Classification of Muscular Dystrophies from MR Images Improves Using the Swin Transformer Deep Learning Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. MRI Protocol
2.3. Dataset
2.4. Implementation of DL Architectures for the Classification Task
2.5. Evaluation of the Classification Performance
2.6. Statistical Analysis
3. Results
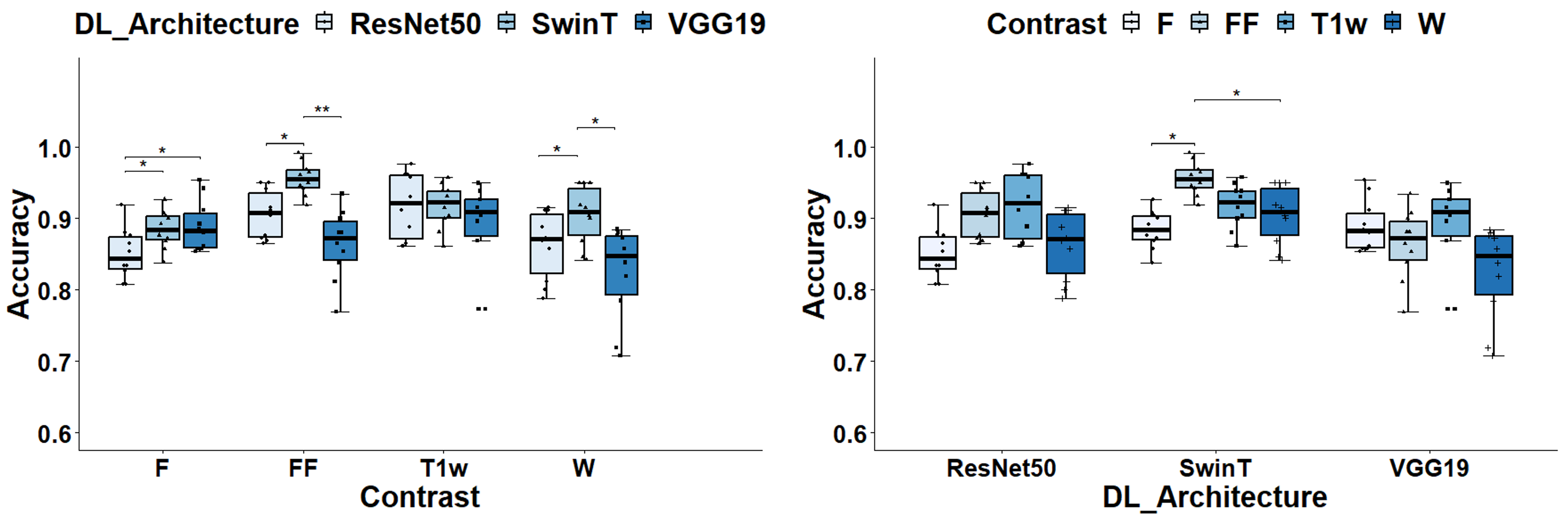
3.1. Global Classification Performances
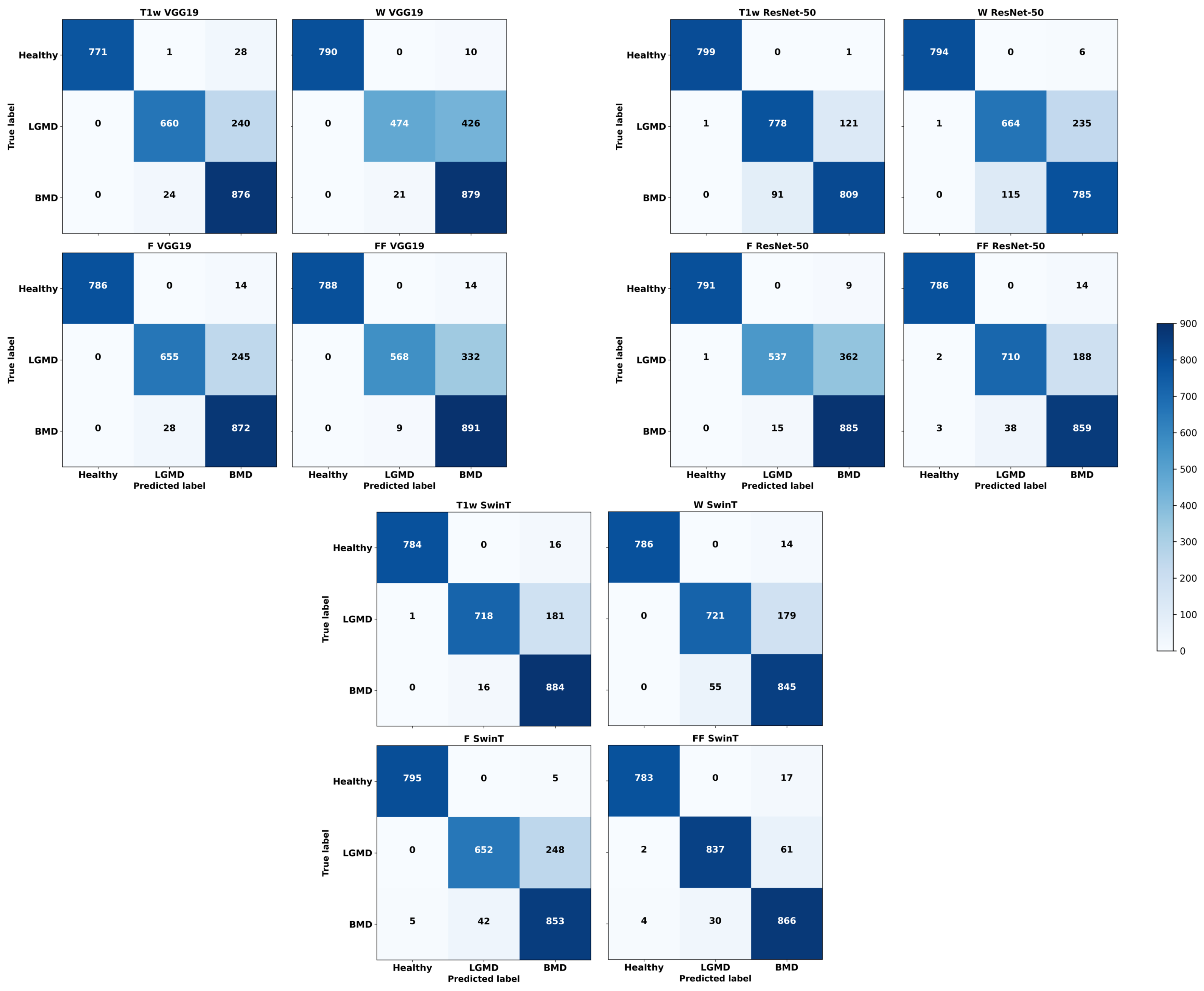
3.2. Class-Specific Classification Performances
3.3. Computational Times
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Emery, A.E. The muscular dystrophies. Lancet 2002, 359, 687–695. [Google Scholar] [CrossRef]
- Lovering, R.M.; Porter, N.C.; Bloch, R.J. The muscular dystrophies: From genes to therapies. Phys. Ther. 2005, 85, 1372–1388. [Google Scholar] [CrossRef]
- Nicolau, S.; Naddaf, E. Muscle MRI for Neuromuscular Disorders. Pract. Neurol. 2020, July/August, 27–32. [Google Scholar]
- Wattjes, M.P.; Kley, R.A.; Fischer, D. Neuromuscular imaging in inherited muscle diseases. Eur. Radiol. 2010, 20, 2447–2460. [Google Scholar] [CrossRef]
- Tasca, G.; Iannaccone, E.; Monforte, M.; Masciullo, M.; Bianco, F.; Laschena, F.; Ottaviani, P.; Pelliccioni, M.; Pane, M.; Mercuri, E.; et al. Muscle MRI in Becker muscular dystrophy. Neuromuscul. Disord. 2012, 22, S100–S106. [Google Scholar] [CrossRef]
- Fanin, M.; Angelini, C. Progress and challenges in diagnosis of dysferlinopathy. Muscle Nerve 2016, 54, 821–835. [Google Scholar] [CrossRef]
- Manzur, A.Y.; Muntoni, F. Diagnosis and new treatments in muscular dystrophies. Postgrad. Med. J. 2009, 85, 622–630. [Google Scholar] [CrossRef]
- Angelini, C. LGMD. Identification, description and classification. Acta Myol. 2020, 39, 207. [Google Scholar]
