Deep Learning for Delineation of the Spinal Canal in Whole-Body Diffusion-Weighted Imaging: Normalising Inter- and Intra-Patient Intensity Signal in Multi-Centre Datasets
Abstract
1. Introduction
2. Material and Methods
2.1. Patient Population and MRI Protocol
2.2. Spinal Canal Delineation
2.2.1. Dataset Description
2.2.2. Image Pre-Processing
2.2.3. U-Net Model Architecture and Hyper-Parameter Selection
2.2.4. Post-Processing
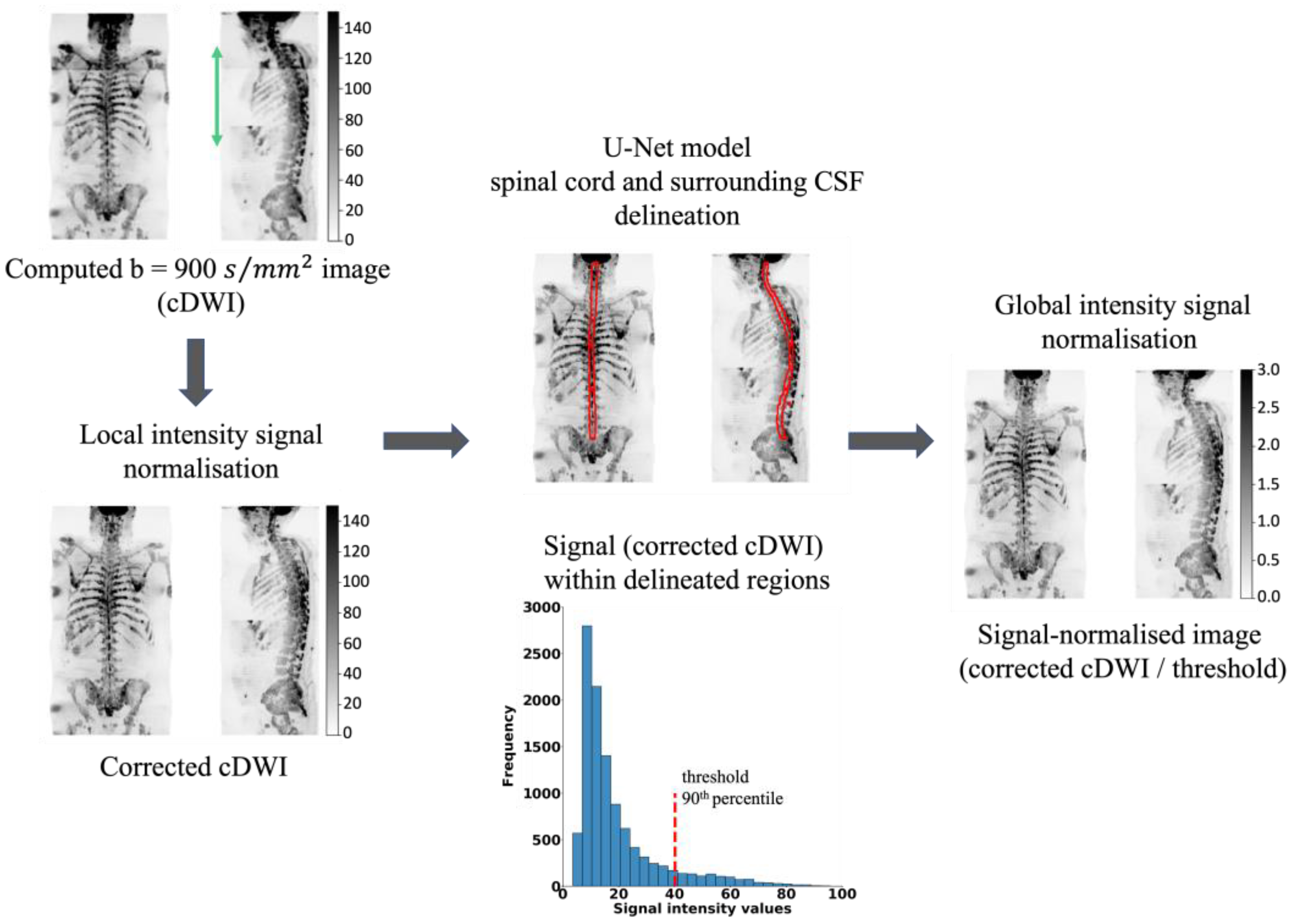
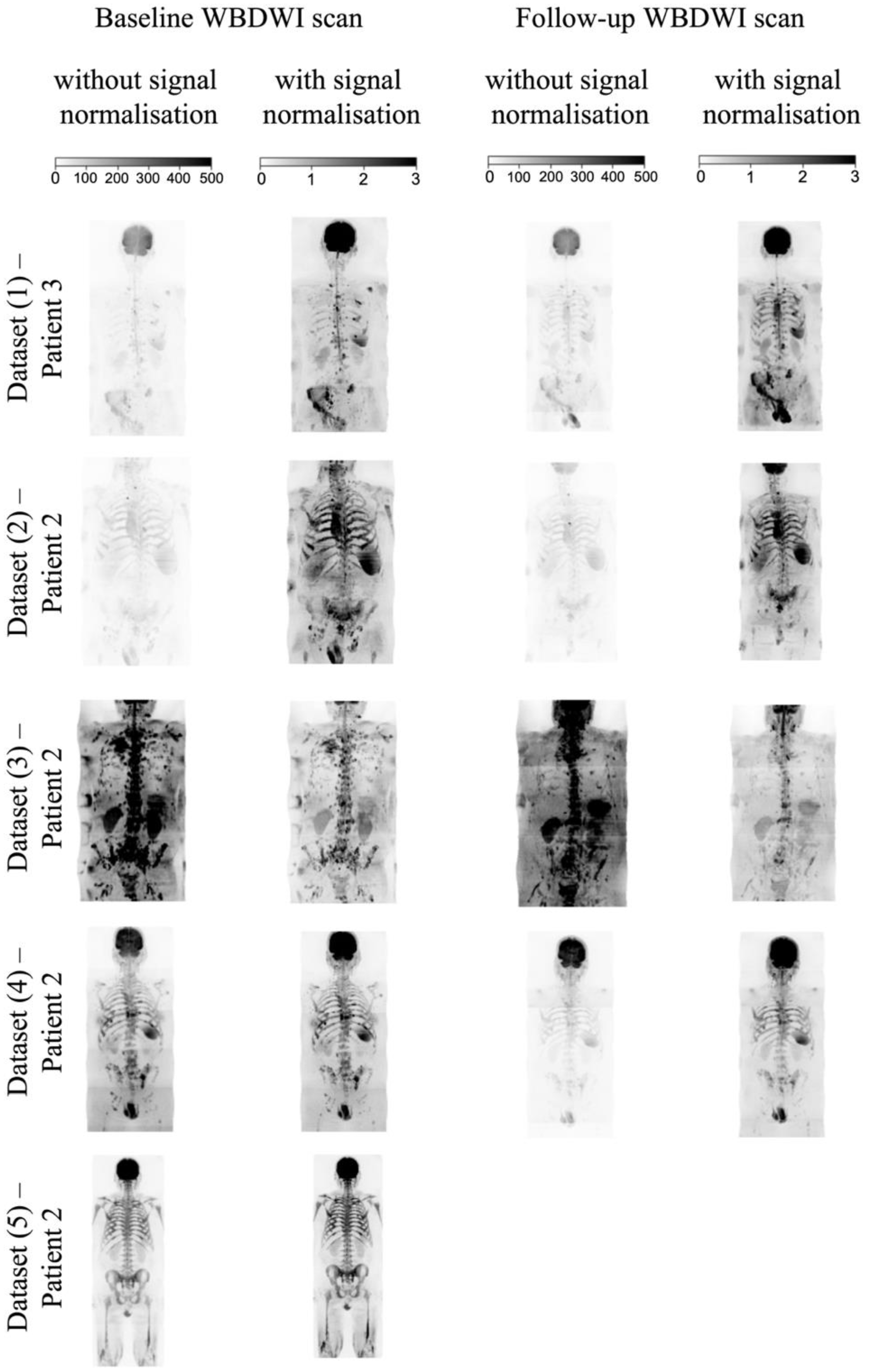
2.3. WBDWI Signal Normalisation
- (1)
- A whole-body bc = 900 s/mm2 image is computed (cDWI) to optimize the SNR and increase the suppression of the background signal for the detection of bone metastases [24]. The cDWI images are computed from the estimated S0 images and ADC maps for each station using:
- (2)
- The signal intensity of sequential stations of derived cDWI volumes is normalised by applying a linear scaling term to the cDWI data that minimises the mean square error between cumulative frequency curves of cDWI intensities from axial images on either side of each station boundary, as previously described [30,31].
- (3)
- The spinal cord and surrounding CSF segmentations derived using the U-Net model (Section 2.2) are transferred to the cDWI images and the voxel values across the entire field of view are standardised to the 90th percentile of the signal within the entire spinal canal.
2.4. Evaluation Criteria
2.4.1. Spinal Canal Segmentation
2.4.2. Intensity Signal WBDWI Normalisation
3. Results
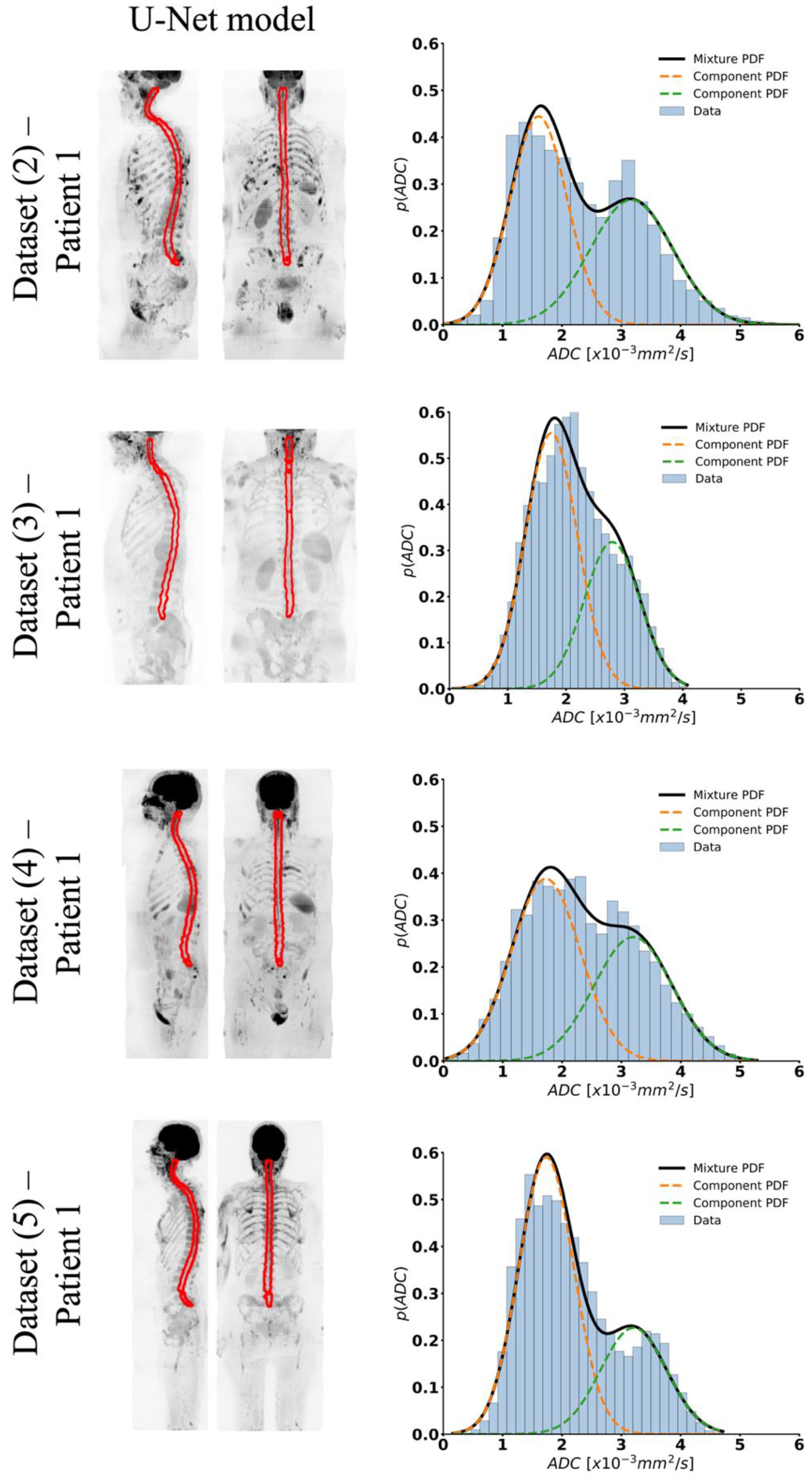
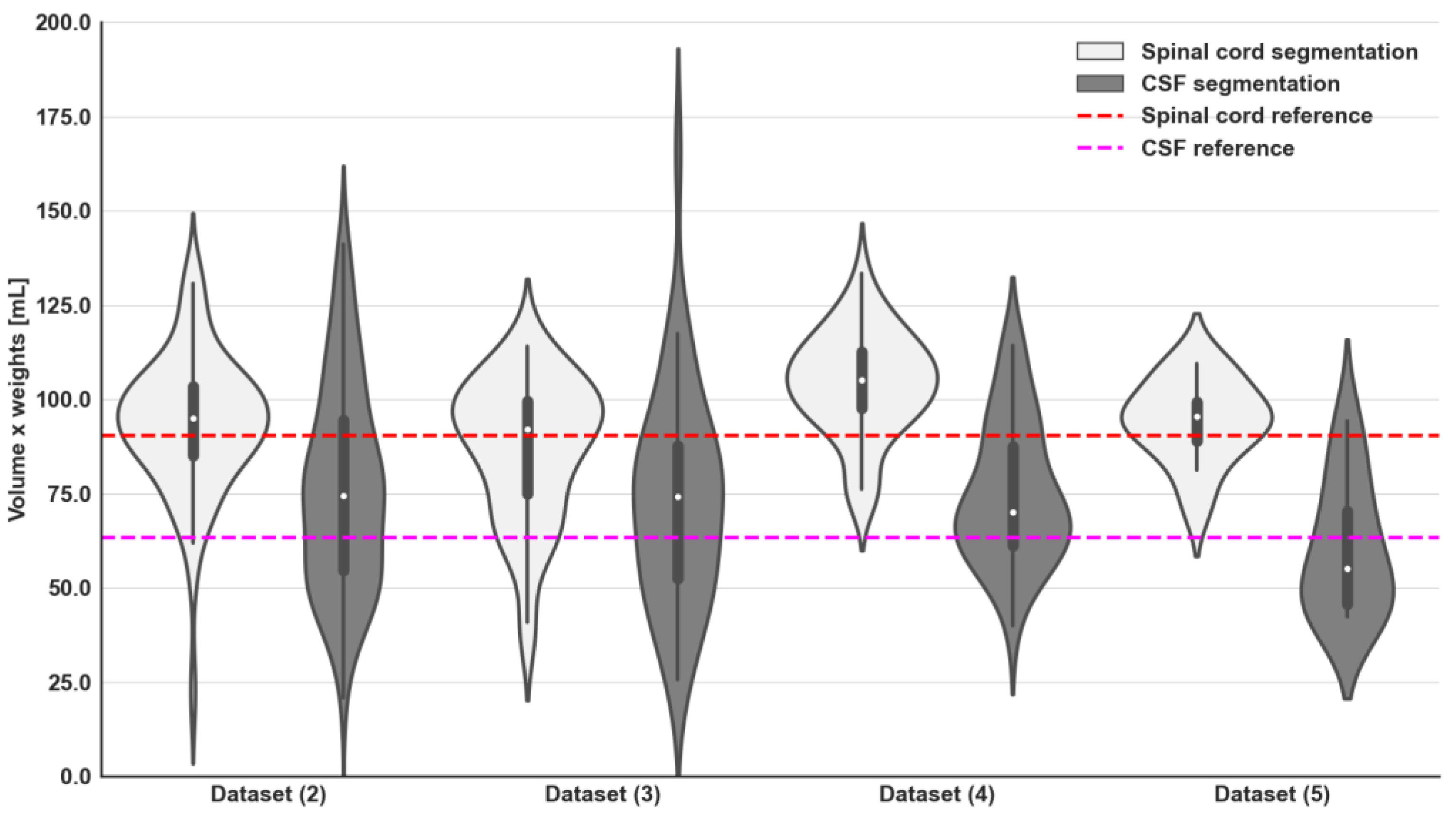
3.1. Spinal Canal Segmentation
3.2. Intensity Signal WBDWI Normalisation
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Padhani, A.R.; Lecouvet, F.E.; Tunariu, N.; Koh, D.-M.; De Keyzer, F.; Collins, D.J.; Sala, E.; Schlemmer, H.P.; Petralia, G.; Vargas, H.A.; et al. METastasis Reporting and Data System for Prostate Cancer: Practical Guidelines for Acquisition, Interpretation, and Reporting of Whole-body Magnetic Resonance Imaging-based Evaluations of Multiorgan Involvement in Advanced Prostate Cancer. Eur. Urol. 2017, 71, 81–92. [Google Scholar] [CrossRef]
- Perez-Lopez, R.; Tunariu, N.; Padhani, A.R.; Oyen, W.J.G.; Fanti, S.; Vargas, H.A.; Omlin, A.; Morris, M.J.; de Bono, J.; Koh, D.-M. Imaging Diagnosis and Follow-up of Advanced Prostate Cancer: Clinical Perspectives and State of the Art. Radiology 2019, 292, 273–286. [Google Scholar] [CrossRef]
