Segmentation of Variants of Nuclei on Whole Slide Images by Using Radiomic Features
Abstract
1. Introduction
- The proposed framework improves the performance of different state-of-the-art (SOTA) segmentation models that learn and segment the type of nuclei regions in the WSIs that exhibit different characteristics.
- The framework exhibits superior performance results for segmentation models, yielding the best score for each nuclei type using different metrics and achieving high metric scores for the public MoNuSAC2020 benchmark.
- We demonstrated the performance of the framework from various perspectives using multiple radiomics types, loss functions, and visualization experiments. The results confirmed that our model was adaptable for diagnosing and segmenting different variants of nuclei.
2. Related Work
3. Proposed Framework and Architecture
3.1. (Stage-A) Processing WSIs Module

3.2. (Stage-B) Patch Retrieval Module
3.2.1. Representation of Radiomics Features
- Gray Level Co-occurrence Matrix (GLCM): This feature describes the spatial distribution of gray-level pixel intensities within a 2D image [37]. Features extracted from GLCM are calculated based on two predefined parameters and d, where 0°, 45°, 90°, and 180°, and d is any integer distance admissible within the image dimensions.
- Gray Level Run Length Matrix (GLRLM): This feature calculates the number of pixels with the same gray and characterize the gray-level run lengths of different gray-level intensities in any direction [38].
- Gray Level Size Zone Matrix (GLSZM): This feature quantifies the gray-level zones, that is the number of connected pixels that share the same gray-level intensity, in a 2D image [39].
- Gray Level Dependence Matrix (GLDM): This feature includes the number of connected pixels within a distance dependent on the center pixel [40].
3.2.2. LASSO Operator
3.2.3. Training Different Classifiers
3.2.4. Selection of the Best Classifier
3.3. (Stage-C) Segmentation Module
3.3.1. User-Defined Selection of Model
3.3.2. Training Different Segmentation Models
4. Dataset
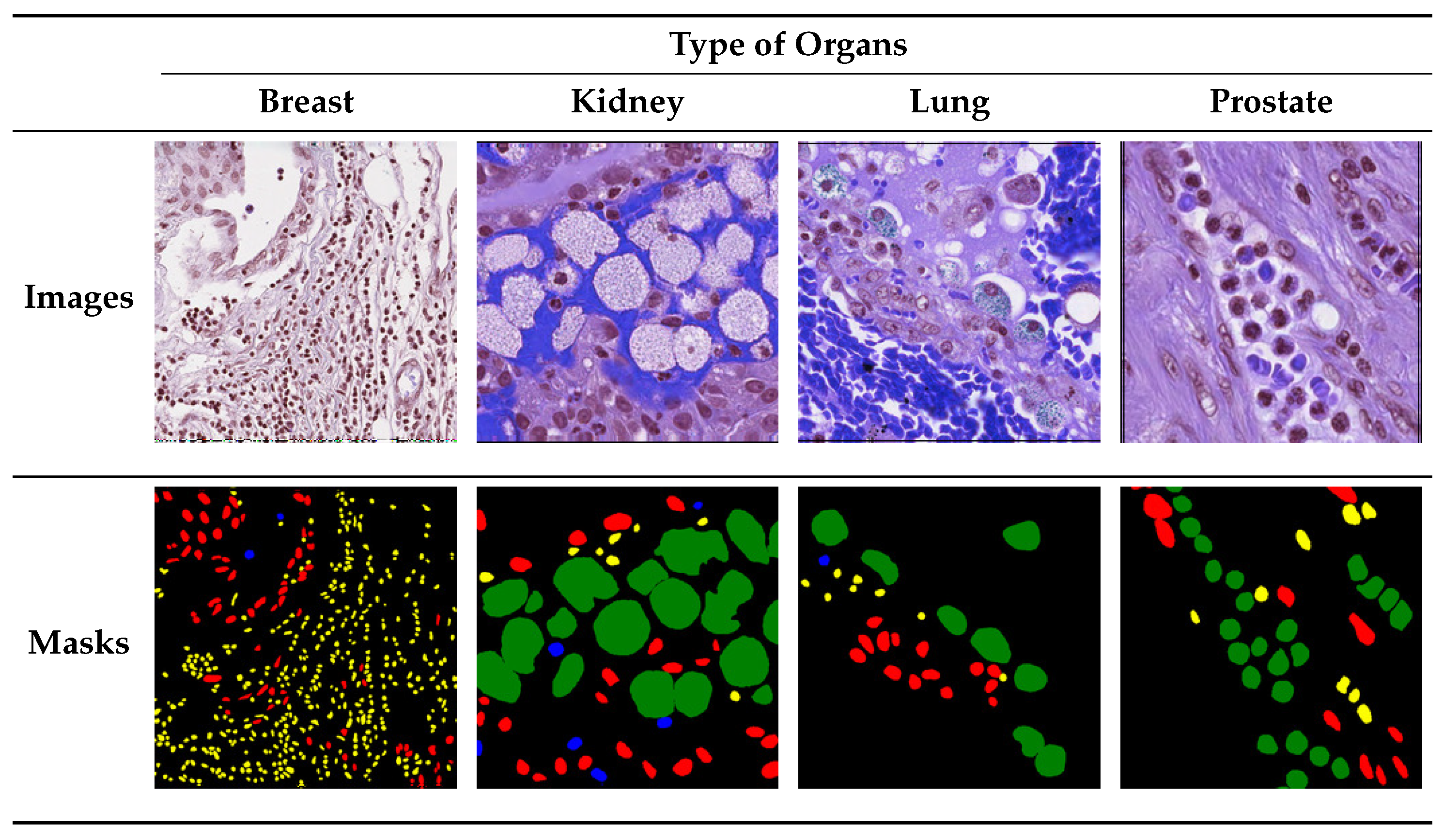
5. Experimental Setup
5.1. Performance Evaluation Criteria
5.2. Best Hyperparameters
5.3. Performance Evaluation Metrics
6. Performance Results and Discussions
6.1. Comparison of Different Classifiers
6.2. Comparison with Different Deep Learning Segmentation Models