- Okubo, M.; Minami, N.; Goto, K.; Goto, Y.; Noguchi, S.; Mitsuhashi, S.; Nishino, I. Genetic diagnosis of Duchenne/Becker muscular dystrophy using next-generation sequencing: Validation analysis of DMD mutations. J. Hum. Genet. 2016, 61, 483–489. [Google Scholar] [CrossRef]
- Nigro, V.; Savarese, M. Next-generation sequencing approaches for the diagnosis of skeletal muscle disorders. Curr. Opin. Neurol. 2016, 29, 621–627. [Google Scholar] [CrossRef]
- Ghaoui, R.; Cooper, S.T.; Lek, M.; Jones, K.; Corbett, A.; Reddel, S.W.; Needham, M.; Liang, C.; Waddell, L.B.; Nicholson, G.; et al. Use of whole-exome sequencing for diagnosis of limb-girdle muscular dystrophy: Outcomes and lessons learned. JAMA Neurol. 2015, 72, 1424–1432. [Google Scholar] [CrossRef]
- Joyce, N.C.; Oskarsson, B.; Jin, L.W. Muscle biopsy evaluation in neuromuscular disorders. Phys. Med. Rehabil. Clin. 2012, 23, 609–631. [Google Scholar] [CrossRef]
- Li, K.; Dortch, R.D.; Welch, E.B.; Bryant, N.D.; Buck, A.K.; Towse, T.F.; Gochberg, D.F.; Does, M.D.; Damon, B.M.; Park, J.H. Multi-parametric MRI characterization of healthy human thigh muscles at 3.0 T–relaxation, magnetization transfer, fat/water, and diffusion tensor imaging. NMR Biomed. 2014, 27, 1070–1084. [Google Scholar] [CrossRef]
- Venturelli, N.; Tordjman, M.; Ammar, A.; Chetrit, A.; Renault, V.; Carlier, R.Y. Contribution of muscle MRI for diagnosis of myopathy. Rev. Neurol. 2023, 179, 61–80. [Google Scholar] [CrossRef]
- Malartre, S.; Bachasson, D.; Mercy, G.; Sarkis, E.; Anquetil, C.; Benveniste, O.; Allenbach, Y. MRI and muscle imaging for idiopathic inflammatory myopathies. Brain Pathol. 2021, 31, e12954. [Google Scholar] [CrossRef]
- Garibaldi, M.; Nicoletti, T.; Bucci, E.; Fionda, L.; Leonardi, L.; Morino, S.; Tufano, L.; Alfieri, G.; Lauletta, A.; Merlonghi, G.; et al. Muscle magnetic resonance imaging in myotonic dystrophy type 1 (DM1): Refining muscle involvement and implications for clinical trials. Eur. J. Neurol. 2022, 29, 843–854. [Google Scholar] [CrossRef]
