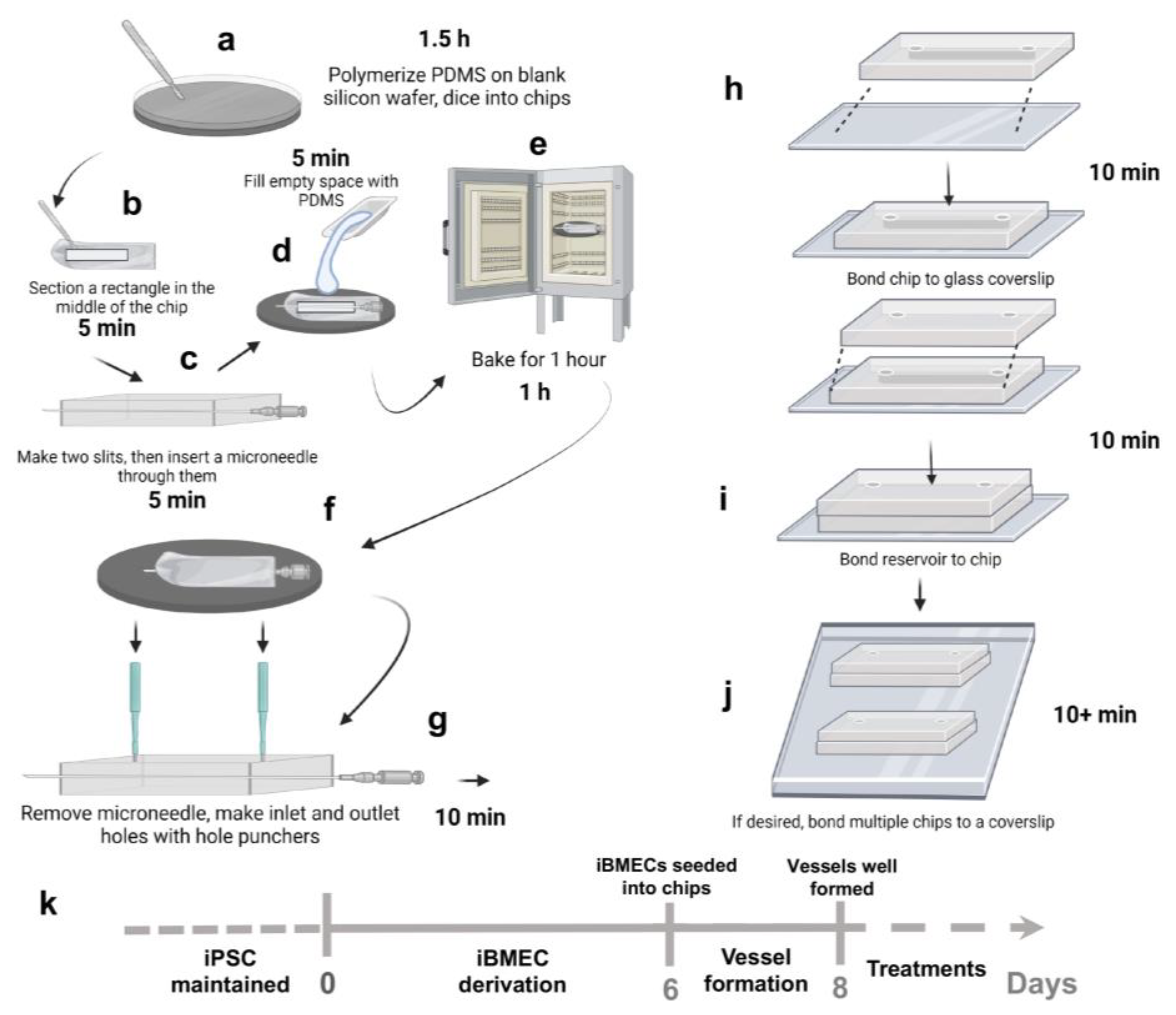
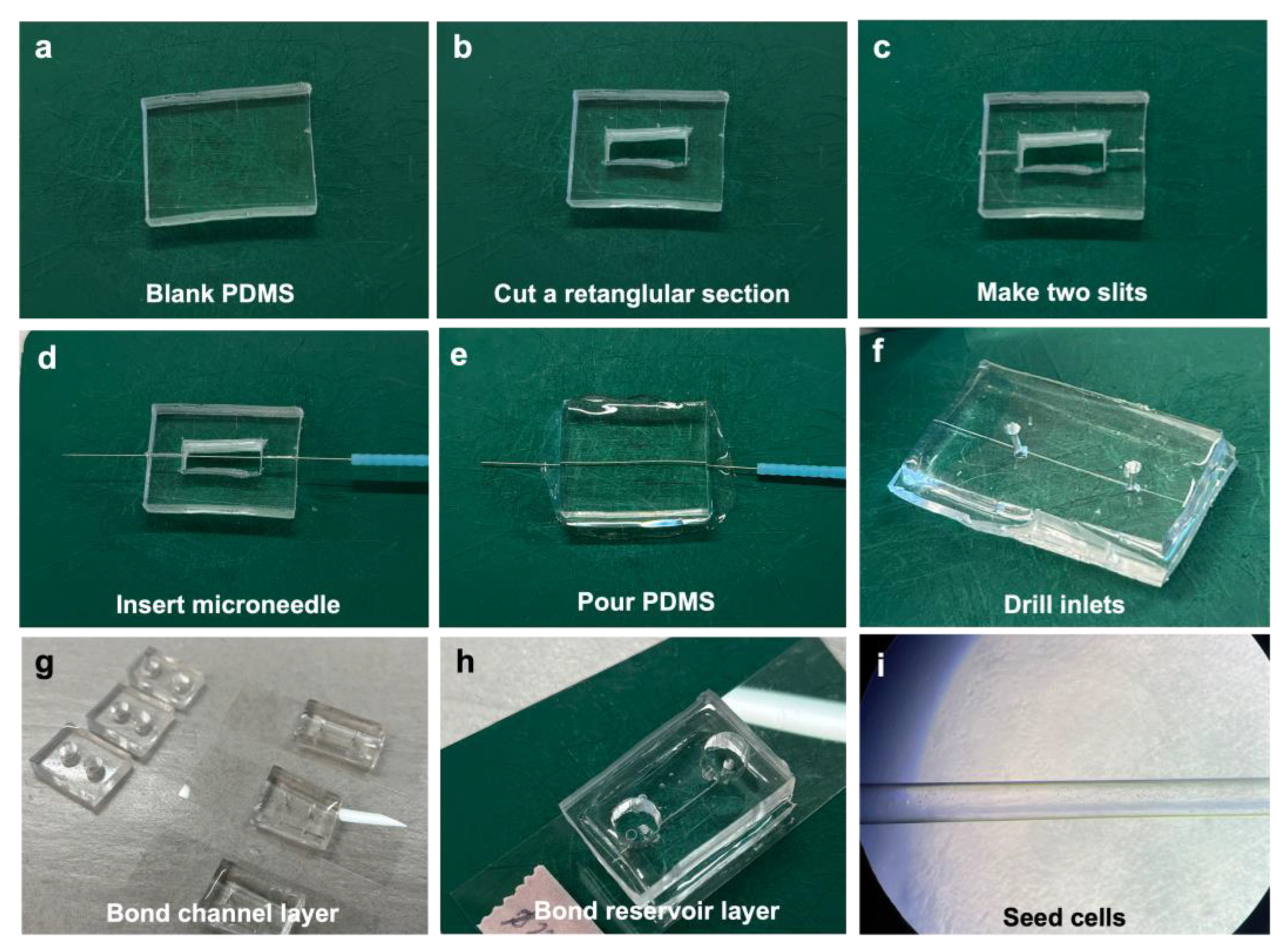
2.2. Vessel-on-Chip Fabrication
2.3. Cell Seeding in Vessel-on-Chips
2.4. Cell Seeding on ECM-Coated PDMS Plates
2.5. TNF-α Treatment in 3D Vessels
2.6. Immunostaining
2.7. Confocal Microscopy
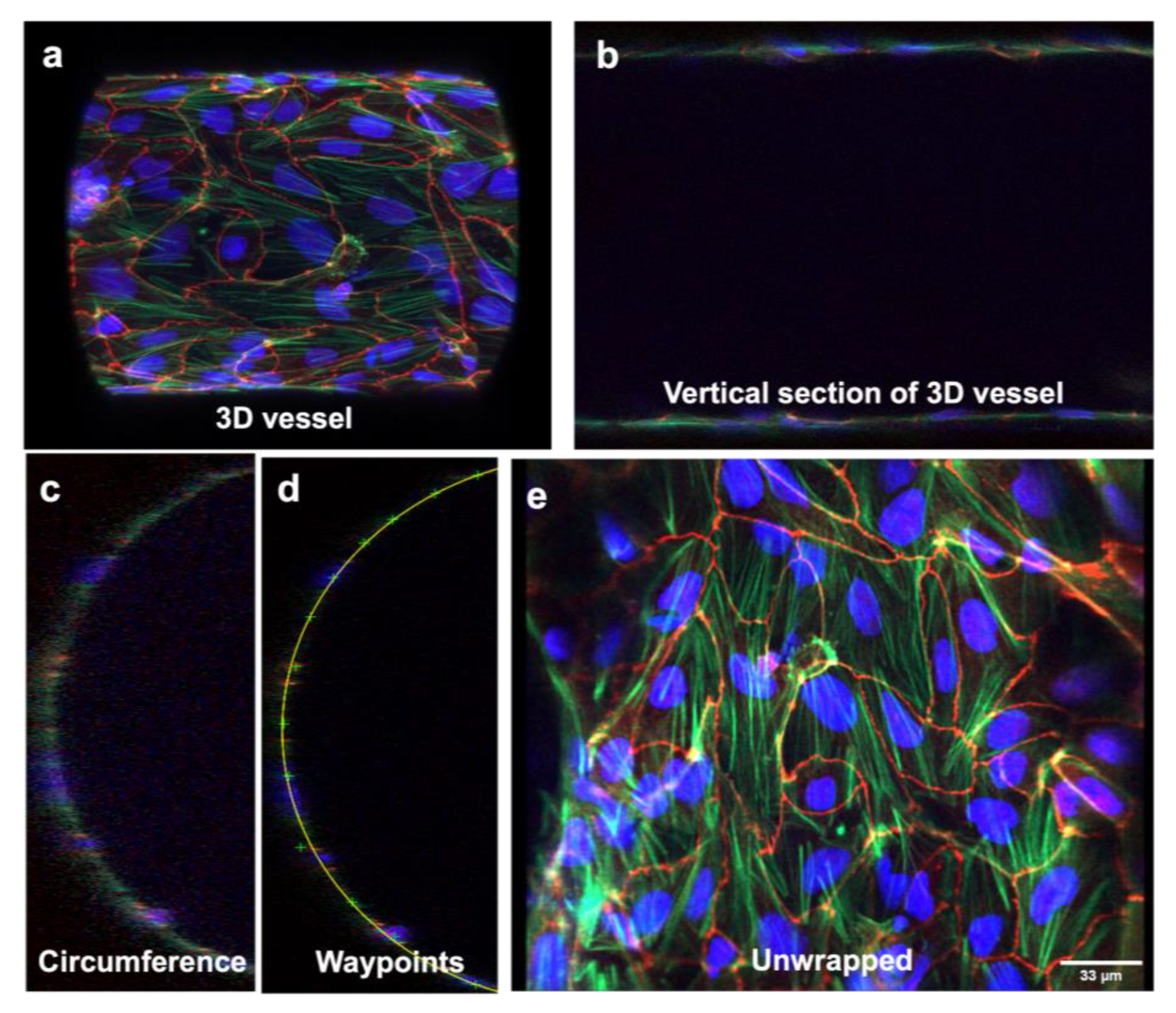
2.8. Microchannel Unwrapping
2.9. JAnaP Analysis
2.10. Statistical Analysis
3. Results
3.1. Fabrication of Chips in Approximately 3 h
3.2. Three-Dimensional Vessel-on-Chip Formation and Imaging
3.3. Three-Dimensional Image Stacks Were Converted to 2D Images for Junction Analysis
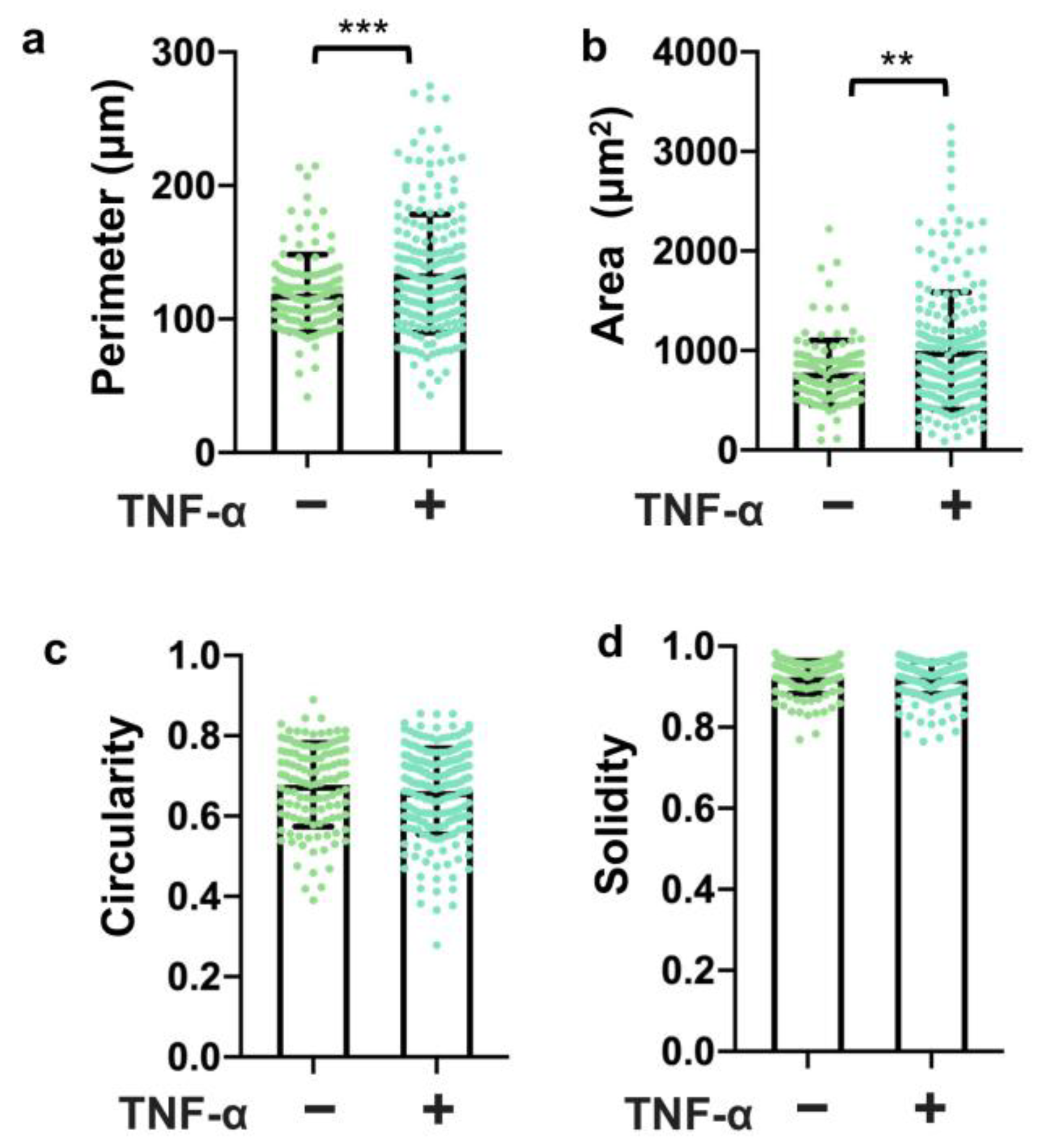
3.4. Three-Dimensional Structure Altered the Expression and Presentation of the Tight Junction Proteins
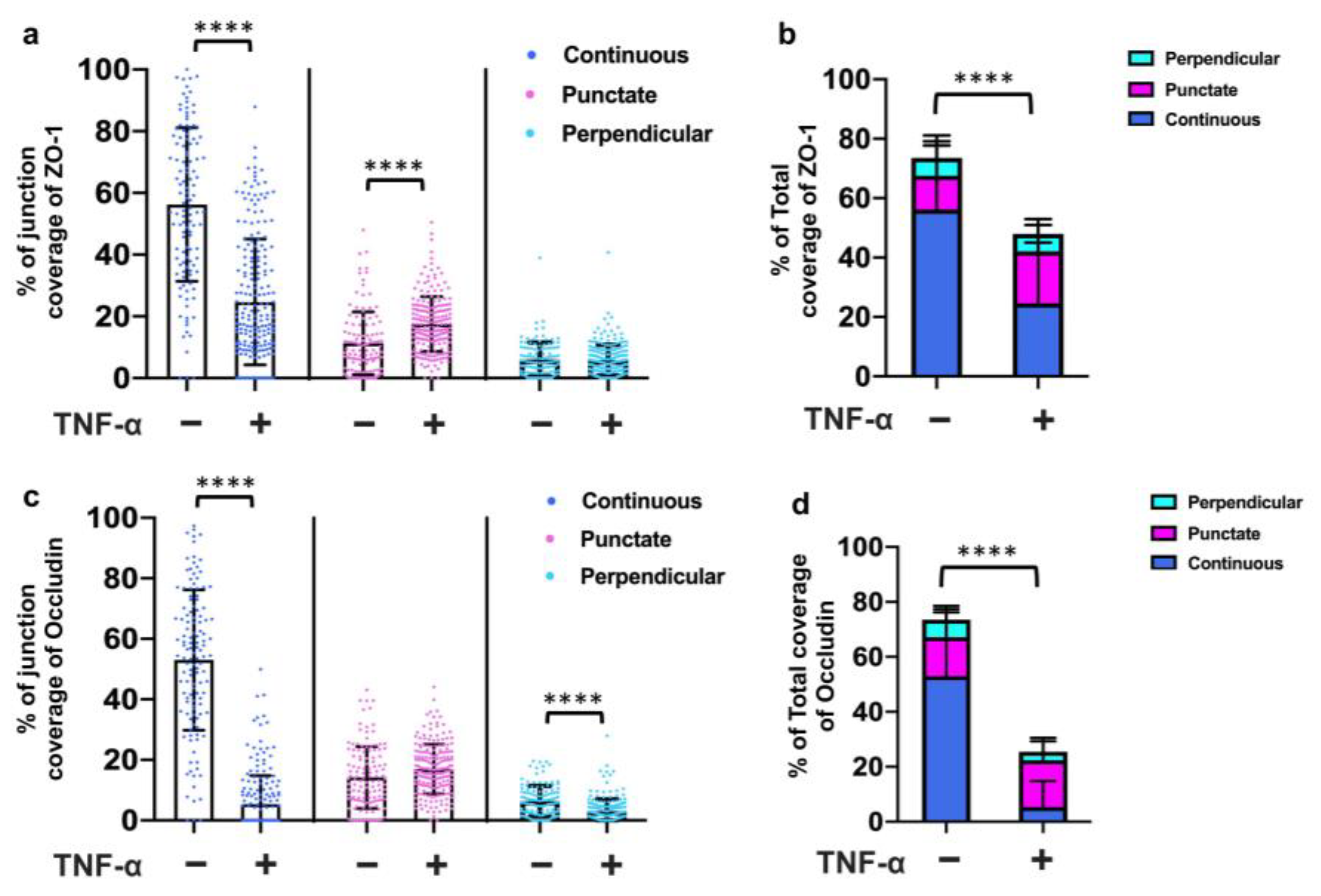
3.5. TNF-α Disrupted the Tight Junction Presentation in 3D Vessels
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hajal, C.; Le Roi, B.; Kamm, R.D.; Maoz, B.M. Biology and Models of the Blood–Brain Barrier. Annu. Rev. Biomed. Eng. 2021, 23, 359–384. [Google Scholar] [CrossRef] [PubMed]
- Wallez, Y.; Huber, P. Endothelial adherens and tight junctions in vascular homeostasis, inflammation and angiogenesis. Biochim. Biophys. Acta-Biomembr. 2008, 1778, 794–809. [Google Scholar] [CrossRef] [PubMed]
- Zenaro, E.; Piacentino, G.; Constantin, G. The blood-brain barrier in Alzheimer’s disease. Neurobiol. Dis. 2017, 107, 41–56. [Google Scholar] [CrossRef]
- Profaci, C.P.; Munji, R.N.; Pulido, R.S.; Daneman, R. The blood–brain barrier in health and disease: Important unanswered questions. J. Exp. Med. 2020, 217, e20190062. [Google Scholar] [CrossRef]
- Workman, M.J.; Svendsen, C.N. Recent advances in human iPSC-derived models of the blood–brain barrier. Fluids Barriers CNS 2020, 17, 30. [Google Scholar] [CrossRef]
- Yan, L.; Moriarty, R.A.; Stroka, K.M. Recent progress and new challenges in modeling of human pluripotent stem cell-derived blood-brain barrier. Theranostics 2021, 11, 10148–10170. [Google Scholar] [CrossRef]