- Zugni, F.; Ruju, F.; Pricolo, P.; Alessi, S.; Iorfida, M.; Colleoni, M.A.; Bellomi, M.; Petralia, G. The added value of whole-body magnetic resonance imaging in the management of patients with advanced breast cancer. PLoS ONE 2018, 13, e0205251. [Google Scholar] [CrossRef] [PubMed]
- Morone, M.; Bali, M.A.; Tunariu, N.; Messiou, C.; Blackledge, M.; Grazioli, L.; Koh, D.-M. Whole-Body MRI: Current Applications in Oncology. Am. J. Roentgenol. 2017, 209, 336–349. [Google Scholar] [CrossRef]
- Messiou, C.; Porta, N.; Sharma, B.; Levine, D.; Koh, D.-M.; Boyd, K.; Pawlyn, C.; Riddell, A.; Downey, K.; Croft, J.; et al. Prospective Evaluation of Whole-Body MRI versus FDG PET/CT for Lesion Detection in Participants with Myeloma. Radiol. Imaging Cancer 2021, 3, e210048. [Google Scholar] [CrossRef]
- Miles, A.; Evans, R.E.; Halligan, S.; Beare, S.; Bridgewater, J.; Goh, V.; Janes, S.M.; Navani, N.; Oliver, A.; Morton, A.; et al. Predictors of patient preference for either whole body magnetic resonance imaging (WB-MRI) or CT/PET-CT for staging colorectal or lung cancer. J. Med. Imaging Radiat. Oncol. 2020, 64, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Isaac, A.; Lecouvet, F.; Dalili, D.; Fayad, L.; Pasoglou, V.; Papakonstantinou, O.; Ahlawat, S.; Messiou, C.; Weber, M.-A.; Padhani, A.R. Detection and Characterization of Musculoskeletal Cancer Using Whole-Body Magnetic Resonance Imaging. Semin. Musculoskelet. Radiol. 2020, 24, 726–750. [Google Scholar] [CrossRef]
- Perez-Lopez, R.; Mateo, J.; Mossop, H.; Blackledge, M.D.; Collins, D.J.; Rata, M.; Morgan, V.A.; Macdonald, A.; Sandhu, S.; Lorente, D.; et al. Diffusion-weighted imaging as a treatment response biomarker for evaluating bone metastases in prostate cancer: A pilot study. Radiology 2017, 283, 168–177. [Google Scholar] [CrossRef]
- Lecouvet, F.E.; Vekemans, M.-C.; Berghe, T.V.D.; Verstraete, K.; Kirchgesner, T.; Acid, S.; Malghem, J.; Wuts, J.; Hillengass, J.; Vandecaveye, V.; et al. Imaging of treatment response and minimal residual disease in multiple myeloma: State of the art WB-MRI and PET/CT. Skelet. Radiol. 2021, 51, 59–80. [Google Scholar] [CrossRef]