- Forbes, S.C.; Willcocks, R.J.; Rooney, W.D.; Walter, G.A.; Vandenborne, K. MRI quantifies neuromuscular disease progression. Lancet Neurol. 2016, 15, 26–28. [Google Scholar] [CrossRef]
- Tobore, I.; Li, J.; Yuhang, L.; Al-Handarish, Y.; Kandwal, A.; Nie, Z.; Wang, L. Deep learning intervention for health care challenges: Some biomedical domain considerations. JMIR MHealth UHealth 2019, 7, e11966. [Google Scholar] [CrossRef]
- Kim, M.; Yun, J.; Cho, Y.; Shin, K.; Jang, R.; Bae, H.j.; Kim, N. Deep learning in medical imaging. Neurospine 2019, 16, 657. [Google Scholar] [CrossRef]
- He, K.; Gan, C.; Li, Z.; Rekik, I.; Yin, Z.; Ji, W.; Gao, Y.; Wang, Q.; Zhang, J.; Shen, D. Transformers in medical image analysis. Intell. Med. 2023, 3, 59–78. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Wei, C.; Ren, S.; Guo, K.; Hu, H.; Liang, J. High-resolution Swin transformer for automatic medical image segmentation. Sensors 2023, 23, 3420. [Google Scholar] [CrossRef]
- Liu, Z.; Hu, H.; Lin, Y.; Yao, Z.; Xie, Z.; Wei, Y.; Ning, J.; Cao, Y.; Zhang, Z.; Dong, L.; et al. Swin transformer v2: Scaling up capacity and resolution. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 12009–12019. [Google Scholar]
- Piñeros-Fernández, M.C. Artificial intelligence applications in the diagnosis of neuromuscular diseases: A narrative review. Cureus 2023, 15, e48458. [Google Scholar] [CrossRef]
- Verdú-Díaz, J.; Alonso-Pérez, J.; Nuñez-Peralta, C.; Tasca, G.; Vissing, J.; Straub, V.; Fernández-Torrón, R.; Llauger, J.; Illa, I.; Díaz-Manera, J. Accuracy of a machine learning muscle MRI-based tool for the diagnosis of muscular dystrophies. Neurology 2020, 94, e1094–e1102. [Google Scholar] [CrossRef]
- Yang, M.; Zheng, Y.; Xie, Z.; Wang, Z.; Xiao, J.; Zhang, J.; Yuan, Y. A deep learning model for diagnosing dystrophinopathies on thigh muscle MRI images. BMC Neurol. 2021, 21, 13. [Google Scholar] [CrossRef]