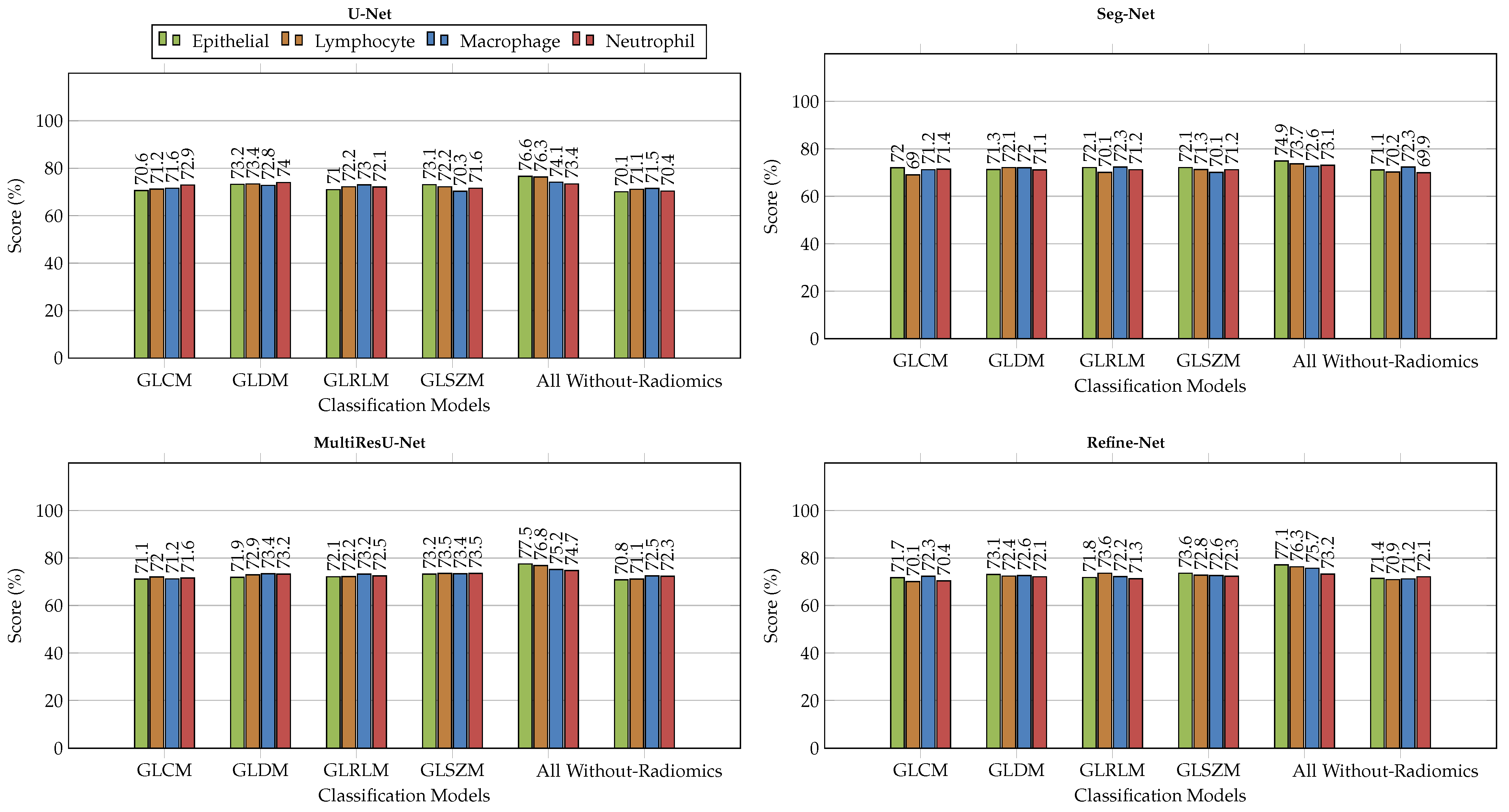
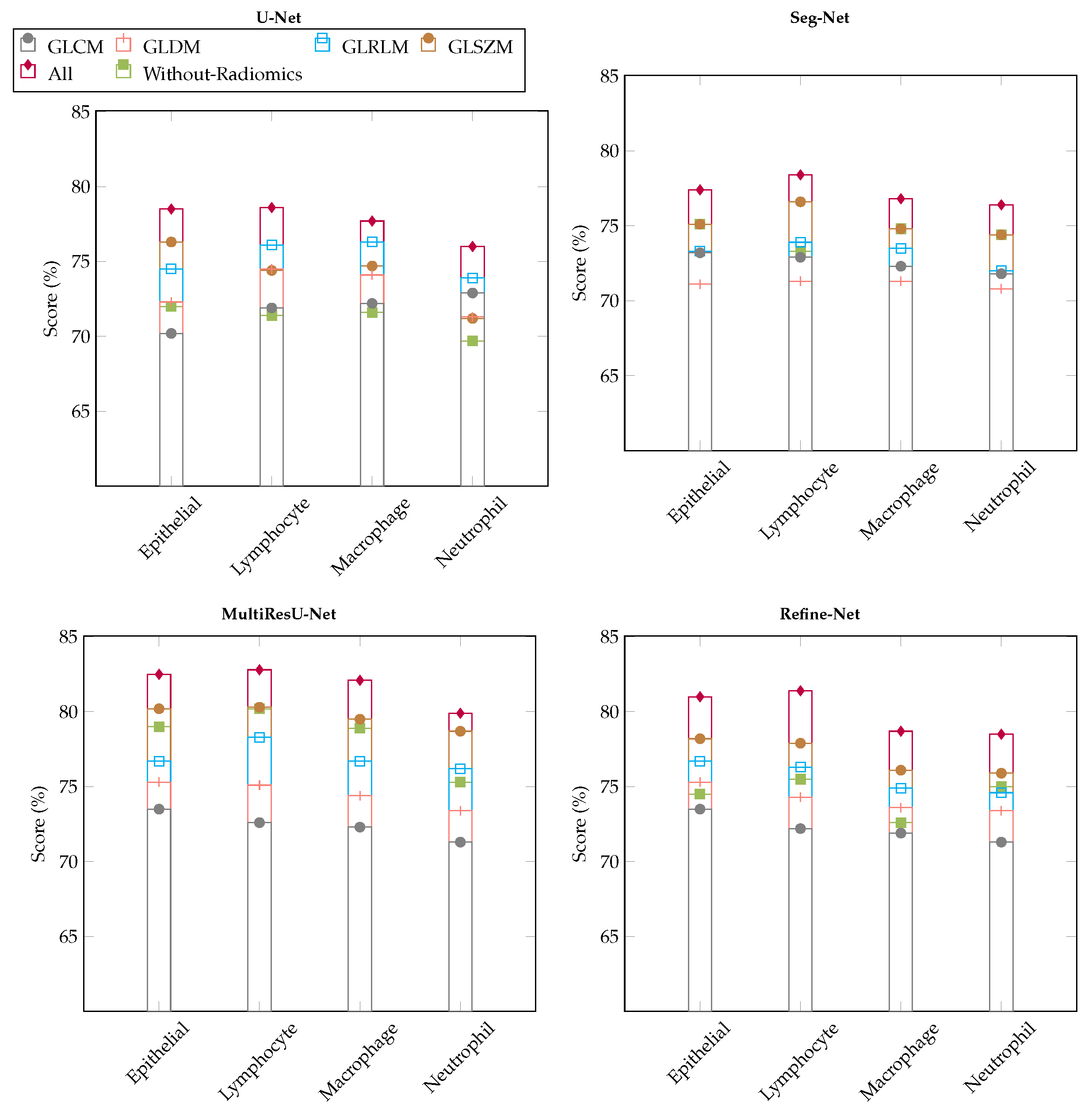
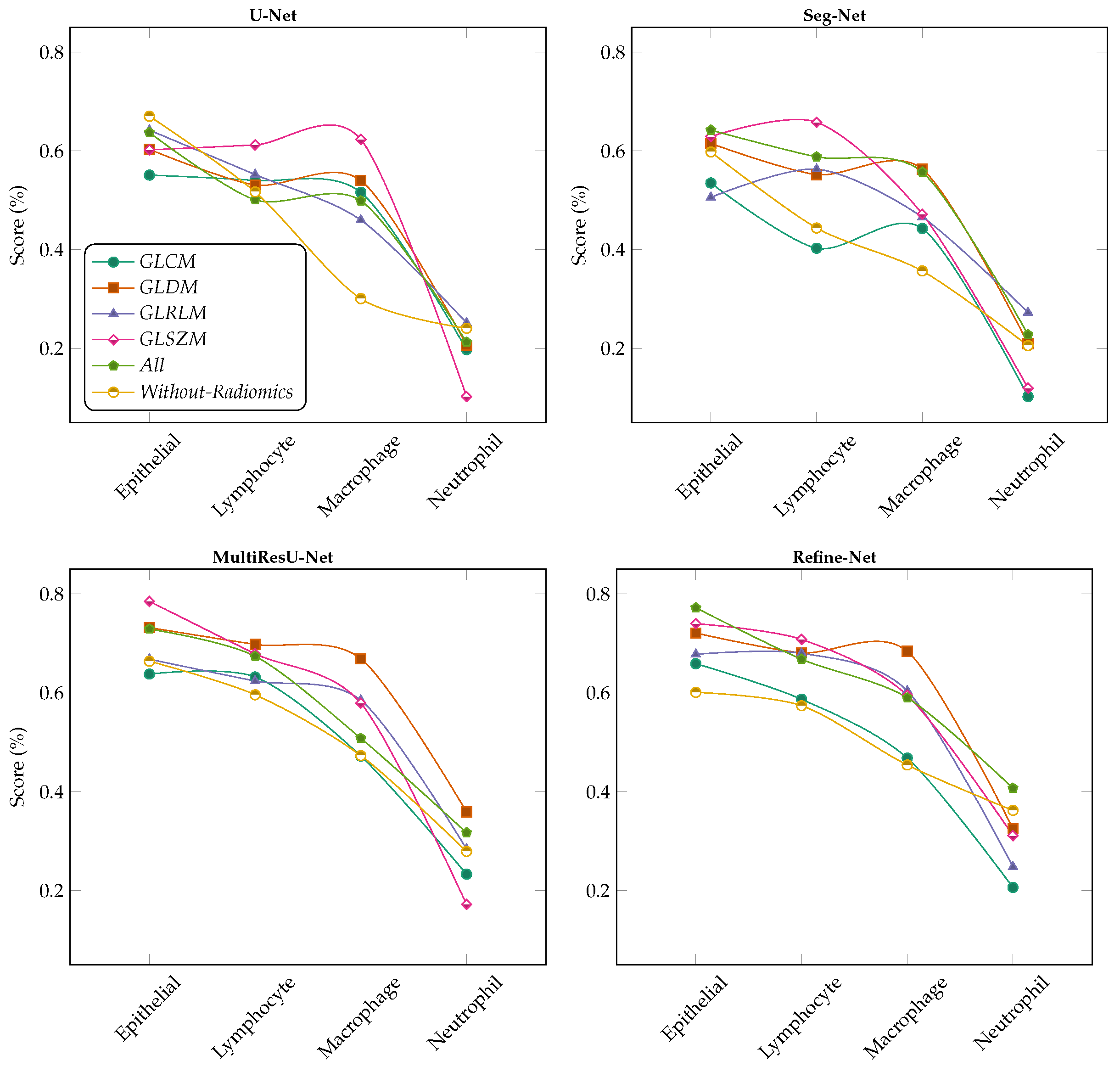
6.3. Visualization of Segmented Samples

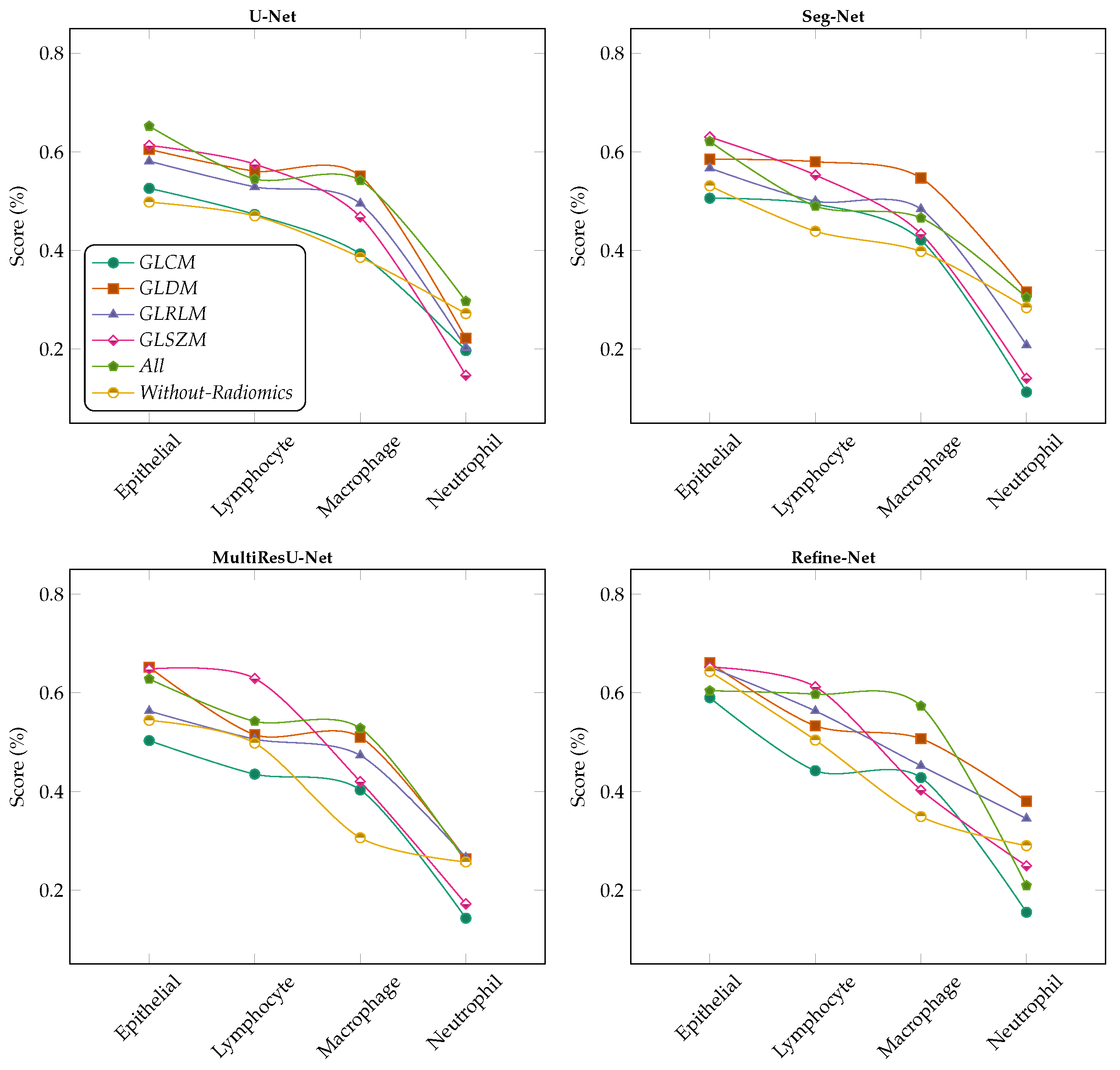
6.4. Ablation Studies
6.4.1. Effect of Lasso Operator
6.4.2. Variants Effect of Radiomic Features Combinations
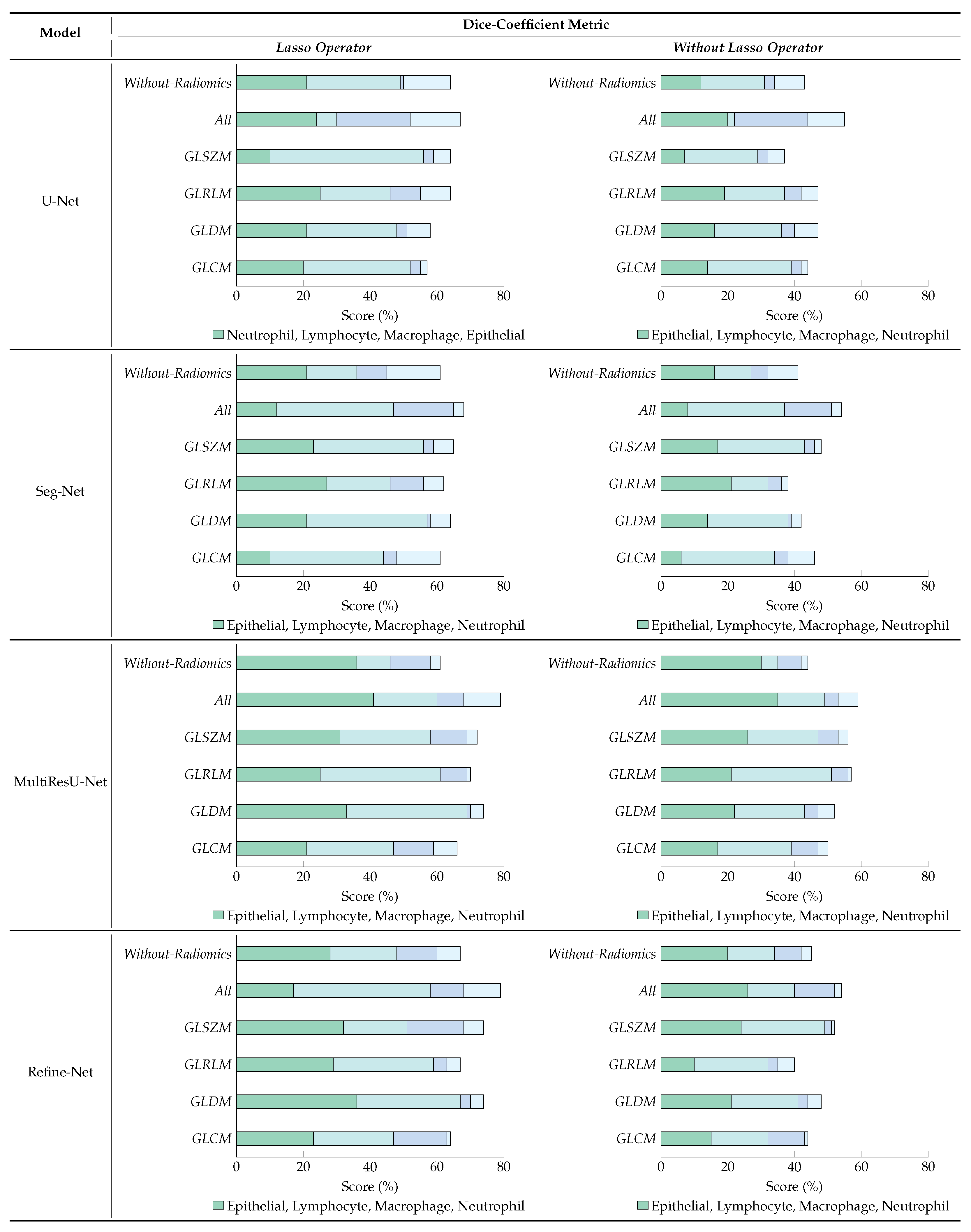

7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nasir, E.S.; Parvaiz, A.; Fraz, M.M. Nuclei and glands instance segmentation in histology images: A narrative review. Artif. Intell. Rev. 2022, 56, 7909–7964. [Google Scholar] [CrossRef]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef]
- Lu, C.; Shiradkar, R.; Liu, Z. Integrating pathomics with radiomics and genomics for cancer prognosis: A brief review. Chin. J. Cancer Res. 2021, 33, 563. [Google Scholar] [CrossRef]
- Wang, K.; Lu, X.; Zhou, H.; Gao, Y.; Zheng, J.; Tong, M.; Wu, C.; Liu, C.; Huang, L.; Jiang, T.; et al. Deep learning Radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: A prospective multicentre study. Gut 2019, 68, 729–741. [Google Scholar] [CrossRef] [PubMed]
- Bera, K.; Braman, N.; Gupta, A.; Velcheti, V.; Madabhushi, A. Predicting cancer outcomes with radiomics and artificial intelligence in radiology. Nat. Rev. Clin. Oncol. 2022, 19, 132–146. [Google Scholar] [CrossRef]
- Vallieres, M.; Kay-Rivest, E.; Perrin, L.J.; Liem, X.; Furstoss, C.; Aerts, H.J.; Khaouam, N.; Nguyen-Tan, P.F.; Wang, C.S.; Sultanem, K.; et al. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci. Rep. 2017, 7, 10117. [Google Scholar] [CrossRef]
- Zhou, M.; Scott, J.; Chaudhury, B.; Hall, L.; Goldgof, D.; Yeom, K.W.; Iv, M.; Ou, Y.; Kalpathy-Cramer, J.; Napel, S.; et al. Radiomics in brain tumor: Image assessment, quantitative feature descriptors, and machine-learning approaches. Am. J. Neuroradiol. 2018, 39, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, A.; Altini, N.; Buongiorno, D.; Garolla, E.; Corallo, F.; Gravina, M.; Bevilacqua, V.; Prencipe, B. A Machine Learning and Radiomics Approach in Lung Cancer for Predicting Histological Subtype. Appl. Sci. 2022, 12, 5829. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Shafiee, M.J.; Chung, A.G.; Khalvati, F.; Haider, M.A.; Wong, A. Discovery radiomics via evolutionary deep radiomic sequencer discovery for pathologically proven lung cancer detection. J. Med. Imaging 2017, 4, 041305. