Artificial Intelligence Models for the Automation of Standard Diagnostics in Sleep Medicine—A Systematic Review
Abstract
1. Introduction
1.1. Training, Testing, and Validation
1.2. Deep Learning and Neural Networks
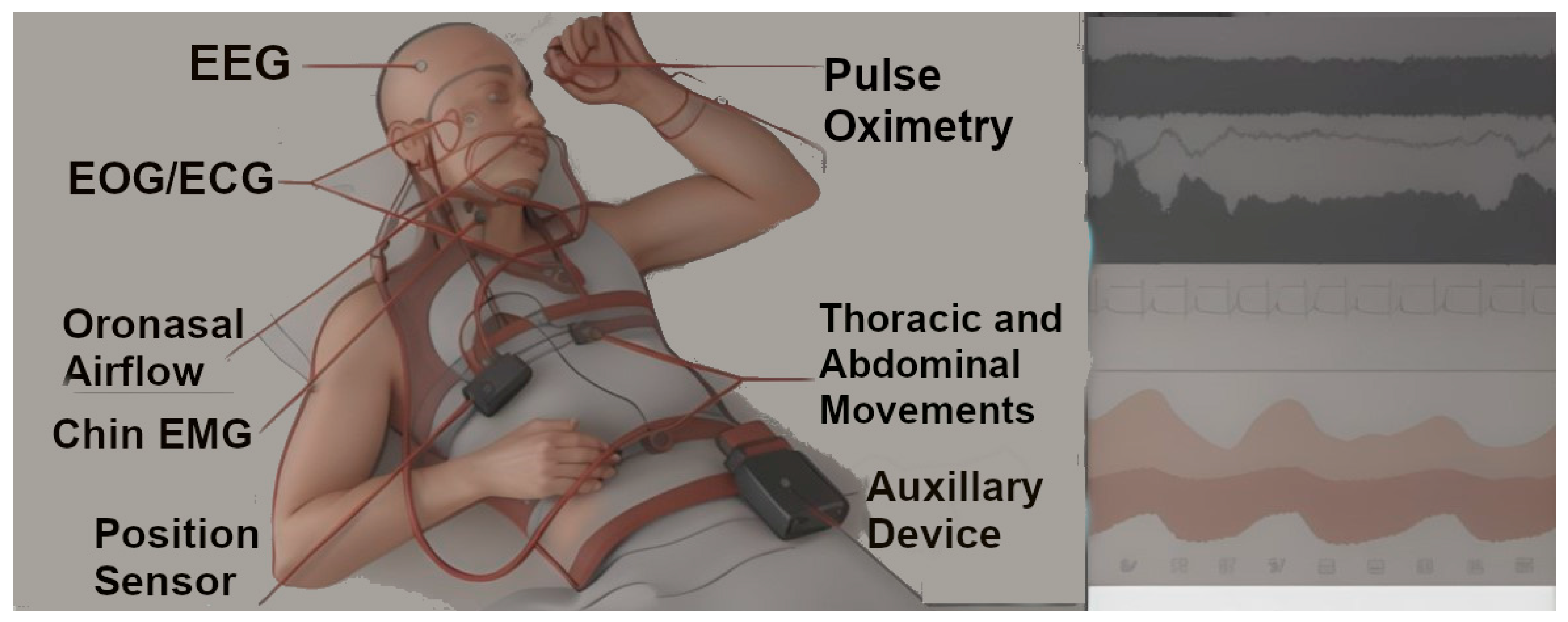
1.3. Sleep Staging and Cortical Arousals
1.4. Sleep Disorders
2. Methods
2.1. Search Strategy
2.2. Selection Criteria
- Trained and validated with data from gold-standard diagnostic modalities for sleep staging or the diagnosis of sleep disorders. Standard diagnostics include PSG-based sleep staging, the detection of OSA using single-lead EKG signals, and a PSG-based detection of sleep disorders using AASM diagnostic criteria.
- Developed using separate training, validation, and testing datasets.
- Internally or externally validated on clinical datasets.
2.3. Screening
2.4. Data Extraction
- Article information including the name of the first author and year of publication.
- Application of the reported AI model, based on which the studies were grouped into two groups (see Section 2.3).
- Specifics of the reported AI model, including model architecture, classification tasks performed by the model, and features used for classifying the input data.
- Composition of the training, validation, and testing datasets, including the proportion of sleep studies used in each of the three datasets, characteristics of the included patients, and the sleep study setup.
- Performance metrics of the model on the testing dataset, including the following:
- Agreement of the model with consensus manual scoring (measured using Cohen’s kappa), and/or accuracy of the model, and/or the F1 score for automated sleep staging models.
- Sensitivity, specificity, and/or other reported metrics for sleep disorder detection models.
- The full-text articles were reviewed, and relevant details about model design, population, and model performance were extracted.
