Pemafibrate Induces a Low Level of PPARα Agonist-Stimulated mRNA Expression of ANGPTL4 in ARPE19 Cell
Abstract
1. Introduction
2. Materials and Methods
2.1. Planar Cultures of ARPE19 Cells
2.2. Gene Expression Analyses
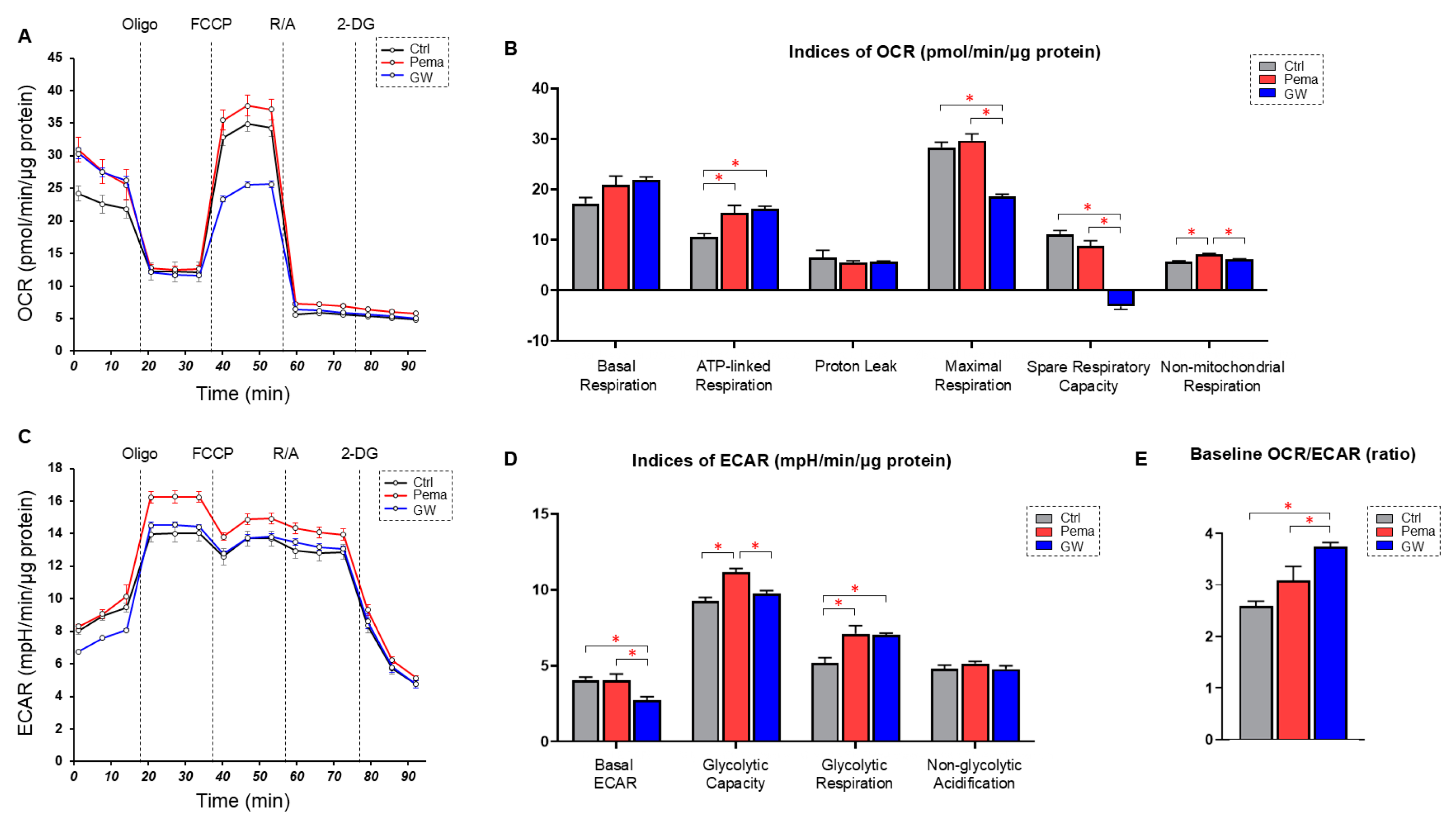
2.3. Real-Time Measurements of Cellular Metabolic Functions
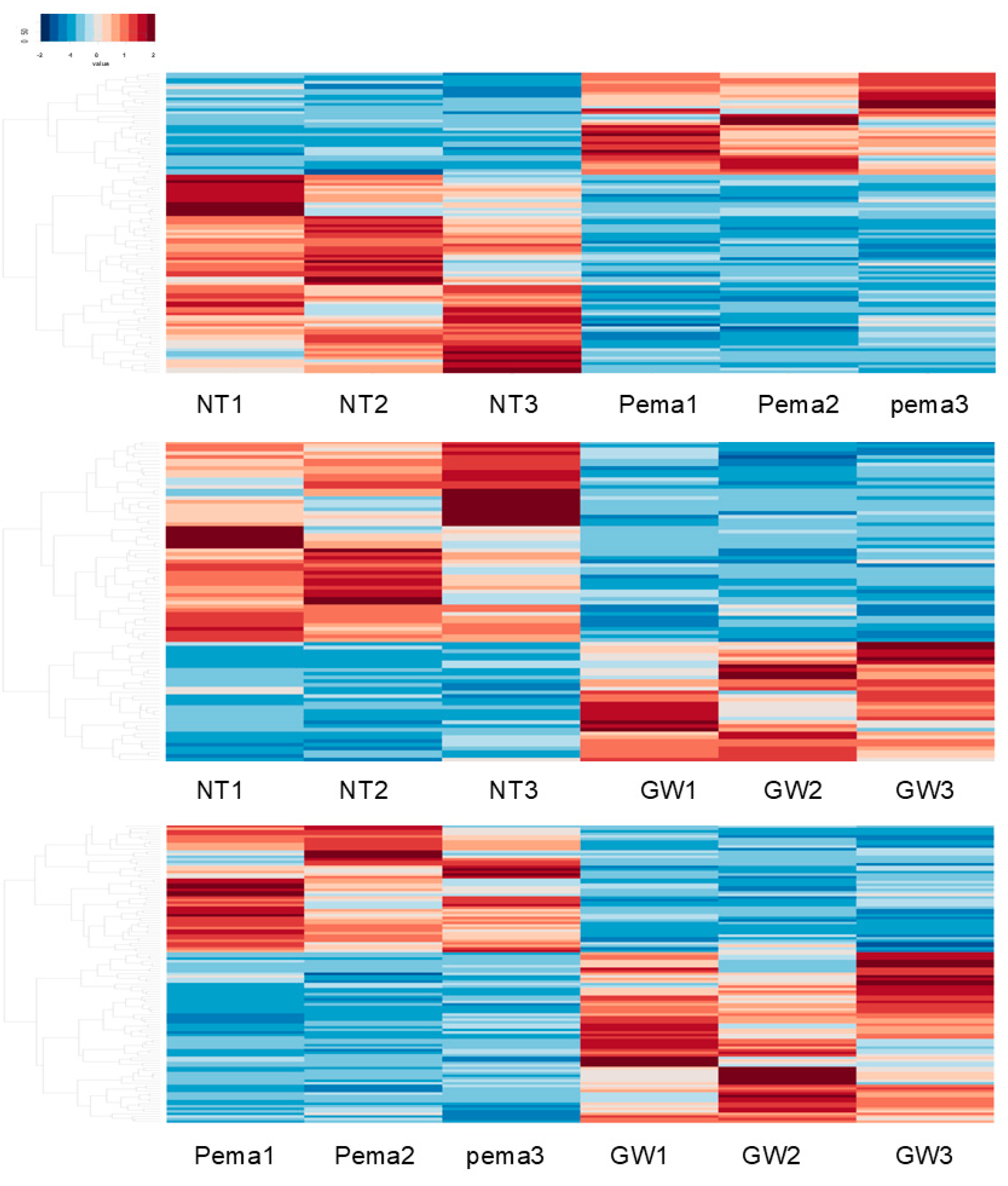
2.4. RNA Sequencing Analysis of Gene Functions and Analysis of Pathways
2.5. Statistical Analysis
3. Results
- (1)
- Group 1: Four downregulated DEGs, namely histone H1.5 (H1-5), NPEPPS pseudo 1 (NPEPPSP1), inhibitor of nuclear factor kappa B kinase subunit gamma pseudogene 1 (IKBKGP1), and Actin-Like 10 (ACTL10); and two upregulated DEGs, namely polycystic kidney disease 1 pseudogene 1 (PKD1P1) and septin 7 pseudogene 3 (SEPTIN7P3), were commonly observed in NT vs. Pema and NT vs. GW, and they were thus most likely to be related to PPARα stimulation in ARPE19 cells.