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, H.; Zhang, S.; Wang, G.; Qi, J. NAS-SCAM: Neural architecture search-based spatial and channel joint attention module for nuclei semantic segmentation and classification. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020; Proceedings, Part I 23. Springer: Cham, Switzerland, 2020; pp. 263–272. [Google Scholar]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Coroller, T.P.; Grossmann, P.; Hou, Y.; Velazquez, E.R.; Leijenaar, R.T.; Hermann, G.; Lambin, P.; Haibe-Kains, B.; Mak, R.H.; Aerts, H.J. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother. Oncol. 2015, 114, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Parmar, C.; Leijenaar, R.T.; Grossmann, P.; Rios Velazquez, E.; Bussink, J.; Rietveld, D.; Rietbergen, M.M.; Haibe-Kains, B.; Lambin, P.; Aerts, H.J. Radiomic feature clusters and prognostic signatures specific for lung and head & neck cancer. Sci. Rep. 2015, 5, 11044. [Google Scholar] [PubMed]
- Yuan, Y.; Qin, W.; Buyyounouski, M.; Ibragimov, B.; Hancock, S.; Han, B.; Xing, L. Prostate cancer classification with multiparametric MRI transfer learning model. Med. Phys. 2019, 46, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Junior, J.R.F.; Koenigkam-Santos, M.; Cipriano, F.E.G.; Fabro, A.T.; de Azevedo-Marques, P.M. Radiomics-based features for pattern recognition of lung cancer histopathology and metastases. Comput. Methods Programs Biomed. 2018, 159, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Song, S.H.; Park, H.; Lee, G.; Lee, H.Y.; Sohn, I.; Kim, H.S.; Lee, S.H.; Jeong, J.Y.; Kim, J.; Lee, K.S.; et al. Imaging phenotyping using radiomics to predict micropapillary pattern within lung adenocarcinoma. J. Thorac. Oncol. 2017, 12, 624–632. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, J.; Nie, D.; Yap, P.T.; Shen, D. Anatomical landmark based deep feature representation for MR images in brain disease diagnosis. IEEE J. Biomed. Health Inform. 2018, 22, 1476–1485. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, B.; Wu, X.; Liu, L.; Fang, J.; Chen, Q.; Li, M.; Chen, Z.; Li, Y.; Dong, D.; et al. Radiomic nomogram improves preoperative T category accuracy in locally advanced laryngeal carcinoma. Front. Oncol. 2019, 9, 1064. [Google Scholar] [CrossRef]