- Barnes, A.; Alonzi, R.; Blackledge, M.; Charles-Edwards, G.; Collins, D.J.; Cook, G.; Coutts, G.; Goh, V.; Graves, M.; Kelly, C.; et al. Guidelines & recommendations: UK quantitative WB-DWI technical workgroup: Consensus meeting recommendations on optimisation, quality control, processing and analysis of quantitative whole-body diffusion-weighted imaging for cancer. Br. J. Radiol. 2018, 91, 1–12. [Google Scholar]
- Petralia, G.; Padhani, A.R. Whole-Body Magnetic Resonance Imaging in Oncology: Uses and Indications. Magn. Reson. Imaging Clin. N. Am. 2018, 26, 495–507. [Google Scholar] [CrossRef]
- Koh, D.M.; Blackledge, M.; Padhani, A.R.; Takahara, T.; Kwee, T.C.; Leach, M.O.; Collins, D.J. Whole-body diffusion-weighted mri: Tips, tricks, and pitfalls. Am. J. Roentgenol. 2012, 199, 252–262. [Google Scholar] [CrossRef]
- Colombo, A.; Saia, G.; Azzena, A.A.; Rossi, A.; Zugni, F.; Pricolo, P.; Summers, P.E.; Marvaso, G.; Grimm, R.; Bellomi, M.; et al. Semi-automated segmentation of bone metastases from whole-body mri: Reproducibility of apparent diffusion coefficient measurements. Diagnostics 2021, 11, 499. [Google Scholar] [CrossRef] [PubMed]
- Wennmann, M.; Thierjung, H.; Bauer, F.; Weru, V.; Hielscher, T.; Grözinger, M.; Gnirs, R.; Sauer, S.; Goldschmidt, H.; Weinhold, N.; et al. Repeatability and Reproducibility of ADC Measurements and MRI Signal Intensity Measurements of Bone Marrow in Monoclonal Plasma Cell Disorders: A Prospective Bi-institutional Multiscanner, Multiprotocol Study. Invest. Radiol. 2022, 57, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.; Porta, N.; Bianchini, D.; McGovern, U.; Elliott, T.; Jones, R.; Stndikus, I.; Ralph, C.; Jin, S.; Varughese, M.; et al. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): A multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020, 21, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Jager, F.; Hornegger, J. Nonrigid registration of joint histograms for intensity standardization in magnetic resonance imaging. IEEE Trans. Med. Imaging 2009, 28, 137–150. [Google Scholar] [CrossRef]
- Digma, L.A.; Feng, C.H.; Conlin, C.C.; Rodríguez-Soto, A.E.; Zhong, A.Y.; Hussain, T.S.; Lui, A.J.; Batra, K.; Simon, A.B.; Karunamuni, R.; et al. Correcting B0 inhomogeneity-induced distortions in whole-body diffusion MRI of bone. Sci. Rep. 2022, 12, 265. [Google Scholar] [CrossRef] [PubMed]
- Blackledge, M.D.; Tunariu, N.; Zugni, F.; Holbrey, R.; Orton, M.R.; Ribeiro, A.; Hughes, J.C.; Scurr, E.D.; Collins, D.J.; Leach, M.O.; et al. Noise-Corrected, Exponentially Weighted, Diffusion-Weighted MRI (niceDWI) Improves Image Signal Uniformity in Whole-Body Imaging of Metastatic Prostate Cancer. Front. Oncol. 2020, 10, 704. [Google Scholar] [CrossRef] [PubMed]