- Bracken, M.B. Why animal studies are often poor predictors of human reactions to exposure. J. R. Soc. Med. 2009, 102, 120–122. [Google Scholar] [CrossRef] [PubMed]
- Sloan, C.D.K.; Nandi, P.; Linz, T.H.; Aldrich, J.V.; Audus, K.L.; Lunte, S.M. Analytical and Biological Methods for Probing the Blood-Brain Barrier. Annu. Rev. Anal. Chem. 2012, 5, 505–531. [Google Scholar] [CrossRef]
- Neal, E.H.; Marinelli, N.A.; Shi, Y.; McClatchey, P.M.; Balotin, K.M.; Gullett, D.R.; Hagerla, K.A.; Bowman, A.B.; Ess, K.C.; Wikswo, J.P.; et al. A Simplified, Fully Defined Differentiation Scheme for Producing Blood-Brain Barrier Endothelial Cells from Human iPSCs. Stem Cell Rep. 2019, 12, 1380–1388. [Google Scholar] [CrossRef]
- Hollmann, E.K.; Bailey, A.K.; Potharazu, A.V.; Neely, M.D.; Bowman, A.B.; Lippmann, E.S. Accelerated differentiation of human induced pluripotent stem cells to blood–brain barrier endothelial cells. Fluids Barriers CNS 2017, 14, 9. [Google Scholar] [CrossRef]
- Bosworth, A.M.; Faley, S.L.; Bellan, L.M.; Lippmann, E.S. Modeling Neurovascular Disorders and Therapeutic Outcomes with Human-Induced Pluripotent Stem Cells. Front. Bioeng. Biotechnol. 2017, 5, 87. [Google Scholar] [CrossRef] [PubMed]
- Lippmann, E.S.; Al-Ahmad, A.; Azarin, S.M.; Palecek, S.P.; Shusta, E.V. A retinoic acid-enhanced, multicellular human blood-brain barrier model derived from stem cell sources. Sci. Rep. 2014, 4, 4160. [Google Scholar] [CrossRef] [PubMed]
- Canfield, S.G.; Stebbins, M.J.; Faubion, M.G.; Gastfriend, B.D.; Palecek, S.P.; Shusta, E.V. An isogenic neurovascular unit model comprised of human induced pluripotent stem cell-derived brain microvascular endothelial cells, pericytes, astrocytes, and neurons. Fluids Barriers CNS 2019, 16, 25. [Google Scholar] [CrossRef]
- Canfield, S.G.; Stebbins, M.J.; Morales, B.S.; Asai, S.W.; Vatine, G.D.; Svendsen, C.N.; Palecek, S.P.; Shusta, E.V. An isogenic blood-brain barrier model comprising brain endothelial cells, astrocytes, and neurons derived from human induced pluripotent stem cells. J. Neurochem. 2017, 140, 874–888. [Google Scholar] [CrossRef] [PubMed]