- (2)
- Group 2: Four downregulated DEGs, namely EF-hand domain containing 2 (EFHC2), FERM and PDZ domain containing 2B pseudogene (FRMPD2B), LINC00910, and phosphodiesterase 7B (PDE7B); and six upregulated DEGs, namely hematopoietic cell-specific Lyn substrate 1 (HCLS1), farnesyl diphosphate synthase pseudogene 2 (FDPSP2), exocyst complex component 5 pseudogene 1 (EXOC5P1), Solute Carrier Family 4 Member 1 Adaptor Protein Pseudogene 1 (SLC4A1APP1), TRPM8 channel-associated factor 2 pseudogene 1 (TCAF2P1), and MIR193BHG, were commonly observed in NT vs. GW and Pema vs. GW, and they were thus related to GW-related specific functions unrelated to PPARα stimulation.
- (3)
- Group 3: Three DEGs, namely small nucleolar RNA H/ACA box 66 (SNORA66), zinc finger protein 890 pseudogene (ZNF890P), and small integral membrane protein 11 (SMIM11), were downregulated in comparison of GW vs. Pema and upregulated in comparison of NT vs. Pema, and these genes were thus thought to be related to Pema-related specific functions unrelated to PPARα stimulation.
- (4)
- Group 4: One DEG, angiopoietin-like 4 (ANGPTL4), was commonly observed as an upregulated DEG in all three comparisons Therefore, upregulation of this gene was induced by both Pema and GW via their PPARα stimulatory activities, with the stimulatory activity of GW being more potent than that of Pema.
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cheng, H.S.; Tan, W.R.; Low, Z.S.; Marvalim, C.; Lee, J.Y.H.; Tan, N.S. Exploration and Development of PPAR Modulators in Health and Disease: An Update of Clinical Evidence. Int. J. Mol. Sci. 2019, 20, 5055. [Google Scholar] [CrossRef] [PubMed]
- Janani, C.; Ranjitha Kumari, B.D. PPAR gamma gene—A review. Diabetes Metab. Syndr. 2015, 9, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zou, Y.; Shen, Z.; Xiong, Y.; Zhang, W.; Liu, C.; Chen, S. Trace Elements, PPARs, and Metabolic Syndrome. Int. J. Mol. Sci. 2020, 21, 2612. [Google Scholar] [CrossRef] [PubMed]
- Arck, P.; Toth, B.; Pestka, A.; Jeschke, U. Nuclear receptors of the peroxisome proliferator-activated receptor (PPAR) family in gestational diabetes: From animal models to clinical trials. Biol. Reprod. 2010, 83, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Cobbina, E.; Akhlaghi, F. Non-alcoholic fatty liver disease (NAFLD)—Pathogenesis, classification, and effect on drug metabolizing enzymes and transporters. Drug Metab. Rev. 2017, 49, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Shen, W.J.; Bittner, S.; Kraemer, F.B.; Azhar, S. PPARs: Regulators of metabolism and as therapeutic targets in cardiovascular disease. Part II: PPAR-β/δ and PPAR-γ. Future Cardiol. 2017, 13, 279–296. [Google Scholar] [CrossRef]
- Escandon, P.; Vasini, B.; Whelchel, A.E.; Nicholas, S.E.; Matlock, H.G.; Ma, J.X.; Karamichos, D. The role of peroxisome proliferator-activated receptors in healthy and diseased eyes. Exp. Eye Res. 2021, 208, 108617. [Google Scholar] [CrossRef]