3. Results
3.1. Number of Screened and Selected Studies
3.2. Datasets Used for Model Development
3.3. Automated Sleep Staging and Sleep Disorder Detection Models
3.4. Deep Learning for Sleep Staging and Cortical Arousals
3.4.1. Sleep Staging
3.4.2. Cortical Arousals
3.5. Deep Learning for Detection of Sleep Disorders
3.5.1. Obstructive Sleep Apnea
3.5.2. REM Sleep Behavior Disorder
3.5.3. Narcolepsy
3.5.4. Periodic Limb Movements of Sleep
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Huyett, P.; Bhattacharyya, N. Incremental Health Care Utilization and Expenditures for Sleep Disorders in the United States. J. Clin. Sleep Med. 2021, 17, 1981–1986. [Google Scholar] [CrossRef] [PubMed]
- National Sleep Foundation. Sleep in America® 2020 Poll. Available online: http://www.thensf.org/wp-content/uploads/2020/03/SIA-2020-Report.pdf (accessed on 12 November 2023).
- Buysse, D.J. Insomnia. JAMA 2013, 309, 706–716. [Google Scholar] [CrossRef] [PubMed]
- Sateia, M.J. International Classification of Sleep Disorders-Third Edition. Chest 2014, 146, 1387–1394. [Google Scholar] [CrossRef]
- Chiao, W.; Durr, M.L. Trends in sleep studies performed for Medicare beneficiaries. Laryngoscope 2017, 127, 2891–2896. [Google Scholar] [CrossRef] [PubMed]
- Nextech Healthcare Data Growth: An Exponential Problem. Available online: https://www.nextech.com/blog/healthcare-data-growth-an-exponential-problem (accessed on 12 November 2023).
- New Scientist. Alan Turing—The Father of Modern Computer Science. Available online: https://www.newscientist.com/people/alan-turing/ (accessed on 5 November 2023).
- McCarthy, J.; Minsky, M.L.; Rochester, N.; Shannon, C.E. A Proposal for the Dartmouth Summer Research Project on Artificial Intelligence, August 31, 1955. AI Mag. 2006, 27, 12. [Google Scholar] [CrossRef]
- Kim, M.-S.; Park, H.-Y.; Kho, B.-G.; Park, C.-K.; Oh, I.-J.; Kim, Y.-C.; Kim, S.; Yun, J.-S.; Song, S.-Y.; Na, K.-J.; et al. Artificial Intelligence and Lung Cancer Treatment Decision: Agreement with Recommendation of Multidisciplinary Tumor Board. Transl. Lung Cancer Res. 2020, 9, 507–514. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.; Goldstein, C. Clinical applications of artificial intelligence in sleep medicine: A sleep clinician’s perspective. Sleep Breath. 2023, 27, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Young, T.; Palta, M.; Dempsey, J.; Peppard, P.; Nieto, F.; Hla, K. Burden of Sleep Apnea: Rationale, Design, and Major Findings of the Wisconsin Sleep Cohort Study. WMJ 2009, 108, 246–249. [Google Scholar]
- Simpson, L.; Hillman, D.R.; Cooper, M.N.; Ward, K.L.; Hunter, M.; Cullen, S.; James, A.; Palmer, L.J.; Mukherjee, S.; Eastwood, P. High Prevalence of Undiagnosed Obstructive Sleep Apnoea in the General Population and Methods for Screening for Representative Controls. Sleep Breath. 2013, 17, 967–973. [Google Scholar] [CrossRef]
- Laugsand, L.E.; Vatten, L.J.; Platou, C.; Janszky, I. Insomnia and the Risk of Acute Myocardial Infarction: A Population Study. Circulation 2011, 124, 2073–2081. [Google Scholar] [CrossRef]
- Sawadogo, W.; Adera, T.; Alattar, M.; Perera, R.; Burch, J.B. Association Between Insomnia Symptoms and Trajectory With the Risk of Stroke in the Health and Retirement Study. Neurology 2023, 101, e475–e488. [Google Scholar] [CrossRef]
- Baglioni, C.; Battagliese, G.; Feige, B.; Spiegelhalder, K.; Nissen, C.; Voderholzer, U.; Lombardo, C.; Riemann, D. Insomnia as a Predictor of Depression: A Meta-Analytic Evaluation of Longitudinal Epidemiological Studies. J. Affect. Disord. 2011, 135, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Watson, N.F.; Rosen, I.M.; Chervin, R.D. The Past Is Prologue: The Future of Sleep Medicine. J. Clin. Sleep Med. 2017, 13, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.B.; Fung, G.; Rosales, R. On the dangers of cross-validation. An experimental evaluation. In Proceedings of the 2008 SIAM International Conference on Data Mining, Atlanta, GA, USA, 24–26 April 2008; pp. 588–596. [Google Scholar]