- Choi, W.; Oh, J.H.; Riyahi, S.; Liu, C.J.; Jiang, F.; Chen, W.; White, C.; Rimner, A.; Mechalakos, J.G.; Deasy, J.O.; et al. Radiomics analysis of pulmonary nodules in low-dose CT for early detection of lung cancer. Med. Phys. 2018, 45, 1537–1549. [Google Scholar] [CrossRef]
- Ning, Z.; Luo, J.; Li, Y.; Han, S.; Feng, Q.; Xu, Y.; Chen, W.; Chen, T.; Zhang, Y. Pattern classification for gastrointestinal stromal tumors by integration of radiomics and deep convolutional features. IEEE J. Biomed. Health Inform. 2018, 23, 1181–1191. [Google Scholar] [CrossRef]
- Urban, G.; Tripathi, P.; Alkayali, T.; Mittal, M.; Jalali, F.; Karnes, W.; Baldi, P. Deep learning localizes and identifies polyps in real time with 96% accuracy in screening colonoscopy. Gastroenterology 2018, 155, 1069–1078. [Google Scholar] [CrossRef]
- Guo, Y.; Song, Q.; Jiang, M.; Guo, Y.; Xu, P.; Zhang, Y.; Fu, C.C.; Fang, Q.; Zeng, M.; Yao, X. Histological subtypes classification of lung cancers on CT images using 3D deep learning and radiomics. Acad. Radiol. 2021, 28, e258–e266. [Google Scholar] [CrossRef]
- Sun, W.; Zheng, B.; Qian, W. Automatic feature learning using multichannel ROI based on deep structured algorithms for computerized lung cancer diagnosis. Comput. Biol. Med. 2017, 89, 530–539. [Google Scholar] [CrossRef]
- Kumar, D.; Chung, A.G.; Shaifee, M.J.; Khalvati, F.; Haider, M.A.; Wong, A. Discovery radiomics for pathologically-proven computed tomography lung cancer prediction. In Proceedings of the Image Analysis and Recognition: 14th International Conference, ICIAR 2017, Montreal, QC, Canada, 5–7 July 2017; Proceedings 14. Springer: Cham, Switzerland, 2017; pp. 54–62. [Google Scholar]
- Avanzo, M.; Wei, L.; Stancanello, J.; Vallieres, M.; Rao, A.; Morin, O.; Mattonen, S.A.; El Naqa, I. Machine and deep learning methods for radiomics. Med. Phys. 2020, 47, e185–e202. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A. Radiomics in nuclear medicine: Robustness, reproducibility, standardization, and how to avoid data analysis traps and replication crisis. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2638–2655. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.; Yosinski, J.; Clune, J. Understanding neural networks via feature visualization: A survey. In Explainable AI: Interpreting, Explaining and Visualizing Deep Learning; Springer: Cham, Switzerland, 2019; pp. 55–76. [Google Scholar]