- Ceranka, J.; Verga, S.; Lecouvet, F.; Metens, T.; De Mey, J.; Vandemeulebroucke, J. Intensity Standardization of Skeleton in Follow-Up Whole-Body MRI. In Proceedings of the Computational Methods and Clinical Applications for Spine Imaging: 5th International Workshop and Challenge, CSI 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 16 September 2018; Volume 2, pp. 77–89. [Google Scholar]
- Padhani, A.R.; van Ree, K.; Collins, D.J.; D’sa, S.; Makris, A. Assessing the relation between bone marrow signal intensity and apparent diffusion coefficient in diffusion-weighted MRI. Am. J. Roentgenol. 2013, 200, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Winfield, J.; Blackledge, M.D.; Tunariu, N.; Koh, D.-M.; Messiou, C. Whole-body MRI: A practical guide for imaging patients with malignant bone disease. Clin. Radiol. 2021, 76, 715–727. [Google Scholar] [CrossRef]
- Petralia, G.; Koh, D.M.; Attariwala, R.; Busch, J.J.; Eeles, R.; Karow, D.; Lo, G.G.; Messiou, C.; Sala, E.; Vargas, H.A.; et al. Oncologically Relevant Findings Reporting and Data System (ONCO-RADS): Guidelines for the Acquisition, Interpretation, and Reporting of Whole-Body MRI for Cancer screening. Radiology 2021, 299, 494–507. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Lecture Notes in Computer Science; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; Volume 9351, pp. 234–241. [Google Scholar]
- Blackledge, M.D.; Leach, M.O.; Collins, D.J.; Koh, D.-M. Computed Diffusion-weighted MR Imaging May Improve Tumor Detection. Radiology 2011, 261, 573–581. [Google Scholar] [CrossRef]
- Jadon, S. A survey of loss functions for semantic segmentation. In Proceedings of the Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Via del Mar, Chile, 27–29 October 2020; pp. 1–7. [Google Scholar]
- Taghanaki, S.A.; Zheng, Y.; Zhou, S.K.; Georgescu, B.; Sharma, P.; Xu, D.; Comaniciu, D.; Hamarneh, G. Combo loss: Handling input and output imbalance in multi-organ segmentation. Comput. Med. Imaging Graph. 2019, 75, 24–33. [Google Scholar] [CrossRef]
- Sadegh, S.; Salehi, M.; Erdogmus, D.; Gholipour, A. Tversky loss function for image segmentation using 3D fully convolutional deep networks. In International Workshop on Machine Learning in Medical Imaging; Springer International Publishing: Cham, Switzerland, 2017; pp. 379–387. [Google Scholar]
