Single-Trial Detection and Classification of Event-Related Optical Signals for a Brain–Computer Interface Application
Abstract
:1. Introduction
- Contributions:
- Conducting a subject-specific evaluation of CNNs for single-trial classification of EROS data.
- Comparing the discriminative potential of phase versus intensity data.
- Exploring various subject-specific model training paradigms using data from multiple recording montages.
- Analyzing the impact of data quality on classification performance.
- Performing a post hoc investigation of relevant time intervals and spatial locations contributing to the model’s predictions.
2. Related Work
2.1. Machine Learning Classification Algorithms for EEG
2.2. Convolutional Neural Networks for EEG
2.3. EROS-Based BCI
3. Materials and Methods
3.1. Participants
3.2. Behavioral Task
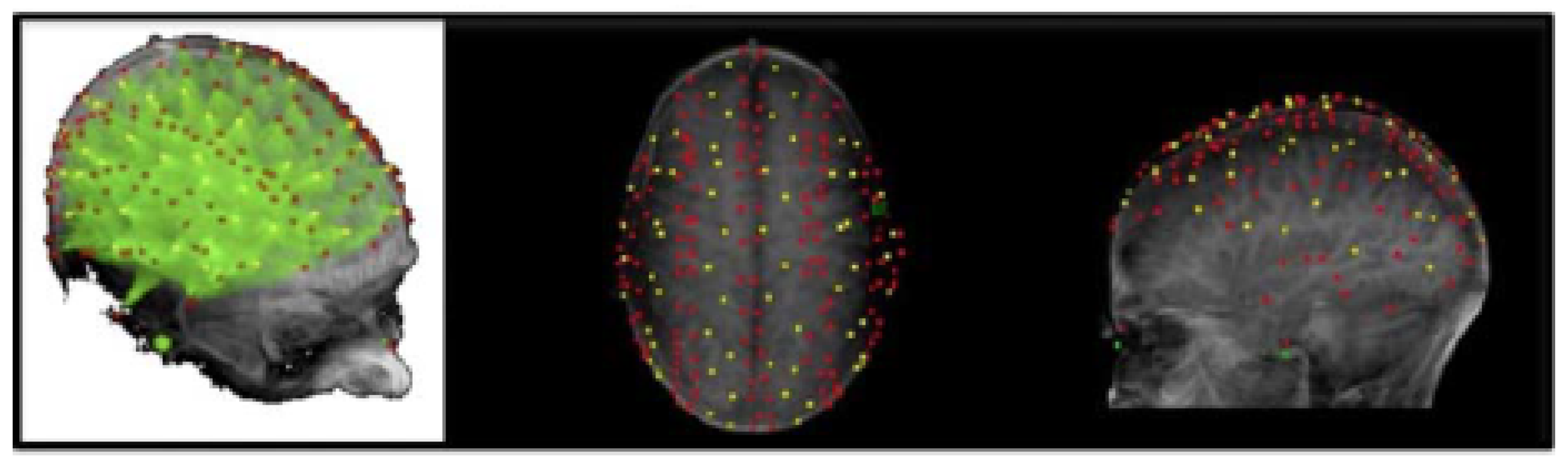
3.3. Optical Imaging Recording
3.4. Fast Optical Signal Pre-Processing
3.5. Machine Learning Approach
3.5.1. Model Architecture
3.5.2. Model Training
- (i)
- Montage-specific classification: A subject-specific model was trained solely on trials obtained from a specific montage and evaluated using the subject’s held-out trials recorded with the same montage. This approach reduced the number of available trials to a maximum of 240 as the data were partitioned into disjoint sets based on the montage configuration used for recording.
- (ii)
- Cross-montage classification: The model was trained using trials recorded by all available montages for the subject while ensuring an equal distribution of trials from each montage in the training, validation, and held-out test splits.
- (iii)
- Montage-specific classification with pre-training: A warm-start initialization of the CNN parameters was obtained by pre-training models on trials corresponding to all available montages, but excluding the specific montage under evaluation. Identically to the cross-montage case, the training and validation splits were stratified based on montage. The pre-trained model was then fine-tuned using montage-specific data from the previously held-out montage and evaluated the final trained model on the subject’s held-out trials for the corresponding montage.