- Ida, S.; Kaneko, R.; Murata, K. Efficacy and safety of pemafibrate administration in patients with dyslipidemia: A systematic review and meta-analysis. Cardiovasc. Diabetol. 2019, 18, 38. [Google Scholar] [CrossRef]
- Tomita, Y.; Lee, D.; Tsubota, K.; Kurihara, T. PPARα Agonist Oral Therapy in Diabetic Retinopathy. Biomedicines 2020, 8, 433. [Google Scholar] [CrossRef]
- Yamashita, S.; Masuda, D.; Matsuzawa, Y. Pemafibrate, a New Selective PPARα Modulator: Drug Concept and Its Clinical Applications for Dyslipidemia and Metabolic Diseases. Curr. Atheroscler. Rep. 2020, 22, 5. [Google Scholar] [CrossRef]
- Das Pradhan, A.; Glynn, R.J.; Fruchart, J.C.; MacFadyen, J.G.; Zaharris, E.S.; Everett, B.M.; Campbell, S.E.; Oshima, R.; Amarenco, P.; Blom, D.J.; et al. Triglyceride Lowering with Pemafibrate to Reduce Cardiovascular Risk. N. Engl. J. Med. 2022, 387, 1923–1934. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, A.D.; Paynter, N.P.; Everett, B.M.; Glynn, R.J.; Amarenco, P.; Elam, M.; Ginsberg, H.; Hiatt, W.R.; Ishibashi, S.; Koenig, W.; et al. Rationale and design of the Pemafibrate to Reduce Cardiovascular Outcomes by Reducing Triglycerides in Patients with Diabetes (PROMINENT) study. Am. Heart J. 2018, 206, 80–93. [Google Scholar] [CrossRef]
- Lee, D.; Tomita, Y.; Allen, W.; Tsubota, K.; Negishi, K.; Kurihara, T. PPARα Modulation-Based Therapy in Central Nervous System Diseases. Life 2021, 11, 1168. [Google Scholar] [CrossRef] [PubMed]
- Tomita, Y.; Lee, D.; Miwa, Y.; Jiang, X.; Ohta, M.; Tsubota, K.; Kurihara, T. Pemafibrate Protects Against Retinal Dysfunction in a Murine Model of Diabetic Retinopathy. Int. J. Mol. Sci. 2020, 21, 6243. [Google Scholar] [CrossRef] [PubMed]
- Shiono, A.; Sasaki, H.; Sekine, R.; Abe, Y.; Matsumura, Y.; Inagaki, T.; Tanaka, T.; Kodama, T.; Aburatani, H.; Sakai, J.; et al. PPARα activation directly upregulates thrombomodulin in the diabetic retina. Sci. Rep. 2020, 10, 10837. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, F.; Campbell, M. The blood-retina barrier in health and disease. FEBS J. 2023, 290, 878–891. [Google Scholar] [CrossRef]
- Ida, Y.; Hikage, F.; Itoh, K.; Ida, H.; Ohguro, H. Prostaglandin F2α agonist-induced suppression of 3T3-L1 cell adipogenesis affects spatial formation of extra-cellular matrix. Sci. Rep. 2020, 10, 7958. [Google Scholar] [CrossRef]
- Itoh, K.; Hikage, F.; Ida, Y.; Ohguro, H. Prostaglandin F2α Agonists Negatively Modulate the Size of 3D Organoids from Primary Human Orbital Fibroblasts. Investig. Ophthalmol. Vis. Sci. 2020, 61, 13. [Google Scholar] [CrossRef]
- Lee, H.Y.; Gao, X.; Barrasa, M.I.; Li, H.; Elmes, R.R.; Peters, L.L.; Lodish, H.F. PPAR-α and glucocorticoid receptor synergize to promote erythroid progenitor self-renewal. Nature 2015, 522, 474–477. [Google Scholar] [CrossRef]
- Bento-Abreu, A.; Tabernero, A.; Medina, J.M. Peroxisome proliferator-activated receptor-alpha is required for the neurotrophic effect of oleic acid in neurons. J. Neurochem. 2007, 103, 871–881. [Google Scholar] [CrossRef]