- Polacheck, W.J.; Kutys, M.L.; Tefft, J.B.; Chen, C.S. Microfabricated blood vessels for modeling the vascular transport barrier. Nat. Protoc. 2019, 14, 1425–1454. [Google Scholar] [CrossRef]
- Gray, K.M.; Stroka, K.M. Vascular endothelial cell mechanosensing: New insights gained from biomimetic microfluidic models. Semin. Cell Dev. Biol. 2017, 71, 106–117. [Google Scholar] [CrossRef]
- DeStefano, J.G.; Xu, Z.S.; Williams, A.J.; Yimam, N.; Searson, P.C. Effect of shear stress on iPSC-derived human brain microvascular endothelial cells (dhBMECs). Fluids Barriers CNS 2017, 14, 20. [Google Scholar] [CrossRef]
- Reinitz, A.; DeStefano, J.; Ye, M.; Wong, A.D.; Searson, P.C. Human brain microvascular endothelial cells resist elongation due to shear stress. Microvasc. Res. 2015, 99, 8–18. [Google Scholar] [CrossRef]
- Ye, M.; Sanchez, H.M.; Hultz, M.; Yang, Z.; Bogorad, M.; Wong, A.D.; Searson, P.C. Brain microvascular endothelial cells resist elongation due to curvature and shear stress. Sci. Rep. 2014, 4, 4681. [Google Scholar] [CrossRef]
- Bosworth, A.M.; Kim, H.; O’grady, K.P.; Richter, I.; Lee, L.; O’grady, B.J.; Lippmann, E.S. Influence of Substrate Stiffness on Barrier Function in an iPSC-Derived In Vitro Blood-Brain Barrier Model. Cell. Mol. Bioeng. 2022, 15, 31–42. [Google Scholar] [CrossRef]
- Linville, R.M.; DeStefano, J.G.; Sklar, M.B.; Xu, Z.; Farrell, A.M.; Bogorad, M.I.; Chu, C.; Walczak, P.; Cheng, L.; Mahairaki, V.; et al. Human iPSC-derived blood-brain barrier microvessels: Validation of barrier function and endothelial cell behavior. Biomaterials 2019, 190–191, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Saunders, N.R.; Dziegielewska, K.M.; Møllgård, K.; Habgood, M.D. Recent Developments in Understanding Barrier Mechanisms in the Developing Brain: Drugs and Drug Transporters in Pregnancy, Susceptibility or Protection in the Fetal Brain? Annu. Rev. Pharmacol. Toxicol. 2019, 59, 487–505. [Google Scholar] [CrossRef] [PubMed]