- Nowotny, T. Two Challenges of Correct Validation in Pattern Recognition. Front. Robot. AI 2014, 1, 5. [Google Scholar] [CrossRef]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Phan, H.; Andreotti, F.; Cooray, N.; Chén, O.Y.; De Vos, M. SeqSleepNet: End-to-End Hierarchical Recurrent Neural Network for Sequence-to-Sequence Automatic Sleep Staging. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Kryger, M.; Roth, T.; Dement, W.C. (Eds.) Principles and Practice of Sleep Medicine, 6th ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 15–17. [Google Scholar]
- Thomas, R.J. Cyclic alternating pattern and positive airway pressure titration. Sleep Med. 2002, 3, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Eckert, D.J.; White, D.P.; Jordan, A.S.; Malhotra, A.; Wellman, A. Defining Phenotypic Causes of Obstructive Sleep Apnea. Identification of Novel Therapeutic Targets. Am. J. Respir. Crit. Care Med. 2013, 188, 996–1004. [Google Scholar] [CrossRef]
- Drazen, E.; Mann, N.; Borun, R.; Laks, M.; Bersen, A. Survey of computer-assisted electrocardiography in the United States. J. Electrocardiol. 1988, 21, S98–S104. [Google Scholar] [CrossRef]
- Galbiati, A.; Verga, L.; Giora, E.; Zucconi, M.; Ferini-Strambi, L. The Risk of Neurodegeneration in REM Sleep Behavior Disorder: A Systematic Review and Meta-Analysis of Longitudinal Studies. Sleep Med. Rev. 2019, 43, 37–46. [Google Scholar] [CrossRef]
- Barateau, L.; Pizza, F.; Plazzi, G.; Dauvilliers, Y. Narcolepsy. J. Sleep Res. 2022, 31, e13631. [Google Scholar] [CrossRef]
- Trotti, L.M. Twice Is Nice? Test-Retest Reliability of the Multiple Sleep Latency Test in the Central Disorders of Hypersomnolence. J. Clin. Sleep Med. 2020, 16, 17–18. [Google Scholar] [CrossRef]
- Koo, B.B.; Sillau, S.; Dean, D.A.; Lutsey, P.L.; Redline, S. Periodic Limb Movements During Sleep and Prevalent Hypertension in the Multi-Ethnic Study of Atherosclerosis. Hypertension 2015, 65, 70–77. [Google Scholar] [CrossRef]
- Cesari, M.; Stefani, A.; Penzel, T.; Ibrahim, A.; Hackner, H.; Heidbreder, A.; Szentkirályi, A.; Stubbe, B.; Völzke, H.; Berger, K.; et al. Interrater Sleep Stage Scoring Reliability between Manual Scoring from Two European Sleep Centers and Automatic Scoring Performed by the Artificial Intelligence-Based Stanford-STAGES Algorithm. J. Clin. Sleep Med. 2021, 17, 1237–1247. [Google Scholar] [CrossRef]
- Wallis, P.; Yaeger, D.; Kain, A.; Song, X.; Lim, M. Automatic Event Detection of REM Sleep Without Atonia From Polysomnography Signals Using Deep Neural Networks. In Proceedings of the ICASSP 2020—2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 4112–4116. [Google Scholar]
- Kuan, Y.C.; Hong, C.T.; Chen, P.C.; Liu, W.T.; Chung, C.C. Logistic Regression and Artificial Neural Network-Based Simple Predicting Models for Obstructive Sleep Apnea by Age, Sex, and Body Mass Index. Math. Biosci. Eng. 2022, 19, 11409–11421. [Google Scholar] [CrossRef] [PubMed]
- Stephansen, J.B.; Olesen, A.N.; Olsen, M.; Ambati, A.; Leary, E.B.; Moore, H.E.; Carrillo, O.; Lin, L.; Han, F.; Yan, H.; et al. Neural Network Analysis of Sleep Stages Enables Efficient Diagnosis of Narcolepsy. Nat. Commun. 2018, 9, 5229. [Google Scholar] [CrossRef] [PubMed]
- Biswal, S.; Sun, H.; Goparaju, B.; Westover, M.B.; Sun, J.; Bianchi, M.T. Expert-level sleep scoring with deep neural networks. J. Am. Med. Inform. Assoc. 2018, 25, 1643–1650. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.; Lorenzen, K.P.; Heremans, E.; Chén, O.Y.; Tran, M.C.; Koch, P.; Mertins, A.; Baumert, M.; Mikkelsen, K.B.; Vos, M.D. L-SeqSleepNet: Whole-Cycle Long Sequence Modelling for Automatic Sleep Staging. IEEE J. Biomed. Health Inform. 2023, 27, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bakker, J.P.; Ross, M.; Cerny, A.; Vasko, R.; Shaw, E.; Kuna, S.; Magalang, U.J.; Punjabi, N.M.; Anderer, P. Scoring Sleep with Artificial Intelligence Enables Quantification of Sleep Stage Ambiguity: Hypnodensity Based on Multiple Expert Scorers and Auto-Scoring. Sleep 2023, 46, zsac154. [Google Scholar] [CrossRef] [PubMed]
- Brink-Kjaer, A.; Olesen, A.N.; Peppard, P.E.; Stone, K.L.; Jennum, P.; Mignot, E.; Sorensen, H.B.D. Automatic Detection of Cortical Arousals in Sleep and Their Contribution to Daytime Sleepiness. Clin. Neurophysiol. 2020, 131, 1187–1203. [Google Scholar] [CrossRef]