- Nielsen, G.; Heiger-Bernays, W.J.; Schlezinger, J.J.; Webster, T.F. Predicting the effects of per- and polyfluoroalkyl substance mixtures on peroxisome proliferator-activated receptor alpha activity in vitro. Toxicology 2022, 465, 153024. [Google Scholar] [CrossRef] [PubMed]
- Ichioka, H.; Hirohashi, Y.; Sato, T.; Furuhashi, M.; Watanabe, M.; Ida, Y.; Hikage, F.; Torigoe, T.; Ohguro, H. G-Protein-Coupled Receptors Mediate Modulations of Cell Viability and Drug Sensitivity by Aberrantly Expressed Recoverin 3 within A549 Cells. Int. J. Mol. Sci. 2023, 24, 771. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Wang, K.; Zhang, S.; Yuan, J.; Liao, X.; Wu, C.; Zou, Y.; Ha, Y.; Shen, Z.; Guo, J.; et al. Ingenuity pathway analysis of differentially expressed genes involved in signaling pathways and molecular networks in RhoE gene-edited cardiomyocytes. Int. J. Mol. Med. 2020, 46, 1225–1238. [Google Scholar] [CrossRef]
- Alimadadi, A.; Aryal, S.; Manandhar, I.; Joe, B.; Cheng, X. Identification of Upstream Transcriptional Regulators of Ischemic Cardiomyopathy Using Cardiac RNA-Seq Meta-Analysis. Int. J. Mol. Sci. 2020, 21, 3472. [Google Scholar] [CrossRef]
- Kamata, S.; Honda, A.; Kashiwagi, N.; Shimamura, A.; Yashiro, S.; Komori, Y.; Hosoda, A.; Akahoshi, N.; Ishii, I. Different Coactivator Recruitment to Human PPARα/δ/γ Ligand-Binding Domains by Eight PPAR Agonists to Treat Nonalcoholic Fatty Liver Disease. Biomedicines 2024, 12, 624. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, Y.; Qiao, P.F.; Zhao, F.L.; Yan, Y. PPAR-α agonist regulates amyloid-β generation via inhibiting BACE-1 activity in human neuroblastoma SH-SY5Y cells transfected with APPswe gene. Mol. Cell Biochem. 2015, 408, 37–46. [Google Scholar] [CrossRef]
- Aryal, B.; Price, N.L.; Suarez, Y.; Fernández-Hernando, C. ANGPTL4 in Metabolic and Cardiovascular Disease. Trends Mol. Med. 2019, 25, 723–734. [Google Scholar] [CrossRef]
- Hato, T.; Tabata, M.; Oike, Y. The role of angiopoietin-like proteins in angiogenesis and metabolism. Trends Cardiovasc. Med. 2008, 18, 6–14. [Google Scholar] [CrossRef]
- Santulli, G. Angiopoietin-like proteins: A comprehensive look. Front. Endocrinol. 2014, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Goh, Y.Y.; Chin, H.F.; Kersten, S.; Tan, N.S. Angiopoietin-like 4: A decade of research. Biosci. Rep. 2012, 32, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Desai, U.; Lee, E.C.; Chung, K.; Gao, C.; Gay, J.; Key, B.; Hansen, G.; Machajewski, D.; Platt, K.A.; Sands, A.T.; et al. Lipid-lowering effects of anti-angiopoietin-like 4 antibody recapitulate the lipid phenotype found in angiopoietin-like 4 knockout mice. Proc. Natl. Acad. Sci. USA 2007, 104, 11766–11771. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.C.; Chickering, T.W.; Rosen, E.D.; Dussault, B.; Qin, Y.; Soukas, A.; Friedman, J.M.; Holmes, W.E.; Spiegelman, B.M. Peroxisome proliferator-activated receptor gamma target gene encoding a novel angiopoietin-related protein associated with adipose differentiation. Mol. Cell. Biol. 2000, 20, 5343–5349. [Google Scholar] [CrossRef] [PubMed]