- Otani, T.; Furuse, M. Tight Junction Structure and Function Revisited: (Trends in Cell Biology 30, 805-817, 2020). Trends Cell Biol. 2020, 30, 1014. [Google Scholar] [CrossRef] [PubMed]
- Lochhead, J.J.; Yang, J.; Ronaldson, P.T.; Davis, T.P. Structure, Function, and Regulation of the Blood-Brain Barrier Tight Junction in Central Nervous System Disorders. Front. Physiol. 2020, 11, 914. [Google Scholar] [CrossRef]
- Gray, K.M.; Katz, D.B.; Brown, E.G.; Stroka, K.M. Quantitative Phenotyping of Cell–Cell Junctions to Evaluate ZO-1 Presentation in Brain Endothelial Cells. Ann. Biomed. Eng. 2019, 47, 1675–1687. [Google Scholar] [CrossRef]
- Pranda, M.A.; Gray, K.M.; DeCastro, A.J.L.; Dawson, G.M.; Jung, J.W.; Stroka, K.M. Tumor Cell Mechanosensing During Incorporation into the Brain Microvascular Endothelium. Cell. Mol. Bioeng. 2019, 12, 455–480. [Google Scholar] [CrossRef]
- Gray, K.M.; Jung, J.W.; Inglut, C.T.; Huang, H.-C.; Stroka, K.M. Quantitatively relating brain endothelial cell–cell junction phenotype to global and local barrier properties under varied culture conditions via the Junction Analyzer Program. Fluids Barriers CNS 2020, 17, 16. [Google Scholar] [CrossRef]
- Inglut, C.T.; Gray, K.M.; Vig, S.; Jung, J.W.; Stabile, J.; Zhang, Y.; Stroka, K.M.; Huang, H.C. Photodynamic Priming Modulates Endothelial Cell–Cell Junction Phenotype for Light-activated Remote Control of Drug Delivery. IEEE J. Sel. Top. Quantum Electron. 2021, 27, 7200311. [Google Scholar] [CrossRef]
- Gagliardi, T.B.; Goldstein, M.E.; Song, D.; Gray, K.M.; Jung, J.W.; Ignacio, M.A.; Stroka, K.M.; Duncan, G.A.; Scull, M.A. Rhinovirus C replication is associated with the endoplasmic reticulum and triggers cytopathic effects in an in vitro model of human airway epithelium. PLoS Pathog. 2022, 18, e1010159. [Google Scholar] [CrossRef]
- Yan, L.; Dwiggins, C.W.; Moriarty, R.A.; Jung, J.W.; Gupta, U.; Brandon, K.D.; Stroka, K.M. Matrix stiffness regulates the tight junction phenotypes and local barrier properties in tricellular regions in an iPSC-derived BBB model. Acta Biomater. 2023, 167, 109–120. [Google Scholar] [CrossRef]