- Abraham, N.; Khan, N.M. A novel focal tversky loss function with improved attention u-net for lesion segmentation. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 683–687. [Google Scholar]
- Chen, M.; Carass, A.; Oh, J.; Nair, G.; Pham, D.L.; Reich, D.S.; Prince, J.L. Automatic Magnetic Resonance Spinal Cord Segmentation with Topology Constraints for Variable Fields of View. Neuroimage 2013, 23, 1051–1062. [Google Scholar] [CrossRef][Green Version]
- Blackledge, M.D.; Koh, D.M.; Orton, M.R.; Collins, D.J.; Leach, M.O. A Fast and Simple Post-Processing Procedure for the Correction of Mis-Registration between Sequentially Acquired Stations in Whole-Body Diffusion Weighted MRI; International Society for Magnetic Resonance in Medicine: Australia, Melbourne, 2012; p. 4111. [Google Scholar]
- Ceranka, J.; Polfliet, M.; Lecouvet, F.; Michoux, N.; De Mey, J.; Vandemeulebroucke, J. Registration strategies for multi-modal whole-body MRI mosaicking. Magn. Reson. Med. 2018, 79, 1684–1695. [Google Scholar] [CrossRef] [PubMed]
- Lemay, A.; Gros, C.; Zhuo, Z.; Zhang, J.; Duan, Y.; Cohen-Adad, J.; Liu, Y. Automatic multiclass intramedullary spinal cord tumor segmentation on MRI with deep learning. NeuroImage Clin. 2021, 31, 102766. [Google Scholar] [CrossRef] [PubMed]
- McCoy, D.; Dupont, S.; Gros, C.; Cohen-Adad, J.; Huie, R.; Ferguson, A.; Duong-Fernandez, X.; Thomas, L.; Singh, V.; Narvid, J.; et al. Convolutional neural network–based automated segmentation of the spinal cord and contusion injury: Deep learning biomarker correlates of motor impairment in acute spinal cord injury. Am. J. Neuroradiol. 2019, 40, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Gros, C.; De Leener, B.; Badji, A.; Maranzano, J.; Eden, D.; Dupont, S.M.; Talbott, J.; Zhuoquiong, R.; Liu, Y.; Granberg, T.; et al. Automatic segmentation of the spinal cord and intramedullary multiple sclerosis lesions with convolutional neural networks. NeuroImage 2019, 184, 901–915. [Google Scholar] [CrossRef]
- Candito, A.; Blackledge, M.D.; Holbrey, R.; Koh, D.M. Automated tool to quantitatively assess bone disease on Whole-Body Diffusion Weighted Imaging for patients with Advanced Prostate Cancer. In Proceedings of the Medical Imaging with Deep Learning, Zurich, Switzerland, 6–8 July 2022; pp. 2–4. [Google Scholar]