- Kersten, S.; Mandard, S.; Tan, N.S.; Escher, P.; Metzger, D.; Chambon, P.; Gonzalez, F.J.; Desvergne, B.; Wahli, W. Characterization of the fasting-induced adipose factor FIAF, a novel peroxisome proliferator-activated receptor target gene. J. Biol. Chem. 2000, 275, 28488–28493. [Google Scholar] [CrossRef]
- Kim, I.; Kim, H.G.; Kim, H.; Kim, H.H.; Park, S.K.; Uhm, C.S.; Lee, Z.H.; Koh, G.Y. Hepatic expression, synthesis and secretion of a novel fibrinogen/angiopoietin-related protein that prevents endothelial-cell apoptosis. Biochem. J. 2000, 346 Pt 3, 603–610. [Google Scholar] [CrossRef]
- Belanger, A.J.; Lu, H.; Date, T.; Liu, L.X.; Vincent, K.A.; Akita, G.Y.; Cheng, S.H.; Gregory, R.J.; Jiang, C. Hypoxia up-regulates expression of peroxisome proliferator-activated receptor gamma angiopoietin-related gene (PGAR) in cardiomyocytes: Role of hypoxia inducible factor 1alpha. J. Mol. Cell. Cardiol. 2002, 34, 765–774. [Google Scholar] [CrossRef]
- Koliwad, S.K.; Kuo, T.; Shipp, L.E.; Gray, N.E.; Backhed, F.; So, A.Y.; Farese, R.V., Jr.; Wang, J.C. Angiopoietin-like 4 (ANGPTL4, fasting-induced adipose factor) is a direct glucocorticoid receptor target and participates in glucocorticoid-regulated triglyceride metabolism. J. Biol. Chem. 2009, 284, 25593–25601. [Google Scholar] [CrossRef]
- Xin, X.; Rodrigues, M.; Umapathi, M.; Kashiwabuchi, F.; Ma, T.; Babapoor-Farrokhran, S.; Wang, S.; Hu, J.; Bhutto, I.; Welsbie, D.S.; et al. Hypoxic retinal Muller cells promote vascular permeability by HIF-1-dependent up-regulation of angiopoietin-like 4. Proc. Natl. Acad. Sci. USA 2013, 110, E3425–E3434. [Google Scholar] [CrossRef]
- Kwon, S.H.; Shin, J.P.; Kim, I.T.; Park, D.H. Aqueous Levels of Angiopoietin-like 4 and Semaphorin 3E Correlate with Nonperfusion Area and Macular Volume in Diabetic Retinopathy. Ophthalmology 2015, 122, 968–975. [Google Scholar] [CrossRef]
- Sodhi, A.; Ma, T.; Menon, D.; Deshpande, M.; Jee, K.; Dinabandhu, A.; Vancel, J.; Lu, D.; Montaner, S. Angiopoietin-like 4 binds neuropilins and cooperates with VEGF to induce diabetic macular edema. J. Clin. Investig. 2019, 129, 4593–4608. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Zou, W.; Chen, B.; Zou, C.; Zhao, M.; Zheng, Z. ANGPTL-4 correlates with vascular endothelial growth factor in patients with proliferative diabetic retinopathy. Graefe’s Arch. Clin. Exp. Ophthalmol. Albrecht Graefes Arch. Klin. Exp. Ophthalmol. 2016, 254, 1281–1288. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Lu, P.; Chen, W.; Lu, L.; Zheng, Z. ANGPTL-4 induces diabetic retinal inflammation by activating Profilin-1. Exp. Eye Res. 2018, 166, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Cao, J.; Du, Y.; Gong, Q.; Cheng, Y.; Su, G. Angiopoietin-Like Protein 4 (ANGPTL4) Induces Retinal Pigment Epithelial Barrier Breakdown by Activating Signal Transducer and Activator of Transcription 3 (STAT3): Evidence from ARPE-19 Cells Under Hypoxic Condition and Diabetic Rats. Med. Sci. Monit. 