- Katt, M.E.; Xu, Z.S.; Gerecht, S.; Searson, P.C. Human Brain Microvascular Endothelial Cells Derived from the BC1 iPS Cell Line Exhibit a Blood-Brain Barrier Phenotype. PLoS ONE 2016, 11, e0152105. [Google Scholar] [CrossRef]
- Sweeney, M.D.; Zhao, Z.; Montagne, A.; Nelson, A.R.; Zlokovic, B.V. Blood-Brain Barrier: From Physiology to Disease and Back. Physiol. Rev. 2019, 99, 21–78. [Google Scholar] [CrossRef]
- DeStefano, J.G.; Jamieson, J.J.; Linville, R.M.; Searson, P.C. Benchmarking in vitro tissue-engineered blood–brain barrier models. Fluids Barriers CNS 2018, 15, 32. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, J.J.; Searson, P.C.; Gerecht, S. Engineering the human blood-brain barrier in vitro. J. Biol. Eng. 2017, 11, 37. [Google Scholar] [CrossRef] [PubMed]
- Park, T.-E.; Mustafaoglu, N.; Herland, A.; Hasselkus, R.; Mannix, R.; FitzGerald, E.A.; Prantil-Baun, R.; Watters, A.; Henry, O.; Benz, M.; et al. Hypoxia-enhanced Blood-Brain Barrier Chip recapitulates human barrier function and shuttling of drugs and antibodies. Nat. Commun. 2019, 10, 2621. [Google Scholar] [CrossRef] [PubMed]
- Faley, S.L.; Neal, E.H.; Wang, J.X.; Bosworth, A.M.; Weber, C.M.; Balotin, K.M.; Lippmann, E.S.; Bellan, L.M. iPSC-Derived Brain Endothelium Exhibits Stable, Long-Term Barrier Function in Perfused Hydrogel Scaffolds. Stem Cell Rep. 2019, 12, 474–487. [Google Scholar] [CrossRef]
- Vatine, G.D.; Barrile, R.; Workman, M.J.; Sances, S.; Barriga, B.K.; Rahnama, M.; Barthakur, S.; Kasendra, M.; Lucchesi, C.; Kerns, J.; et al. Human iPSC-Derived Blood-Brain Barrier Chips Enable Disease Modeling and Personalized Medicine Applications. Cell Stem Cell 2019, 24, 995–1005.e6. [Google Scholar] [CrossRef]
- DeCastro, A.J.L.; Pranda, M.A.; Gray, K.M.; Merlo-Coyne, J.; Girma, N.; Hurwitz, M.; Zhang, Y.; Stroka, K.M. Morphological Phenotyping of Organotropic Brain- and Bone-Seeking Triple Negative Metastatic Breast Tumor Cells. Front. Cell Dev. Biol. 2022, 10, 790410. [Google Scholar] [CrossRef]