- Traverso, A.; Wee, L.; Dekker, A.; Gillies, R. Repeatability and reproducibility of radiomic features: A systematic review. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1143–1158. [Google Scholar] [CrossRef]
- Kong, Y.; Genchev, G.Z.; Wang, X.; Zhao, H.; Lu, H. Nuclear segmentation in histopathological images using two-stage stacked U-nets with attention mechanism. Front. Bioeng. Biotechnol. 2020, 8, 573866. [Google Scholar] [CrossRef]
- Zaki, G.; Gudla, P.R.; Lee, K.; Kim, J.; Ozbun, L.; Shachar, S.; Gadkari, M.; Sun, J.; Fraser, I.D.; Franco, L.M.; et al. A deep learning pipeline for nucleus segmentation. Cytom. Part A 2020, 97, 1248–1264. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, G.; Liu, Z.; Xiong, Z.; Hu, J. A deep learning algorithm for one-step contour aware nuclei segmentation of histopathology images. Med. Biol. Eng. Comput. 2019, 57, 2027–2043. [Google Scholar] [CrossRef]
- Zhang, Z. Introduction to machine learning: K-nearest neighbors. Ann. Transl. Med. 2016, 4, 218. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Thawani, R.; McLane, M.; Beig, N.; Ghose, S.; Prasanna, P.; Velcheti, V.; Madabhushi, A. Radiomics and radiogenomics in lung cancer: A review for the clinician. Lung Cancer 2018, 115, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Nanni, L.; Brahnam, S.; Ghidoni, S.; Menegatti, E.; Barrier, T. Different approaches for extracting information from the co-occurrence matrix. PLoS ONE 2013, 8, e83554. [Google Scholar] [CrossRef] [PubMed]
- Tang, X. Texture information in run-length matrices. IEEE Trans. Image Process. 1998, 7, 1602–1609. [Google Scholar] [PubMed]
- Thibault, G.; Fertil, B.; Navarro, C.; Pereira, S.; Cau, P.; Levy, N.; Sequeira, J.; Mari, J. Texture indexes and gray level size zone matrix. Application to Cell Nuclei Classification. In Proceedings of the 10th International Conference on Pattern Recognition and Information Processing (PRIP 2009), Minsk, Belarus, 19–21 May 2009; pp. 140–145. [Google Scholar]
- Sun, C.; Wee, W.G. Neighboring gray level dependence matrix for texture classification. Comput. Graph. Image Process. 1982, 20, 297. [Google Scholar] [CrossRef]
- Verma, R.; Kumar, N.; Patil, A.; Kurian, N.C.; Rane, S.; Graham, S.; Vu, Q.D.; Zwager, M.; Raza, S.E.A.; Rajpoot, N.; et al. MoNuSAC2020: A multi-organ nuclei segmentation and classification challenge. IEEE Trans. Med. Imaging 2021, 40, 3413–3423. [Google Scholar] [CrossRef]