- Pourbabaee, B.; Patterson, M.H.; Patterson, M.R.; Benard, F. SleepNet: Automated Sleep Analysis via Dense Convolutional Neural Network Using Physiological Time Series. Physiol. Meas. 2019, 40, 84005. [Google Scholar] [CrossRef]
- Li, A.; Chen, S.; Quan, S.F.; Powers, L.S.; Roveda, J.M. A Deep Learning-Based Algorithm for Detection of Cortical Arousal during Sleep. Sleep 2020, 43, zsaa120. [Google Scholar] [CrossRef]
- Patanaik, A.; Ong, J.L.; Gooley, J.J.; Ancoli-Israel, S.; Chee, M.W.L. An End-to-End Framework for Real-Time Automatic Sleep Stage Classification. Sleep 2018, 41, zsy041. [Google Scholar] [CrossRef] [PubMed]
- Olesen, A.N.; Jennum, P.J.; Mignot, E.; Sorensen, H.B.D. Automatic Sleep Stage Classification with Deep Residual Networks in a Mixed-Cohort Setting. Sleep 2021, 44, zsaa161. [Google Scholar] [CrossRef] [PubMed]
- Abou Jaoude, M.; Sun, H.; Pellerin, K.R.; Pavlova, M.; Sarkis, R.A.; Cash, S.S.; Westover, M.B.; Lam, A.D. Expert-Level Automated Sleep Staging of Long-Term Scalp Electroencephalography Recordings Using Deep Learning. Sleep 2020, 43, zsaa112. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xu, M.; Li, Y.; Su, M.; Xu, Z.; Wang, C.; Kang, D.; Li, H.; Mu, X.; Ding, X.; et al. Automated Multi-Model Deep Neural Network for Sleep Stage Scoring with Unfiltered Clinical Data. Sleep Breath. 2020, 24, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Estevez, D.; Rijsman, R.M. Inter-Database Validation of a Deep Learning Approach for Automatic Sleep Scoring. PLoS ONE 2021, 16, e0256111. [Google Scholar] [CrossRef] [PubMed]
- Guillot, A.; Thorey, V. RobustSleepNet: Transfer Learning for Automated Sleep Staging at Scale. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 1441–1451. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Fujiwara, K.; Nakayama, C.; Sumi, Y.; Kano, M.; Nagamoto, T.; Kadotani, H. R-R Interval-Based Sleep Apnea Screening by a Recurrent Neural Network in a Large Clinical Polysomnography Dataset. Clin. Neurophysiol. 2022, 139, 80–89. [Google Scholar] [CrossRef]
- Carvelli, L.; Olesen, A.N.; Brink-Kjær, A.; Leary, E.B.; Peppard, P.E.; Mignot, E.; Sørensen, H.B.D.; Jennum, P. Design of a Deep Learning Model for Automatic Scoring of Periodic and Non-Periodic Leg Movements during Sleep Validated against Multiple Human Experts. Sleep Med. 2020, 69, 109–119. [Google Scholar] [CrossRef]
- Quan, S.F.; Howard, B.V.; Iber, C.; Kiley, J.P.; Nieto, F.J.; O’Connor, G.T.; Rapoport, D.M.; Redline, S.; Robbins, J.; Samet, J.M.; et al. The Sleep Heart Health Study: Design, Rationale, and Methods. Sleep 1997, 20, 1077–1085. [Google Scholar] [CrossRef] [PubMed]
- Kemp, B.; Zwinderman, A.H.; Tuk, B.; Kamphuisen, H.A.C.; Oberye, J.J.L. Analysis of a Sleep-Dependent Neuronal Feedback Loop: The Slow-Wave Microcontinuity of the EEG. IEEE Trans. Biomed. Eng. 2000, 47, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Andlauer, O.; Moore, H., IV; Jouhier, L.; Drake, C.; Peppard, P.E.; Han, F.; Hong, S.-C.; Poli, F.; Plazzi, G.; O’Hara, R.; et al. Nocturnal Rapid Eye Movement Sleep Latency for Identifying Patients With Narcolepsy/Hypocretin Deficiency. JAMA Neurol. 2013, 70, 891–902. [Google Scholar] [CrossRef] [PubMed]
- Khalighi, S.; Sousa, T.; Santos, J.M.; Nunes, U. ISRUC-Sleep: A Comprehensive Public Dataset for Sleep Researchers. Comput. Methods Programs Biomed. 2016, 124, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Anderer, P.; Gruber, G.; Parapatics, S.; Woertz, M.; Miazhynskaia, T.; Klösch, G.; Saletu, B.; Zeitlhofer, J.; Barbanoj, M.J.; Danker-Hopfe, H.; et al. An E-Health Solution for Automatic Sleep Classification According to Rechtschaffen and Kales: Validation Study of the Somnolyzer 24 × 7 Utilizing the Siesta Database. Neuropsychobiology 2005, 51, 115–133. [Google Scholar] [CrossRef]
- Mainali, S.; Park, S. Artificial Intelligence and Big Data Science in Neurocritical Care. Crit. Care Clin. 2023, 39, 235–242. [Google Scholar] [CrossRef]
- Mainali, S.; Darsie, M.E.; Smetana, K.S. Machine Learning in Action: Stroke Diagnosis and Outcome Prediction. Front. Neurol. 2021, 12, 734345. [Google Scholar] [CrossRef]