- Qaiser, T.; Winzeck, S.; Barfoot, T.; Barwick, T.; Doran, S.J.; Kaiser, M.F.; Glocker, B. Multiple Instance Learning with Auxiliary Task Weighting for Multiple Myeloma Classification. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI, Strasbourg, France, 27 September–1 October 2021. [Google Scholar]
- Wennmann, M.; Neher, P.; Stanczyk, N.; Kahl, K.C.; Kächele, J.; Weru, V.; Hielscher, T.; Grozinger, M.; Chmelik, J.; Zhang, K.; et al. Deep Learning for Automatic Bone Marrow Apparent Diffusion Coefficient Measurements from Whole-Body Magnetic Resonance Imaging in Patients with Multiple Myeloma: A Retrospective Multicenter Study. Invest. Radiol. 2023, 58, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Donners, R.; Blackledge, M.; Tunariu, N.; Messiou, C.; Merkle, E.M.; Koh, D.-M. Quantitative Whole-Body Diffusion-Weighted MR Imaging. Magn. Reson. Imaging Clin. N. Am. 2018, 26, 479–495. [Google Scholar] [CrossRef]


| APC Cohort Dataset (1) (40 Patients; 80 WBDWI Scans) | APC Cohort Dataset (2) (33 Patients; 115 WBDWI Scans) | APC Cohort Dataset (3) (22 Patients; 44 WBDWI Scans) | APC Cohort Dataset (4) (18 Patients; 36 WBDWI Scans) | MM Cohort Dataset (5) (12 Patients; 12 WBDWI Scans) | |
|---|---|---|---|---|---|
| MR scanner | 1.5T Siemens Aera | 1.5T Siemens Aera/Avanto | 1.5T Siemens Aera | 1.5T Siemens Aera | 1.5T Siemens Avanto |
| Sequence | Diffusion-Weighted SS-EPI | Diffusion-Weighted SS-EPI | Diffusion-Weighted SS-EPI | Diffusion-Weighted SS-EPI | Diffusion-Weighted SS-EPI |
| Acquisition plane | Axial | Axial | Axial | Axial | Axial |
| Breathing mode | Free breathing | Free breathing | Free breathing | Free breathing | Free breathing |
| b-values [s/mm2] | b50/b600/b900 for all patients | b50/b600/b900 for all patients | b50/b900 for 7 patients; B50/b600/b900 for 15 patients | b50/b600/b900 for all patients | b50/b900 for 9 patients; B50/b600/b900 for 3 patients |
| Number of averages (per b-value) | (2,2,4)–(3,3,5) | (3,3,5) | (3,5)–(3,3,5) | (3,6,6) | (4,4)–(2,2,4) |
| Reconstructed resolution [mm2] | [1.56 × 1.56–1.68 × 1.68] | [1.68 × 1.68–2.5 × 2.5] | [1.68 × 1.68–3.12 × 3.12] | [3.21 × 3.21] | [1.54 × 1.54–1.68 × 1.68] |
| Slice thickness [mm] | 5 | 5 | 6 | 5 | 5 |
| Repetition time [ms] | [6150–12,700] | [5490–10,700] | 12,003 | 6320 | [6150–14,500] |
| Echo time [ms] | [60–79] | 69 | [69–72] | 76 | [66.4–69.6] |
| Inversion time (STIR fat suppression) [ms] | 180 | 180 | 180 | 180 | 180 |