- Kumar, N.; Verma, R.; Sharma, S.; Bhargava, S.; Vahadane, A.; Sethi, A. A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans. Med. Imaging 2017, 36, 1550–1560. [Google Scholar] [CrossRef]
- Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [CrossRef]
- Lin, G.; Milan, A.; Shen, C.; Reid, I. Refinenet: Multi-path refinement networks for high-resolution semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1925–1934. [Google Scholar]
| GLCM (24) | GLDM (14) | GLRLM (16) | GLSZM (16) |
|---|---|---|---|
| Autocorrelation | Dependence Entropy | Gray Level Non-Uniformity | Gray Level Non-Uniformity |
| Cluster Prominence | Dependence Non-Uniformity | Gray Level Non-Uniformity Normalized | Gray Level Non-Uniformity Normalized |
| Cluster Shade, Joint Entropy | Dependence Non-Uniformity Normalized | Gray Level Variance | Gray Level Variance |
| Cluster Tendency, Joint Average | Dependence Variance | High Gray Level Run Emphasis | High Gray Level Zone Emphasis |
| Contrast, Joint Energy | Gray Level Non-Uniformity | Long Run High Gray Level Emphasis | Large Area Emphasis |
| Correlation, Inverse Variance | Gray Level Variance | Long Run Low Gray Level Emphasis | Large Area High Gray Level Emphasis |
| Difference Average, Sum Average | High Gray Level Emphasis | Low Gray Level Run Emphasis | Large Area Low Gray Level Emphasis |
| Difference Entropy, Sum Entropy | Large Dependence Emphasis | Run Entropy | Low Gray Level Zone Emphasis |
| Difference Variance, Maximum Probability | Large Dependence High Gray Level Emphasis | Run Length Non-Uniformity | Size Zone Non-Uniformity |
| Inverse Difference | Large Dependence Low Gray Level Emphasis | Run Length Non-Uniformity Normalized | Size Zone Non-Uniformity Normalized |
| Inverse Difference Moment, Sum Squares | Low Gray Level Emphasis | Run Percentage | Small Area Emphasis |
| Inverse Difference Moment Normalized | Small Dependence Emphasis | Run Variance | Small Area High Gray Level Emphasis |
| Inverse Difference Normalized | Small Dependence High Gray Level Emphasis | Short Run Emphasis | Small Area Low Gray Level Emphasis |
| Informational Measure of Correlation 1 | Small Dependence Low Gray Level Emphasis | Short Run High Gray Level Emphasis | Zone Entropy |
| Informational Measure of Correlation 2 | - | Short Run Low Gray Level Emphasis | Zone Percentage |
| Maximal Correlation Coefficient | - | Long Run Emphasis | Zone Variance |