2019, 25, 6742–6754. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, Y.; Cheng, L.; Kuang, H. Regulation of Long Noncoding RNA NEAT1/miR-320a/HIF-1α Competitive Endogenous RNA Regulatory Network in Diabetic Retinopathy. Investig. Ophthalmol. Vis. Sci. 2023, 64, 11. [Google Scholar] [CrossRef]
- Raza-Iqbal, S.; Tanaka, T.; Anai, M.; Inagaki, T.; Matsumura, Y.; Ikeda, K.; Taguchi, A.; Gonzalez, F.J.; Sakai, J.; Kodama, T. Transcriptome Analysis of K-877 (a Novel Selective PPARα Modulator (SPPARMα))-Regulated Genes in Primary Human Hepatocytes and the Mouse Liver. J. Atheroscler. Thromb. 2015, 22, 754–772. [Google Scholar] [CrossRef]
- Sefried, S.; Häring, H.U.; Weigert, C.; Eckstein, S.S. Suitability of hepatocyte cell lines HepG2, AML12 and THLE-2 for investigation of insulin signalling and hepatokine gene expression. Open Biol. 2018, 8, 180147. [Google Scholar] [CrossRef]
- Mimura, I.; Nangaku, M.; Kanki, Y.; Tsutsumi, S.; Inoue, T.; Kohro, T.; Yamamoto, S.; Fujita, T.; Shimamura, T.; Suehiro, J.; et al. Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Mol. Cell. Biol. 2012, 32, 3018–3032. [Google Scholar] [CrossRef]
- Inoue, T.; Kohro, T.; Tanaka, T.; Kanki, Y.; Li, G.; Poh, H.M.; Mimura, I.; Kobayashi, M.; Taguchi, A.; Maejima, T.; et al. Cross-enhancement of ANGPTL4 transcription by HIF1 alpha and PPAR beta/delta is the result of the conformational proximity of two response elements. Genome Biol. 2014, 15, R63. [Google Scholar] [CrossRef]
- Wang, Y.; Lam, K.S.; Lam, J.B.; Lam, M.C.; Leung, P.T.; Zhou, M.; Xu, A. Overexpression of angiopoietin-like protein 4 alters mitochondria activities and modulates methionine metabolic cycle in the liver tissues of db/db diabetic mice. Mol. Endocrinol. 2007, 21, 972–986. [Google Scholar] [CrossRef]
- Zheng, X.; Liu, R.; Zhou, C.; Yu, H.; Luo, W.; Zhu, J.; Liu, J.; Zhang, Z.; Xie, N.; Peng, X.; et al. ANGPTL4-Mediated Promotion of Glycolysis Facilitates the Colonization of Fusobacterium nucleatum in Colorectal Cancer. Cancer Res. 2021, 81, 6157–6170. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Teng, L.; Lai, Y.; Yao, X.; Fang, Y.; Wang, Z.; Lin, S.; Zhang, H.; Li, Q.; Li, Y.; et al. Adipose-derived stem cells promote glycolysis and peritoneal metastasis via TGF-β1/SMAD3/ANGPTL4 axis in colorectal cancer. Cell. Mol. Life Sci. CMLS 2024, 81, 189. [Google Scholar] [CrossRef] [PubMed]


| NT vs. Pema | ||
| ID | Associated Network Functions | Score |
| 1 | Connective tissue disorders, developmental disorder, hereditary disorder | 47 |
| 2 | Developmental disorder, hereditary disorder, organismal injury and abnormalities | 40 |
| 3 | Developmental disorder, endocrine system disorders, hereditary disorder | 36 |
| 4 | Cancer, neurological disease, organismal injury and abnormalities | 32 |
| 5 | Cell-to-cell signaling and interaction, cellular response to therapeutics | 32 |