- Goldstein, C.A.; Berry, R.B.; Kent, D.T.; Kristo, D.A.; Seixas, A.A.; Redline, S.; Westover, M.B. Artificial Intelligence in Sleep Medicine: Background and Implications for Clinicians. J. Clin. Sleep Med. 2020, 16, 609–618. [Google Scholar] [CrossRef]
- Goldstein, C.A.; Berry, R.B.; Kent, D.T.; Kristo, D.A.; Seixas, A.A.; Redline, S.; Westover, M.B.; Abbasi-Feinberg, F.; Aurora, R.N.; Carden, K.A.; et al. Artificial intelligence in sleep medicine: An American Academy of Sleep Medicine position statement. J. Clin. Sleep Med. 2020, 16, 605–607. [Google Scholar] [CrossRef]


| Term | Definition | Example |
|---|---|---|
| Iterations | Repetitive cycles where an ML model practices making predictions using training data. | Like a basketball player practicing free throws multiple times to improve. |
| Features | Traits or details in the data that help in categorizing or analyzing the data. | In a smartphone camera’s photo-sorting app, features might include color, brightness, or the presence of faces to categorize images into different albums. |
| Cross-validation | It is technique in statistical analysis and Machine Learning with which a dataset is divided into multiple parts. These parts are then used interchangeably as training and testing sets to validate the accuracy and generalizability of a model. This process helps in assessing how well a model will perform on an independent dataset and in preventing overfitting. | Like a coach dividing the team into several groups and having each group play both roles of the main team and the opponent in different matches. This helps the coach understand how well the team adapts to different scenarios. However, this does not guarantee that the team will perform equally well against actual external opponents |
| Overfitting | When an ML model learns the training data too well but struggles with new data. | A student who memorizes answers for a specific set of questions for a history test cannot apply the knowledge to new, unseen questions on the same topic. |
| Underfitting | This occurs when an ML model is not complex enough to capture the underlying patterns and relationships in the training data, leading to poor performances on both training and new, unseen data. | Like trying to understand a complex novel by only reading the summary, and as a result, being unable to grasp the full story or discuss it in detail. |
| Weight | The importance given to a message passed between neurons in a neural network. | Similar to adjusting the balance on a music mixer to control how much each instrument contributes to the overall sound of a band. |
| Biases | In a neural network, “bias” is an adjustable parameter that enables the model to modify its output independently of the input data, playing a crucial role in determining neuron activation and thus influencing the model’s overall behavior and accuracy. | In a job application screening system trained mostly on resumes from a few top universities, there may be a bias favoring applicants from those schools, potentially overlooking equally qualified candidates from other institutions. |
| Activation Function | An activation function in a neural network is a mathematical formula that determines whether and to what extent a neuron should be activated, based on the input it receives. It helps the network make non-linear decisions, allowing it to handle complex data patterns. | In a photo-filtering app, an activation function might decide how strongly a certain feature, like brightness or color saturation, should influence whether a photo is categorized as “outdoor” or “indoor”. |
| Loss Function | How much the model’s prediction differs from the actual result. | The difference between a GPS’s estimated time of arrival and the actual time you reach your destination. |
| Gradient | The gradient refers to the measure of change in the network’s error (or loss) in response to adjustments in its weights and biases. | How changing the amount of sugar in a cake recipe affects its sweetness when the goal is to find the ideal sweetness (accuracy) of the product. |
| Backpropagation | A process that calculates how each part of the network contributed to the error. | Analyzing which step in a baking recipe went wrong when the cake does not rise. |