- Cai, J.; Xing, F.; Batra, A.; Liu, F.; Walter, G.A.; Vandenborne, K.; Yang, L. Texture analysis for muscular dystrophy classification in MRI with improved class activation mapping. Pattern Recognit. 2019, 86, 368–375. [Google Scholar] [CrossRef]
- Al-Wesabi, F.N.; Obayya, M.; Hilal, A.M.; Castillo, O.; Gupta, D.; Khanna, A. Multi-objective quantum tunicate swarm optimization with deep learning model for intelligent dystrophinopathies diagnosis. Soft Comput. 2022, 27, 13077–13092. [Google Scholar] [CrossRef]
- Gopalakrishnan, T.; Sudhakaran, P.; Ramya, K.; Sathesh Kumar, K.; Al-Wesabi, F.N.; Alohali, M.A.; Hilal, A.M. An Automated Deep Learning Based Muscular Dystrophy Detection and Classification Model. Comput. Mater. Contin. 2022, 71, 305–320. [Google Scholar]
- Eggers, H.; Börnert, P. Chemical shift encoding-based water–fat separation methods. J. Magn. Reson. Imaging 2014, 40, 251–268. [Google Scholar] [CrossRef]
- Sayak, P. Implementation of Swin Transformers in TensorFlow along with Converted Pre-Trained Models, Code for off-the-Shelf Classification and Fine-Tuning. 2022. Available online: https://github.com/sayakpaul/swin-transformers-tf (accessed on 21 April 2024).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Ieee, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Chollet, F. Keras. 2023. Available online: https://keras.io/ (accessed on 4 April 2024).
- Tang, Y. YanTang’s Blog. 2023. Available online: https://ehehe.cn/2023/08/09/Swin/ (accessed on 5 April 2024).
- Rastogi, A. ResNet50. 2022. Available online: https://blog.devgenius.io/resnet50-6b42934db431 (accessed on 5 April 2024).
- Varshney, P. VGG19. 2020. Available online: https://www.kaggle.com/code/blurredmachine/vggnet-16-architecture-a-complete-guide (accessed on 5 April 2024).