| Flip angle [°] | 90 | 90 | 90 | 90 | 180 |
| Encoding code | 3-scan Trace | 3-scan Trace | 3-scan Trace | 3-scan Trace | 3-scan Trace |
| Field of view [mm] | [98 × 128–256 × 256] | [130 × 160–208 × 256] | [208 × 256–216 × 257] | [108–134] | [208 × 256–224 × 280] |
| Receive bandwidth [Hz/Px] | [1955–2330] | 1955 | 1955 | 2195 | [1984–2330] |
| Loss Name | Definition | Discussion |
|---|---|---|
| Log-cosh Dice | This univariate transformation of the Dice loss, DL, has been suggested for improving medical image segmentation in the context of imbalanced distributions of labels [25]. | |
| Combo | A weighted sum of Dice and binary cross-entropy losses [26]. To identify the optimal weight between these two losses, training/validation of the U-Net model was compared using values of ω from 0 to 1 at increments of 0.1. | |
| Tversky | A generalised version of the Dice loss , this loss provides more nuanced balancing between a requirement for high sensitivity or precision . The best trade-off was investigated by varying the values of α and β, from 0 to 1 with an increment of 0.1 [27]. | |
| Focal Tversky | A further generalisation of the Tversky loss, this loss employs a third parameter γ, which controls the non-linearity of the loss. In class-imbalanced data, small-scale segmentations might result in a high TL score; however, γ > 0 causes a higher gradient loss, forcing the model to focus on harder examples (small regions of interest that do not contribute to the loss significantly) [28]. We varied γ from 1 to 3 with an increment of 0.1 to determine the optimal value. |
| Loss Function | Dice Score | Precision | Recall |
|---|---|---|---|
| Log cosh Dice | 0.865 ± 0.04 | 0.898 ± 0.03 | 0.839 ± 0.08 |
| Combo (ω = 0.8) | 0.858 ± 0.06 | 0.912 ± 0.02 | 0.819 ± 0.104 |
| Tversky (α = 0.7, β = 0.3) | 0.860 ± 0.04 | 0.844 ± 0.03 | 0.883 ± 0.08 |
| Focal Tversky (α = 0.7, β = 0.3, γ = 1.1) | 0.871 ± 0.04 | 0.870 ± 0.03 | 0.878 ± 0.081 |
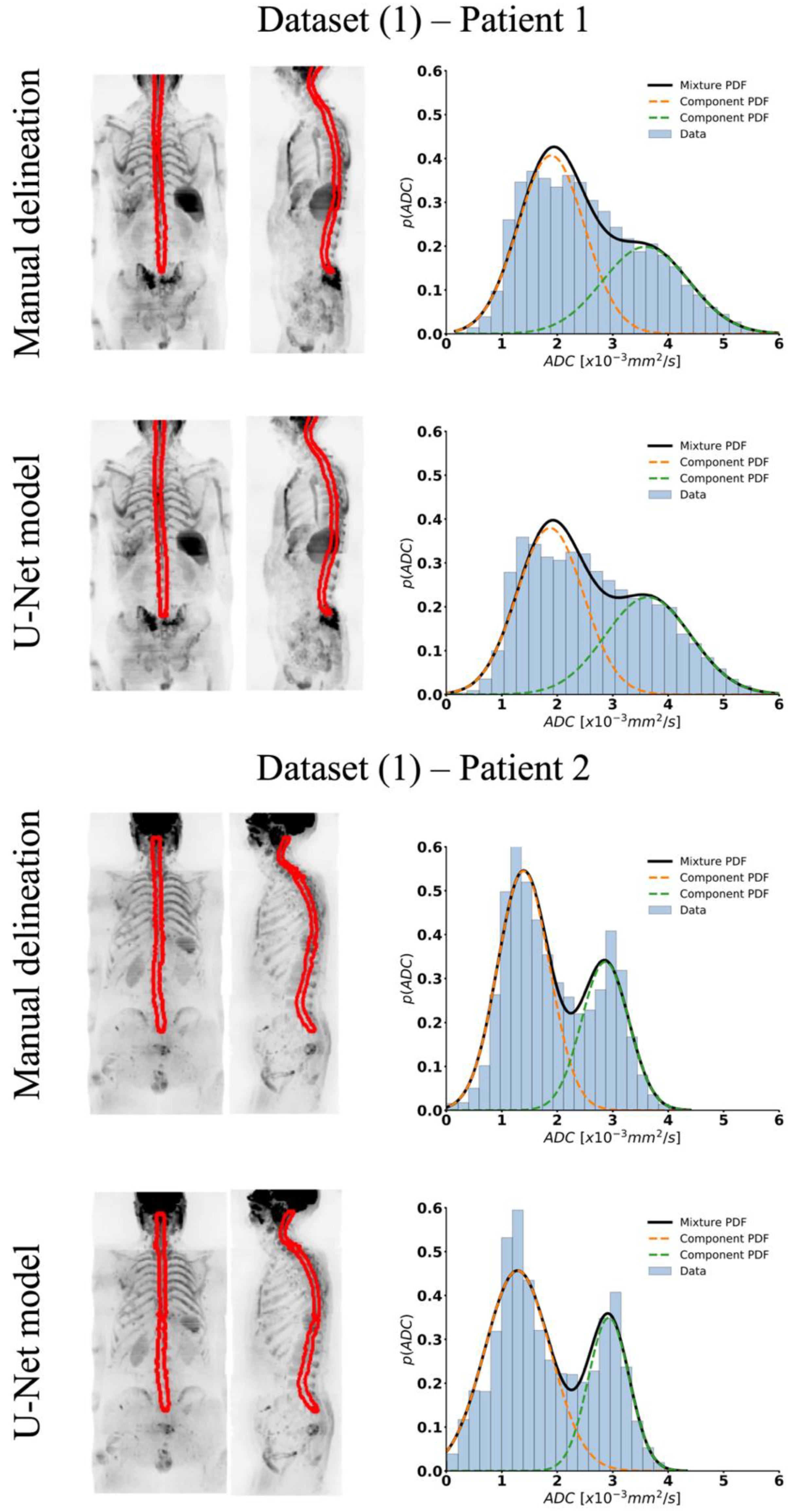
| Volume [mL] | Average Cross-Section Area [mm2] | GMM—Weights | GMM—Means [×10−3 mm2/s] | GMM—Variance | ||||
|---|---|---|---|---|---|---|---|---|
| Validation set (8 patients; 16 WBDWI scans) | 1st comp PDF (spinal cord) | 2nd comp PDF (CSF) | 1st comp PDF (spinal cord) | 2nd comp PDF (CSF) | 1st comp PDF (spinal cord) | 2nd comp PDF (CSF) | ||