- Tourovskaia, A.; Fauver, M.; Kramer, G.; Simonson, S.; Neumann, T. Tissue-engineered microenvironment systems for modeling human vasculature. Exp. Biol. Med. 2014, 239, 1264–1271. [Google Scholar] [CrossRef]
- Katt, M.E.; Linville, R.M.; Mayo, L.N.; Xu, Z.S.; Searson, P.C. Functional brain-specific microvessels from iPSC-derived human brain microvascular endothelial cells: The role of matrix composition on monolayer formation. Fluids Barriers CNS 2018, 15, 7. [Google Scholar] [CrossRef]
- Grifno, G.N.; Farrell, A.M.; Linville, R.M.; Arevalo, D.; Kim, J.H.; Gu, L.; Searson, P.C. Tissue-engineered blood-brain barrier models via directed differentiation of human induced pluripotent stem cells. Sci. Rep. 2019, 9, 13957. [Google Scholar] [CrossRef] [PubMed]
- Bang, S.; Lee, S.-R.; Ko, J.; Son, K.; Tahk, D.; Ahn, J.; Im, C.; Jeon, N.L. A Low Permeability Microfluidic Blood-Brain Barrier Platform with Direct Contact between Perfusable Vascular Network and Astrocytes. Sci. Rep. 2017, 7, 8083. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Han, S.; Jeon, J.S.; Yamamoto, K.; Zervantonakis, I.K.; Sudo, R.; Kamm, R.D.; Chung, S. Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels. Nat. Protoc. 2012, 7, 1247–1259. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Sudo, R.; Mack, P.J.; Wan, C.-R.; Vickerman, V.; Kamm, R.D. Cell migration into scaffolds under co-culture conditions in a microfluidic platform. Lab A Chip 2009, 9, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Tran, M.; Heo, C.; Lee, L.P.; Cho, H. Human mini-blood–brain barrier models for biomedical neuroscience research: A review. Biomater. Res. 2022, 26, 82. [Google Scholar] [CrossRef]
- Wolburg, H.; Lippoldt, A. Tight junctions of the blood–brain barrier: Development, composition and regulation. Vasc. Pharmacol. 2002, 38, 323–337. [Google Scholar] [CrossRef]