3.6. Evaluation
- Input signal: Phase delay and intensity data were recorded for each participant. The following inputs were explored to train the proposed method: (i) only phase signals, (ii) only intensity signals, and (iii) both phase and intensity signals, recorded simultaneously.
- Frequency band: This study identifies informative frequency ranges for event-related activity occurring at specific frequencies (see Table 4 for frequency ranges of interest). Additionally, it assesses the CNN’s capability to automatically learn temporal filters compared to the conventional practice of manually selecting frequency bands, which has been commonly employed in the existing literature. The narrow frequency band analysis focuses on intensity in dual-input experiments using both phase and intensity input signals, given that intensity is particularly susceptible to the masking of discriminative information by low-frequency noise contamination.
- Training paradigm: Three distinct neural network training paradigms were used, and each was evaluated separately on the 0.1–12 Hz filtered phase input and intensity input: (i) montage-specific, (ii) cross-montage, and (iii) montage-specific with pre-training. In the case of dual-input experiments, separate spatiotemporal CNN architectures were initialized for each input type. These parallel models were trained jointly, and the flattened outputs from each model were combined and passed through a final dense layer for prediction.
- Viable voxel count: The quality of montage placement was quantified by determining the number of voxels that contain viable channels mapping to that voxel. This measure provides insight into how well the recording montage covers the region of interest, resulting in signal recording focused around the discriminative region.