| Manual delineation | 152 [138–188] | 176 [151–184] | 0.58 ± 0.09 | 0.41 ± 0.09 | 1.67 ± 0.20 | 3.17 ± 0.31 | 0.38 ± 0.09 | 0.59 ± 0.11 |
| U-Net model | 169 [141–192] | 17 9 [158–192] | 0.61 ± 0.08 | 0.40 ± 0.08 | 1.72 ± 0.13 | 3.17 ± 0.27 | 0.37 ± 0.1 | 0.58 ± 0.12 |
| p-value | 0.91 | 0.94 | 0.31 | 0.31 | 0.04 | 0.12 | 0.69 | 0.16 |
| Volume [mL] | Average Cross-Section Area [mm2] | GMM—Weights | GMM—Means [×10−3 mm2/s] | GMM—Variance | ||||
| Holdout set (8 patients; 16 WBDWI scans) | 1st comp PDF (spinal cord) | 2nd comp PDF (CSF) | 1st comp PDF (spinal cord) | 2nd comp PDF (CSF) | 1st comp PDF (spinal cord) | 2nd comp PDF (CSF) | ||
| Manual delineation | 160 [147–184] | 199 [184–226] | 0.59 ± 0.06 | 0.41 ± 0.06 | 1.70 ± 0.15 | 3.33 ± 0.28 | 0.35 ± 0.07 | 0.56 ± 0.20 |
| U-Net model | 164 [153–175] | 204 [192–213] | 0.60 ± 0.06 | 0.40 ± 0.06 | 1.73 ± 0.13 | 3.36 ± 0.28 | 0.35 ± 0.10 | 0.60 ± 0.19 |
| p-value | 0.26 | 0.28 | 0.67 | 0.67 | 0.14 | 0.30 | 0.73 | 0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Candito, A.; Holbrey, R.; Ribeiro, A.; Messiou, C.; Tunariu, N.; Koh, D.-M.; Blackledge, M.D. Deep Learning for Delineation of the Spinal Canal in Whole-Body Diffusion-Weighted Imaging: Normalising Inter- and Intra-Patient Intensity Signal in Multi-Centre Datasets. Bioengineering 2024, 11, 130. https://doi.org/10.3390/bioengineering11020130
Candito A, Holbrey R, Ribeiro A, Messiou C, Tunariu N, Koh D-M, Blackledge MD. Deep Learning for Delineation of the Spinal Canal in Whole-Body Diffusion-Weighted Imaging: Normalising Inter- and Intra-Patient Intensity Signal in Multi-Centre Datasets. Bioengineering. 2024; 11(2):130. https://doi.org/10.3390/bioengineering11020130
Chicago/Turabian StyleCandito, Antonio, Richard Holbrey, Ana Ribeiro, Christina Messiou, Nina Tunariu, Dow-Mu Koh, and Matthew D. Blackledge. 2024. "Deep Learning for Delineation of the Spinal Canal in Whole-Body Diffusion-Weighted Imaging: Normalising Inter- and Intra-Patient Intensity Signal in Multi-Centre Datasets" Bioengineering 11, no. 2: 130. https://doi.org/10.3390/bioengineering11020130
APA StyleCandito, A., Holbrey, R., Ribeiro, A., Messiou, C., Tunariu, N., Koh, D.-M., & Blackledge, M. D. (2024). Deep Learning for Delineation of the Spinal Canal in Whole-Body Diffusion-Weighted Imaging: Normalising Inter- and Intra-Patient Intensity Signal in Multi-Centre Datasets. Bioengineering, 11(2), 130. https://doi.org/10.3390/bioengineering11020130