| Gradient Descent | A method to find the best weights and biases for the lowest error in a network. | Searching for the perfect oven temperature and baking time for the ideal cake texture. |
| Dataset | Characteristics of Included Subjects | Number of Recordings | PSG Setup |
|---|---|---|---|
| Massachusetts General Hospital (MGH)-PSG [33] | Symptomatic | 10,000 (10,000 subjects) | Six-channel EEG, EOG, chin and leg EMG, EKG, SaO2, chest and abdomen movement sensors, nasal airflow, pressure transducer (PTAF), position |
| Sleep Heart Health Study (SHHS) [47] | Adults over 40 with and without a history of snoring who were previously recruited to epidemiological cohort studies on cardiovascular health | 9376 (5804 subjects) | Two-channel EEG, EOG, EKG, SpO2, chest and abdomen movement sensors, nasal airflow |
| Sleep EDF (expanded) [48] | Symptomatic (difficulty falling asleep) subjects and healthy controls | 197 (96 subjects) | Two-channel EEG, EOG, chin EMG, SaO2, chest and abdomen movement sensors, nasal airflow |
| Stanford Sleep Cohort (SSC) [49] | Symptomatic | 760 (760 subjects) | Six-channel EEG, EOG, chin and leg EMG, EKG, SaO2, chest and abdomen movement sensors, nasal airflow |
| Wisconsin Sleep Cohort (WSC) [11] | Combination of subjects at high and low risk for OSA based on questionnaire responses | 2570 (1549 subjects at baseline, ongoing follow-up) | Two-channel EEG (six channels in select recordings), EOG, chin and leg EMG, EKG, SaO2, chest and abdomen movement sensors, nasal and oral airflow, nasal pressure, position |
| Institute of Systems and Robotics—University of Coimbra (ISRUC) [50] | Symptomatic patients with diagnosed sleep disorders and healthy controls | 126 (118 subjects) | Six-channel EEG, EOG, chin and leg EMG, SaO2, chest movement sensor, nasal airflow |
| First Author, Year | Type of Neural Network | Training and Validation Datasets | Testing Datasets | Comments |
|---|---|---|---|---|
| Patanaik, 2018 [39] | Deep CNN | 1046 PSGs of healthy adolescents (DS1) * 284 PSGs of healthy adults (DS2) 75% for training and 25% for validation | 210 PSGs of adolescent and adult patients with suspected sleep disorders (DS3) 77 PSGs of Parkinson’s disease patients, 42% of whom were classified as having REM sleep behavior disorder (RBD) and 28% as probably having RBD (DS4) | Trained on data from healthy subjects, tested on data from patients with suspected sleep disorders and RBD |
| Biswal, 2018 [33] | RCNN | 9000 training and validation PSGs from the MGH dataset 5224 from the SHHS dataset | 1000 held out PSGs from MGH 580 held out PSGs from SHSS | |
| Stephansen, 2018 [32] | Ensemble of Cross-Correlation (CC) encoded CNN models (Stanford STAGES model) | 3507 (90% training, 10% validation) from the WSC, SSC, and KHC datasets | 70 PSGs scored manually by six scorers from the IS-RC cohort (staging) | |
| Olesen, 2020 [40] | CNN (feature extraction) + RNN (staging) | 15,684 PSGs from ISRUC, MrOS, SHHS, SSC, and WSC datasets (87.5% training, 2.5% validation, 10% testing) | 10% PSGs held out from the same datasets | Same accuracy obtained using 75% training data (four out of five cohorts) as 100% (five out of five cohorts) |
| Abou Jaoude, 2020 [41] | Multi-modal DNN | 5041 PSGs for training 650 for validation From the MGH-PSG dataset | HomePAP—243 PSGs ABC—129 PSGs MGH-PSG—650 PSGs | Developed a PSG-based staging model (CRNN-PSG) and fine-tuned it into a scalp EEG-based staging model (CRNN-EEG); only the performance of the PSG-based model is considered |
| Zhang, 2020 [42] | LSTM-RNN | 122 training and 20 validation PSGs of adults with a history of snoring performed at Beijing Tongren Hospital | 152 PSGs from the study dataset 40 PSGs from 20 SC subjects from the SleepEDF dataset | Compared performance of staging model between participants with and without OSA |
| Alvarez-Estevez, 2021 [43] | CNN | 354 PSGs (80% of a total 443) from six datasets further split into 80% for training and 20% for validation | 89 PSGs (20% of a total 443) held out from the six study datasets that included ISRUC and SHHS | |
| Cesari, 2021 [29] | Ensemble of (CC) encoded CNN models (Stanford STAGES model) | 1066 PSGs from the Study of Health in Pomerania-TREND | Externally validated, previously developed Stanford STAGES model (see above) | |