- Krishnapriya, S.; Karuna, Y. Pre-trained deep learning models for brain MRI image classification. Front. Hum. Neurosci. 2023, 17, 1150120. [Google Scholar] [CrossRef]
- Swati, Z.N.K.; Zhao, Q.; Kabir, M.; Ali, F.; Ali, Z.; Ahmed, S.; Lu, J. Brain tumor classification for MR images using transfer learning and fine-tuning. Comput. Med Imaging Graph. 2019, 75, 34–46. [Google Scholar] [CrossRef]
- Gaeta, M.; Messina, S.; Mileto, A.; Vita, G.L.; Ascenti, G.; Vinci, S.; Bottari, A.; Vita, G.; Settineri, N.; Bruschetta, D.; et al. Muscle fat-fraction and mapping in Duchenne muscular dystrophy: Evaluation of disease distribution and correlation with clinical assessments: Preliminary experience. Skelet. Radiol. 2012, 41, 955–961. [Google Scholar] [CrossRef]
- Morrow, J.M.; Sinclair, C.D.; Fischmann, A.; Machado, P.M.; Reilly, M.M.; Yousry, T.A.; Thornton, J.S.; Hanna, M.G. MRI biomarker assessment of neuromuscular disease progression: A prospective observational cohort study. Lancet Neurol. 2016, 15, 65–77. [Google Scholar] [CrossRef]


| Total | Healthy Subjects | BMD Patients | LGMD2 Patients | |
|---|---|---|---|---|
| N | 75 | 17 | 27 | 31 |
| Age (mean ± SD) | 42.4 ± 11.8 yo | 39.1 ± 11.5 yo | 39.3 ± 10.3 yo | 47.0 ± 11.9 yo |
| Gender (M/F) | 50/25 | 10/7 | 27/0 | 13/18 |
| Disease duration | 25.7 ± 9.4 y (pat. only) | n.a | 23.3 ± 8.3 yo | 28.0 ± 10.1 yo |
| D1 (MFM) (mean ± SD) | 17.3 ± 18.6 (pat. only) | n.a. | 22.7 ± 18.6 | 12.9 ± 17.9 |
| Ambulation (Y/N) | 29/29 (pat. only) | n.a. | 19/8 | 10/21 |
| T1W | DIXON | |
|---|---|---|
| N° of slices | 30/36/50 * | 30/36/50 * |
| Slice thickness | 6 mm | 6 mm |
| In-plane voxel size | 1 × 1 mm | 1.6 × 1.6 mm |
| Acquired matrix | 256 × 256 | 160 × 160 |
| N° of echoes | 1 | 12 |
| Echo Time | 2.025 ms | 1.48–14.68 ms |
| Echo spacing | - | 1.2 ms |
| Repetition Time | 647 ms | 16.11 ms |
| Flip Angle | 90 deg | 3 deg |
| Percentage sampling | 100% | 78% |
| Averages | 2 | 2 |
| Architecture | Training Time (min) | Classification Time (s) |
|---|---|---|
| ResNet50 | 11 ± 0.36 | 0.423 ± 0.014 |
| SwinT | 17 ± 0.08 | 1.087 ± 0.026 |
| VGG19 | 22 ± 3.85 | 0.606 ± 0.006 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mastropietro, A.; Casali, N.; Taccogna, M.G.; D’Angelo, M.G.; Rizzo, G.; Peruzzo, D. Classification of Muscular Dystrophies from MR Images Improves Using the Swin Transformer Deep Learning Model. Bioengineering 2024, 11, 580. https://doi.org/10.3390/bioengineering11060580
Mastropietro A, Casali N, Taccogna MG, D’Angelo MG, Rizzo G, Peruzzo D. Classification of Muscular Dystrophies from MR Images Improves Using the Swin Transformer Deep Learning Model. Bioengineering. 2024; 11(6):580. https://doi.org/10.3390/bioengineering11060580
Chicago/Turabian StyleMastropietro, Alfonso, Nicola Casali, Maria Giovanna Taccogna, Maria Grazia D’Angelo, Giovanna Rizzo, and Denis Peruzzo. 2024. "Classification of Muscular Dystrophies from MR Images Improves Using the Swin Transformer Deep Learning Model" Bioengineering 11, no. 6: 580. https://doi.org/10.3390/bioengineering11060580
APA StyleMastropietro, A., Casali, N., Taccogna, M. G., D’Angelo, M. G., Rizzo, G., & Peruzzo, D. (2024). Classification of Muscular Dystrophies from MR Images Improves Using the Swin Transformer Deep Learning Model. Bioengineering, 11(6), 580. https://doi.org/10.3390/bioengineering11060580