| Nuclei Type | Training Subset | |||
|---|---|---|---|---|
| Breast | Kidney | Lung | Prostate | |
| N = 10 (7) | N = 12 (11) | N = 10 (12) | N = 14 (8) | |
| 670 (2055) | 566 (2477) | 506 (2647) | 570 (1021) | |
| Epithelial | 4566 (540) | 3547 (1501) | 2981 (474) | 3445 (575) |
| Lymphocytes | 4689 (946) | 4126 (259) | 3018 (916) | 3821 (275) |
| Macrophoges | 147 (37) | 102 (354) | 186 (887) | 152 (70) |
| Neutrophils | 105 (43) | 149 (250) | 142 (281) | 235 (55) |
| Testing Subset | ||||
| N = 5 (4) | N = 7 (3) | N = 7 (3) | N = 6 (3) | |
| 220 (725) | 310 (860) | 220 (670) | 260 (610) | |
| Epithelial | 1377 (415) | 2248 (510) | 1489 (478) | 2099 (274) |
| Lymphocytes | 1833 (258) | 2141 (256) | 1674 (128) | 2158 (191) |
| Macrophoges | 49 (32) | 84 (56) | 53 (42) | 121 (106) |
| Neutrophils | 26 (20) | 56 (38) | 36 (22) | 54 (39) |
| Classifier | Parameters | Radiomics | ||||
|---|---|---|---|---|---|---|
| GLCM | GLDM | GLRLM | GLSZM | All | ||
| Bagging Classifier | N Estimators | 50 | 50 | 25 | 25 | 50 |
| Max Samples | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| Max Features | 1.0 | 0.5 | 0.5 | 0.5 | 1.0 | |
| Models | Parameters | Radiomics GLCM GLDM GLRLM GLSZM | All | Without-Radiomics |
|---|---|---|---|---|
| U-Net | Number of layers | 41 | ||
| Epochs | 20 | 15 | 15 | |
| Optimizer | Nadam | |||
| Batch Size | 16 | 16 | 32 | |
| Kernel Size | 3, 3 | |||
| Filters | 16, 32, 64, 128, 256 | |||
| # of Parameters | 1.9 M | |||
| Kernel Regularizer | He Normal | |||
| Activation Map | Sigmoid, Relu | |||
| Dropout Rate | 0.5 | |||
| Learning Rate | 10−4 | 10−3 | 10−3 | |
| Seg-Net | Number of layers | 62 | ||
| Epochs | 15 | 20 | 15 | |
| Optimizer | Nadam | |||
| Batch Size | 16 | 16 | 32 | |
| Kernel Size | 3, 3 | |||
| Filters | 64, 128, 256, 512 | |||
| # of Parameters | 11 M | |||
| Kernel Regularizer | He Normal | |||
| Activation Map | Sigmoid, Relu | |||
| Dropout Rate | 0.5 | |||
| Learning Rate | 10−4 | 10−3 | 10−4 | |
| MultiResU-Net | Number of layers | 239 | ||
| Epochs | 15 | 20 | 20 | |
| Optimizer | Adamax | |||
| Batch Size | 16 | 32 | 32 | |
| Kernel Size | 3, 3 | |||
| Filters | 32, 64, 128, 256, 512 | |||
| # of Parameters | 7.2 M | |||
| Kernel Regularizer | l2 | |||
| Activation Map | Sigmoid, Elu | |||
| Dropout Rate | 0.5 | |||
| Learning Rate | 10−3 | 10−3 | 10−3 | |