| Guillot, 2021 [44] | RNN | 5788 total PSGs from eight datasets including the MASS, SleepEDF, MrOS, and SHHS datasets Did not specify the split between the training, validation, and testing sets | Testing dataset was unseen during training in a Direct Transfer (DT) setting Both of the other settings required the use of cross-validation (CV); their performance was not considered for the review | Designed to classify sequences of multiple epochs at once PSG settings varied across datasets and included 2, 3, 5, 8, and 12 lead EEGs with or without EMG. |
| Bakker, 2023 [35] | Bi-directional LSTM-RNN (Somnolyzer) | 588 PSGs from SIESTA dataset | 95 PSGs from three clinical datasets separate from the training set; datasets A (n = 70), B (n = 15), and C (n = 10) were scored by 6, 9, and 12 scorers, respectively. | |
| Phan, 2023 [34] | Hierarchical RNN | 3824 (70% of 5463) PSGs from SHHS dataset with 100 PSGs held out for validation | 1639 (30% of 5463) PSGs from the SHHS dataset |
| First Author, Year | Type of Neural Network | Model Performance on Testing Dataset | ||
|---|---|---|---|---|
| Cohen’s Kappa | Accuracy | F1 Score | ||
| Patanaik, 2018 [39] | Deep CNN | 0.740 (DS3); 0.597 (DS4) | 81.4% (DS3); 72.1% (DS4) | |
| Biswal, 2018 [33] | RCNN | 80.5 (MGH); 73.2 (SHSS) | 87.5% (MGH); 77.7% (SHSS) | |
| Stephansen, 2018 [32] | CNN | 57.7 ± 6.1 (Unbiased overall Cohen’s kappa across six scorers) | 86.8 ± 4.3 | |
| Olesen, 2020 [40] | CNN + RNN | 0.728, 95%CI: 0.726–0.731 (Mean of all combinations of four training and one validation cohorts) | ||
| Abou Jaoude, 2020 [41] | Multi-modal DNN | 0.64 (For CRNN-PSG model on both testing datasets) | ||
| Zhang, 2020 [42] | LSTM-RNN | 0.7276 (study dataset) 0.77 for (SleepEDF) | 0.8181 (study dataset); 0.836 (SleepEDF) | 0.8150 (study dataset); 0.781 (SleepEDF) |
| Alvarez-Estevez, 2021 [43] | CNN | 0.63 (Average kappa on external datasets) | ||
| Cesari, 2021 [29] | CNN | 0.61 ± 0.14 (Overall manual vs. auto scoring for both datasets) | ||
| Guillot, 2021 [44] | RNN | 64.9 to 84.4 across eight datasets 84.4 for DOD-H dataset (12-lead EEG) | ||
| Bakker, 2023 [35] | LSTM-RNN | 0.78 ± 0.01 compared to unbiased consensus of scorers for auto-scoring model (vs. 0.69 ± 0.063 for best manual scorer) | ||
| Phan, 2023 [34] | RNN | 0.838 (SHHS) | ||
| First Author, Year | Type of Neural Network | Disease Classified (Present vs. Absent) | Features | Training and Validation Datasets | Testing Dataset |
|---|---|---|---|---|---|
| Stephansen, 2018 [32] | Ensemble of Cross-Correlation (CC) encoded CNN models (used as biomarker of narcolepsy) | Narcolepsy Type 1 | Hypnodensity of sleep stages, sleep latency, REM latency, SOREMPs *, etc. | 645 training, 445 validation PSGs from seven cohorts: WSC, SSC, KHC, AHC, JCTS, IHC, and DHC | 321 PSGs from two cohorts never seen by the model: FHC and CNC |
| Iwasaki, 2022 [45] | LSTM-RNN | Severe OSA (AHI ≥ 30); Moderate-to-severe OSA (AHI ≥ 15) | Ratio of total apnea to total sleep duration (AS ratio) based on RR intervals labeled as apneic or normal | 938 adolescent (>12 yrs) and adult PSGs (468 training, 470 validation) performed at the Nakamura clinic in Okinawa, Japan | SUMS dataset—N = 59 PhysioNet dataset—N = 35 (34 PSGs used to determine AS ratio threshold) |
| Kuan, 2022 [31] | ANN | Moderate-to-severe OSA | Age, sex, BMI | 7328 manually scored full-night PSGs of adult patients who had not been previously treated for OSA | 2094 held out PSGs from the same lab |
| Pourbabaee, 2019 [37] | Dense Recurrent CNN (DRCNN) with bi-directional LSTM | Primary classification task: non-apnea/hypopnea arousal (target arousal) vs. apnea/hypopnea vs. normal sleep vs. wake | EEG, EMG, EOG, SpO2 | 1985 PSGs from the MGH dataset: 794 training, 100 validation, 100 testing | 989 held out PSGs from the same dataset |
| Brink-Kjaer, 2020 [36] | LSTM-RNN | Cortical arousal vs. sleep; wake vs. sleep | EEG, EOG, EMG | Training: 2889 Home Sleep Test (HST) from MrOS, CFS Validation: 996 PSGs from MrOS, CFS and in-lab PSGs from WSC | 30 unseen PSGs, each from SSC (clinical) and WSC annotated by nine sleep technicians |