| NT vs. GW | ||
| ID | Associated Network Functions | Score |
| 1 | Cancer, endocrine system disorders, organismal injury, and abnormalities | 42 |
| 2 | Connective tissue disorders, developmental disorder, gastrointestinal disease | 38 |
| 3 | Embryonic development, organismal development, tissue morphology | 33 |
| 4 | Behavior, cell cycle, cell death and survival | 31 |
| 5 | Cardiovascular disease, organismal injury and abnormalities, molecular transport | 31 |
| NT vs. Pema | ||
| Name | p-Value | Overlap (%) |
| CREB signaling in neurons | 3.65 × 10−9 | 12.4 |
| Cellular effects of sildenafil (Viagra) | 3.69 × 10−9 | 11.9 |
| Class A/1 (rhodopsin-like receptors) | 4.36 × 10−6 | 15.1 |
| Potassium channels | 4.22 × 10−8 | 22.3 |
| G-protein coupled receptor signaling | 7.40 × 10−7 | 10.8 |
| NT vs. GW | ||
| Name | p-Value | Overlap (%) |
| G alpha (s) signaling events | 4.22 × 10−5 | 13.8 |
| Class A/1 (rhodopsin-like receptors) | 7.12 × 10−5 | 10.4 |
| Activation of matrix metalloproteinases | 1.84 × 10−4 | 24.2 |
| Striated muscle contraction | 3.51 × 10−4 | 22.2 |
| Cardiac hypertrophy signaling (enhanced) | 4.27 × 10−4 | 8.5 |
| Downregulation | Upregulation | ||||
|---|---|---|---|---|---|
| NT vs. Pema | NT vs. GW | Pema vs. GW | NT vs. Pema | NT vs. GW | Pema vs. GW |
| H1-5 | H1-5 | PKD1P1 | PKD1P1 | ||
| NPEPPSP1 | NPEPPSP1 | SEPTIN7P3 | SEPTIN7P3 | ||
| IKBKGP1 | IKBKGP1 | HCLS1 | HCLS1 | ||
| ACTL10 | ACTL10 | FDPSP2 | FDPSP2 | ||
| EFHC2 | EFHC2 | EXOC5P1 | EXOC5P1 | ||
| FRMPD2B | FRMPD2B | SLC4A1APP1 | SLC4A1APP1 | ||
| LINC00910 | LINC00910 | TCAF2P1 | TCAF2P1 | ||
| PDE7B | PDE7B | MIR193BHG | MIR193BHG | ||
| SNORA66 | SNORA66 | ||||
| ZNF890P | ZNF890P | ||||
| SMIM11 | SMIM11 | ||||
| ANGPTL4 | ANGPTL4 | ANGPTL4 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ohguro, H.; Nishikiori, N.; Sato, T.; Watanabe, M.; Higashide, M.; Furuhashi, M. Pemafibrate Induces a Low Level of PPARα Agonist-Stimulated mRNA Expression of ANGPTL4 in ARPE19 Cell. Bioengineering 2024, 11, 1247. https://doi.org/10.3390/bioengineering11121247
Ohguro H, Nishikiori N, Sato T, Watanabe M, Higashide M, Furuhashi M. Pemafibrate Induces a Low Level of PPARα Agonist-Stimulated mRNA Expression of ANGPTL4 in ARPE19 Cell. Bioengineering. 2024; 11(12):1247. https://doi.org/10.3390/bioengineering11121247
Chicago/Turabian StyleOhguro, Hiroshi, Nami Nishikiori, Tatsuya Sato, Megumi Watanabe, Megumi Higashide, and Masato Furuhashi. 2024. "Pemafibrate Induces a Low Level of PPARα Agonist-Stimulated mRNA Expression of ANGPTL4 in ARPE19 Cell" Bioengineering 11, no. 12: 1247. https://doi.org/10.3390/bioengineering11121247
APA StyleOhguro, H., Nishikiori, N., Sato, T., Watanabe, M., Higashide, M., & Furuhashi, M. (2024). Pemafibrate Induces a Low Level of PPARα Agonist-Stimulated mRNA Expression of ANGPTL4 in ARPE19 Cell. Bioengineering, 11(12), 1247. https://doi.org/10.3390/bioengineering11121247