- Ge, S.; Jiang, X.; Paul, D.; Song, L.; Wang, X.; Pachter, J.S. Human ES-derived MSCs correct TNF-α-mediated alterations in a blood–brain barrier model. Fluids Barriers CNS 2019, 16, 18. [Google Scholar] [CrossRef]
- Saunders, N.R.; Dreifuss, J.-J.; Dziegielewska, K.M.; Johansson, P.A.; Habgood, M.D.; Møllgård, K.; Bauer, H.-C. The rights and wrongs of blood-brain barrier permeability studies: A walk through 100 years of history. Front. Neurosci. 2014, 8, 404. [Google Scholar] [CrossRef]
- Srinivasan, B.; Kolli, A.R.; Esch, M.B.; Abaci, H.E.; Shuler, M.L.; Hickman, J.J. TEER Measurement Techniques for In Vitro Barrier Model Systems. JALA J. Assoc. Lab. Autom. 2015, 20, 107–126. [Google Scholar] [CrossRef]
- Odijk, M.; van der Meer, A.D.; Levner, D.; Kim, H.J.; van der Helm, M.W.; Segerink, L.I.; Frimat, J.-P.; Hamilton, G.A.; Ingber, D.E.; Berg, A.v.D. Measuring direct current trans-epithelial electrical resistance in organ-on-a-chip microsystems. Lab Chip 2014, 15, 745–752. [Google Scholar] [CrossRef]
- Zhao, N.; Kulkarni, S.; Zhang, S.; Linville, R.M.; Chung, T.D.; Guo, Z.; Jamieson, J.J.; Norman, D.; Liang, L.; Pessell, A.F.; et al. Modeling angiogenesis in the human brain in a tissue-engineered post-capillary venule. Angiogenesis 2023, 26, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Linville, R.M.; Sklar, M.B.; Grifno, G.N.; Nerenberg, R.F.; Zhou, J.; Ye, R.; DeStefano, J.G.; Guo, Z.; Jha, R.; Jamieson, J.J.; et al. Three-dimensional microenvironment regulates gene expression, function, and tight junction dynamics of iPSC-derived blood–brain barrier microvessels. Fluids Barriers CNS 2022, 19, 87. [Google Scholar] [CrossRef] [PubMed]
- Linville, R.M.; Nerenberg, R.F.; Grifno, G.; Arevalo, D.; Guo, Z.; Searson, P.C. Brain microvascular endothelial cell dysfunction in an isogenic juvenile iPSC model of Huntington’s disease. Fluids Barriers CNS 2022, 19, 54. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.M.; Houghton, S.; Magdeldin, T.; Durán, J.G.B.; Minotti, A.P.; Snead, A.; Sproul, A.; Nguyen, D.-H.T.; Xiang, J.; Fine, H.A.; et al. Pluripotent stem cell-derived epithelium misidentified as brain microvascular endothelium requires ETS factors to acquire vascular fate. Proc. Natl. Acad. Sci. USA 2021, 118, e2016950118. [Google Scholar] [CrossRef] [PubMed]