| Li, 2020 [38] | LSTM-RNN | Cortical arousal detection | Single-lead EKG signal | MESA cohort: 1112 training and 124 test PSGs SHHS cohort: 1058 training and 118 test PSGs | 311 unseen PSGs from MESA and 785 from SHHS |
| Wallis, 2020 [30] | 1D-CNN with residual connections | REM sleep with Atonia | Amplitude and sustained duration of chin and leg EMG compared to predefined baseline for short phasic (P) bursts and longer tonic (T) events; EEG and EOG for staging | 554 training and 60 validation in-hospital PSGs manually scored and annotated for T and P events | 78 unseen PSGs from the same dataset |
| Carvelli, 2020 [46] | LSTM-RNN | Limb movement score calculation | Left/right anterior tibialis (LAT/RAT) EMG signal used to calculate Periodic Limb Movements as per AASM criteria; EKG signal used to filter out artifacts in LAT/RAT signal | 655 training and 53 test PSGs from the MrOS, WSC, and SSC datasets manually scored and annotated for limb movements | 92 unseen PSGs from the same datasets |
| Biswal, 2018 [33] | RCNN | OSA detection Limb Movement Detection | Chest, abdomen movements, SaO2 (for AHI) LAT/RAT EMG (for limb movements) | 9000 training and testing PSGs from the MGH dataset; 5224 from the SHHS dataset | 1000 held out PSGs from MGH 580 held out PSGs from SHSS |
| First Author, Year | Type of Neural Network | Disease Classified (Present vs. Absent) | Model Performance |
|---|---|---|---|
| Stephansen, 2018 [32] | CNN | Narcolepsy Type 1 | Sensitivity = 93%; Specificity = 91%; Accuracy = 0.92; PPV = 0.87; NPV = 0.95; for narcolepsy biomarker in never-seen replication sample Sensitivity = 90%; Specificity = 92%; for narcolepsy biomarker in HPT cohort |
| Iwasaki, 2022 [45] | LSTM-RNN | Severe OSA (AHI ≥ 30); Moderate-to-severe OSA (AHI ≥ 15) | For the detection of moderate-to-severe OSA (AHI ≥ 15): SUMS dataset: AUC = 0.93; Sensitivity = 0.92; Specificity = 0.89; PhysioNet dataset: AUC = 0.95; Sensitivity = 0.95; Specificity = 0.86 |
| Kuan, 2022 [31] | ANN | Moderate-to-severe OSA | For predicting moderate-to-severe OSA: Accuracy = 76.4%; Sensitivity = 87.7%; Specificity = 56.9%; PPV = 77.7%; NPV = 73.0% |
| Pourbabaee, 2019 [37] | Dense Recurrent CNN (DRCNN) with bi-directional LSTM | Primary classification task: non-apnea/hypopnea arousal (target arousal) vs. apnea/hypopnea vs. normal sleep vs. wake | AUROC = 0.931 AUPRC = 0.543 (For non-apnea/hypopnea arousal detection on blind test set) |
| Brink-Kjaer, 2020 [36] | LSTM-RNN | Cortical arousal vs. sleep; wake vs. sleep | Mean F1 score of the model for arousal detection in unseen testing dataset = 0.76 Mean precision = 0.72; Recall = 0.81; AUPRC = 0.82 |
| Li, 2020 [38] | LSTM-RNN | Cortical arousal detection | AUPRC = 0.39 AUROC = 0.86 For model trained on MESA and validated on SHHS |
| Wallis, 2020 [30] | CNN | REM sleep with Atonia | Balanced accuracy (BAC) = 0.91; Cohen’s Kappa 0.68 |
| Carvelli, 2020 [46] | LSTM-RNN | Limb movement score calculation | Maximum F1 score = 0.770 ± 0.049 for LM and 0.757 ± 0.050 for PLMS |
| Biswal, 2018 [33] | RCNN | OSA detection Limb movement detection | r2 for AHI scoring = 0.85; r2 for limb movement detection = 0.7 (MGH dataset only) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alattar, M.; Govind, A.; Mainali, S. Artificial Intelligence Models for the Automation of Standard Diagnostics in Sleep Medicine—A Systematic Review. Bioengineering 2024, 11, 206. https://doi.org/10.3390/bioengineering11030206
Alattar M, Govind A, Mainali S. Artificial Intelligence Models for the Automation of Standard Diagnostics in Sleep Medicine—A Systematic Review. Bioengineering. 2024; 11(3):206. https://doi.org/10.3390/bioengineering11030206
Chicago/Turabian StyleAlattar, Maha, Alok Govind, and Shraddha Mainali. 2024. "Artificial Intelligence Models for the Automation of Standard Diagnostics in Sleep Medicine—A Systematic Review" Bioengineering 11, no. 3: 206. https://doi.org/10.3390/bioengineering11030206
APA StyleAlattar, M., Govind, A., & Mainali, S. (2024). Artificial Intelligence Models for the Automation of Standard Diagnostics in Sleep Medicine—A Systematic Review. Bioengineering, 11(3), 206. https://doi.org/10.3390/bioengineering11030206