| Refine-Net | Number of layers | 480 | ||
| Epochs | 20 | 15 | 15 | |
| Optimizer | Adamax | |||
| Batch Size | 32 | 16 | 16 | |
| Kernel Size | 3, 3 | |||
| Filters | 64, 128, 256, 512, 256, 1024, 2048 | |||
| # of Parameters | 42 M | |||
| Kernel Regularizer | l1 | |||
| Activation Map | Sigmoid, Elu | |||
| Dropout Rate | 0.6 | |||
| Learning Rate | 10−3 | 10−4 | 10−4 | |
| Radiomics Feature | Classifier | Metrics | ||
|---|---|---|---|---|
| Accuracy | Sensitivity | Precision | ||
| GLCM | Decision Tree | 0.756 | 0.757 | 0.759 |
| KNeighbors | 0.760 | 0.760 | 0.769 | |
| Bagging | 0.776 | 0.778 | 0.784 | |
| GradientBoost | 0.779 | 0.779 | 0.788 | |
| GLDM | Decision Tree | 0.772 | 0.774 | 0.779 |
| KNeighbors | 0.794 | 0.793 | 0.799 | |
| Bagging | 0.808 | 0.804 | 0.814 | |
| GradientBoost | 0.810 | 0.809 | 0.816 | |
| GLRLM | Decision Tree | 0.780 | 0.789 | 0.781 |
| KNeighbors | 0.803 | 0.801 | 0.808 | |
| Bagging | 0.805 | 0.809 | 0.807 | |
| GradientBoost | 0.819 | 0.810 | 0.825 | |
| GLSZM | Decision Tree | 0.763 | 0.764 | 0.767 |
| KNeighbors | 0.782 | 0.781 | 0.785 | |
| Bagging | 0.795 | 0.796 | 0.802 | |
| GradientBoost | 0.796 | 0.797 | 0.803 | |
| All | Decision Tree | 0.780 | 0.781 | 0.783 |
| KNeighbors | 0.803 | 0.802 | 0.807 | |
| Bagging | 0.816 | 0.817 | 0.821 | |
| GradientBoost | 0.821 | 0.820 | 0.826 | |
| Without-Radiomics | Decision Tree | 0.774 | 0.787 | 0.749 |
| KNeighbors | 0.796 | 0.759 | 0.765 | |
| Bagging | 0.785 | 0.736 | 0.742 | |
| GradientBoost | 0.784 | 0.691 | 0.764 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheikh, T.S.; Cho, M. Segmentation of Variants of Nuclei on Whole Slide Images by Using Radiomic Features. Bioengineering 2024, 11, 252. https://doi.org/10.3390/bioengineering11030252
Sheikh TS, Cho M. Segmentation of Variants of Nuclei on Whole Slide Images by Using Radiomic Features. Bioengineering. 2024; 11(3):252. https://doi.org/10.3390/bioengineering11030252
Chicago/Turabian StyleSheikh, Taimoor Shakeel, and Migyung Cho. 2024. "Segmentation of Variants of Nuclei on Whole Slide Images by Using Radiomic Features" Bioengineering 11, no. 3: 252. https://doi.org/10.3390/bioengineering11030252
APA StyleSheikh, T. S., & Cho, M. (2024). Segmentation of Variants of Nuclei on Whole Slide Images by Using Radiomic Features. Bioengineering, 11(3), 252. https://doi.org/10.3390/bioengineering11030252