- Lippmann, E.S.; Azarin, S.M.; Palecek, S.P.; Shusta, E.V. Commentary on human pluripotent stem cell-based blood–brain barrier models. Fluids Barriers CNS 2020, 17, 64. [Google Scholar] [CrossRef]
- Lu, T.M.; Durán, J.G.B.; Houghton, S.; Rafii, S.; Redmond, D.; Lis, R. Human Induced Pluripotent Stem Cell-Derived Brain Endothelial Cells: Current Controversies. Front. Physiol. 2021, 12, 642812. [Google Scholar] [CrossRef]
- Ferro, M.P.; Heilshorn, S.C.; Owens, R.M. Materials for blood brain barrier modeling in vitro. Mater. Sci. Eng. R Rep. 2020, 140, 100522. [Google Scholar] [CrossRef]
- Shin, W.; Hinojosa, C.D.; Ingber, D.E.; Kim, H.J. Human Intestinal Morphogenesis Controlled by Transepithelial Morphogen Gradient and Flow-Dependent Physical Cues in a Microengineered Gut-on-a-Chip. iScience 2019, 15, 391–406. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, L.; Dwiggins, C.W.; Gupta, U.; Stroka, K.M. A Rapid-Patterning 3D Vessel-on-Chip for Imaging and Quantitatively Analyzing Cell–Cell Junction Phenotypes. Bioengineering 2023, 10, 1080. https://doi.org/10.3390/bioengineering10091080
Yan L, Dwiggins CW, Gupta U, Stroka KM. A Rapid-Patterning 3D Vessel-on-Chip for Imaging and Quantitatively Analyzing Cell–Cell Junction Phenotypes. Bioengineering. 2023; 10(9):1080. https://doi.org/10.3390/bioengineering10091080
Chicago/Turabian StyleYan, Li, Cole W. Dwiggins, Udit Gupta, and Kimberly M. Stroka. 2023. "A Rapid-Patterning 3D Vessel-on-Chip for Imaging and Quantitatively Analyzing Cell–Cell Junction Phenotypes" Bioengineering 10, no. 9: 1080. https://doi.org/10.3390/bioengineering10091080
APA StyleYan, L., Dwiggins, C. W., Gupta, U., & Stroka, K. M. (2023). A Rapid-Patterning 3D Vessel-on-Chip for Imaging and Quantitatively Analyzing Cell–Cell Junction Phenotypes. Bioengineering, 10(9), 1080. https://doi.org/10.3390/bioengineering10091080