- Channels per voxel: The average number of channels mapping to each voxel, averaged over all viable voxels within a montage, was computed. This measure evaluates the robustness of the recorded signal to noise disturbances, as voxels with a higher number of averaged channels tend to have a higher signal-to-noise ratio.
- These signal quality measures are summarized for each subject and montage in Table 2.
3.7. Feature Explainability
4. Results
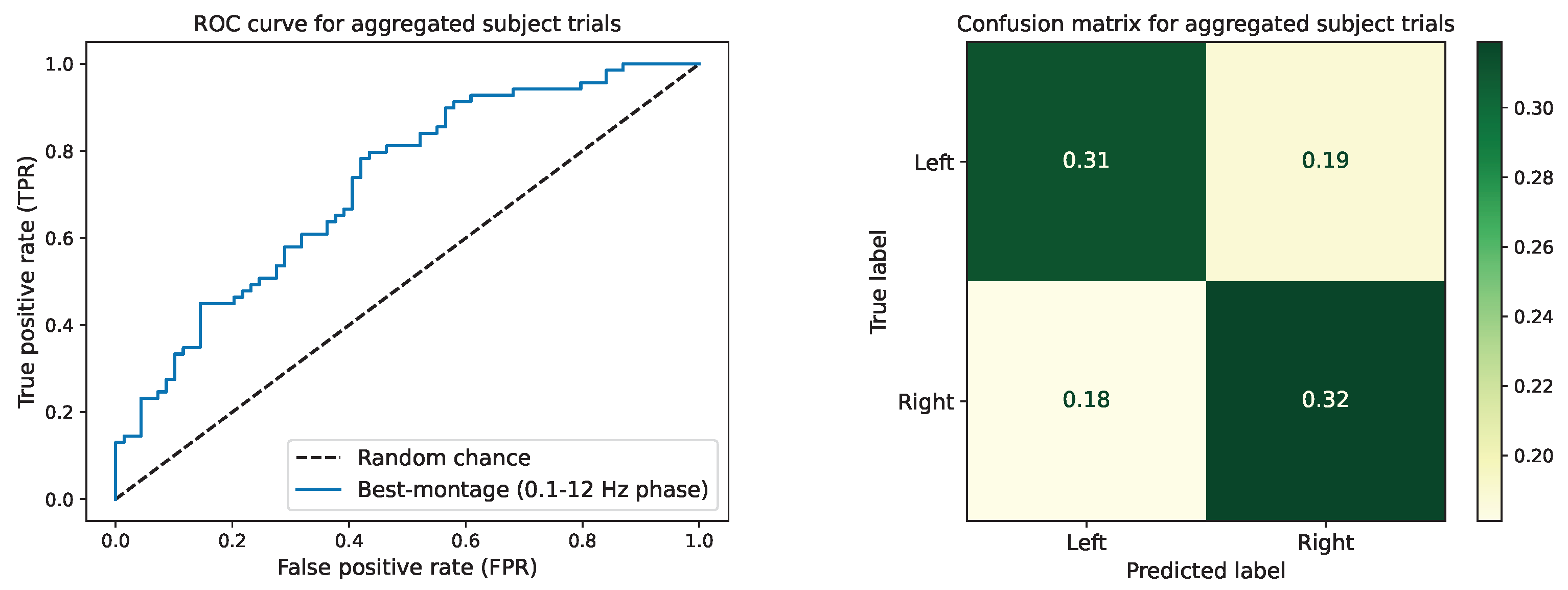
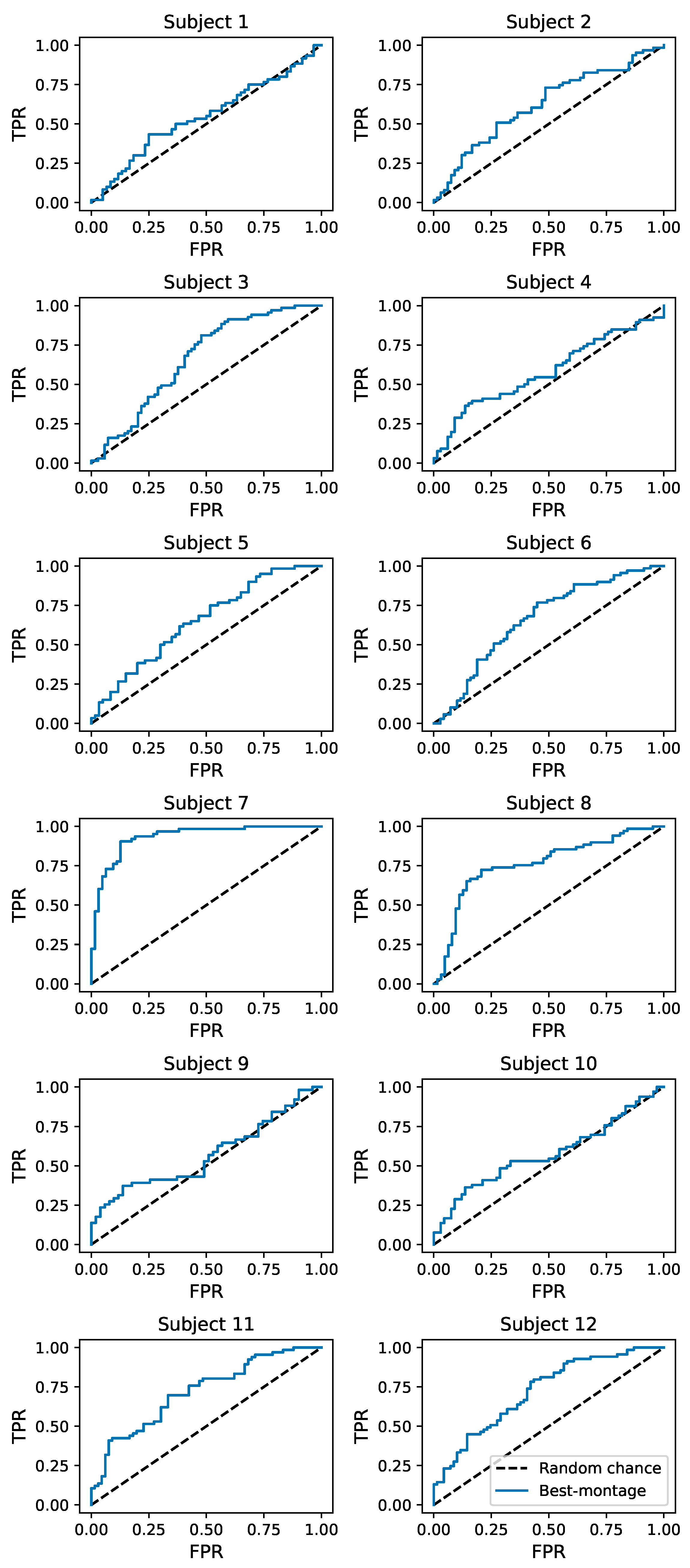
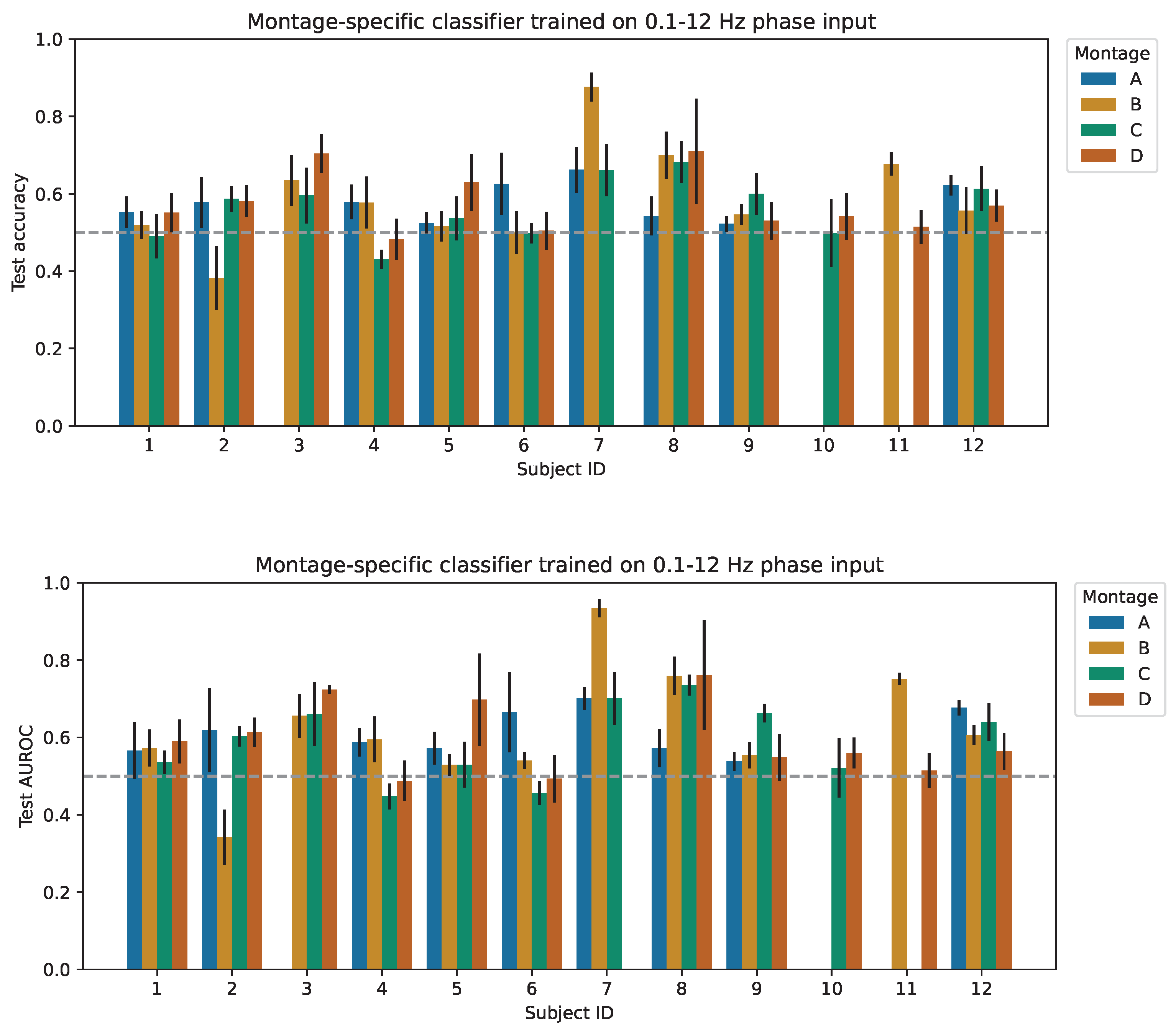
4.1. Subject-Specific Classification Performance
4.2. Correlation between Data Quality and Montage-Specific Performance
4.3. Correlation between Data Quality and Cross-Montage Performance
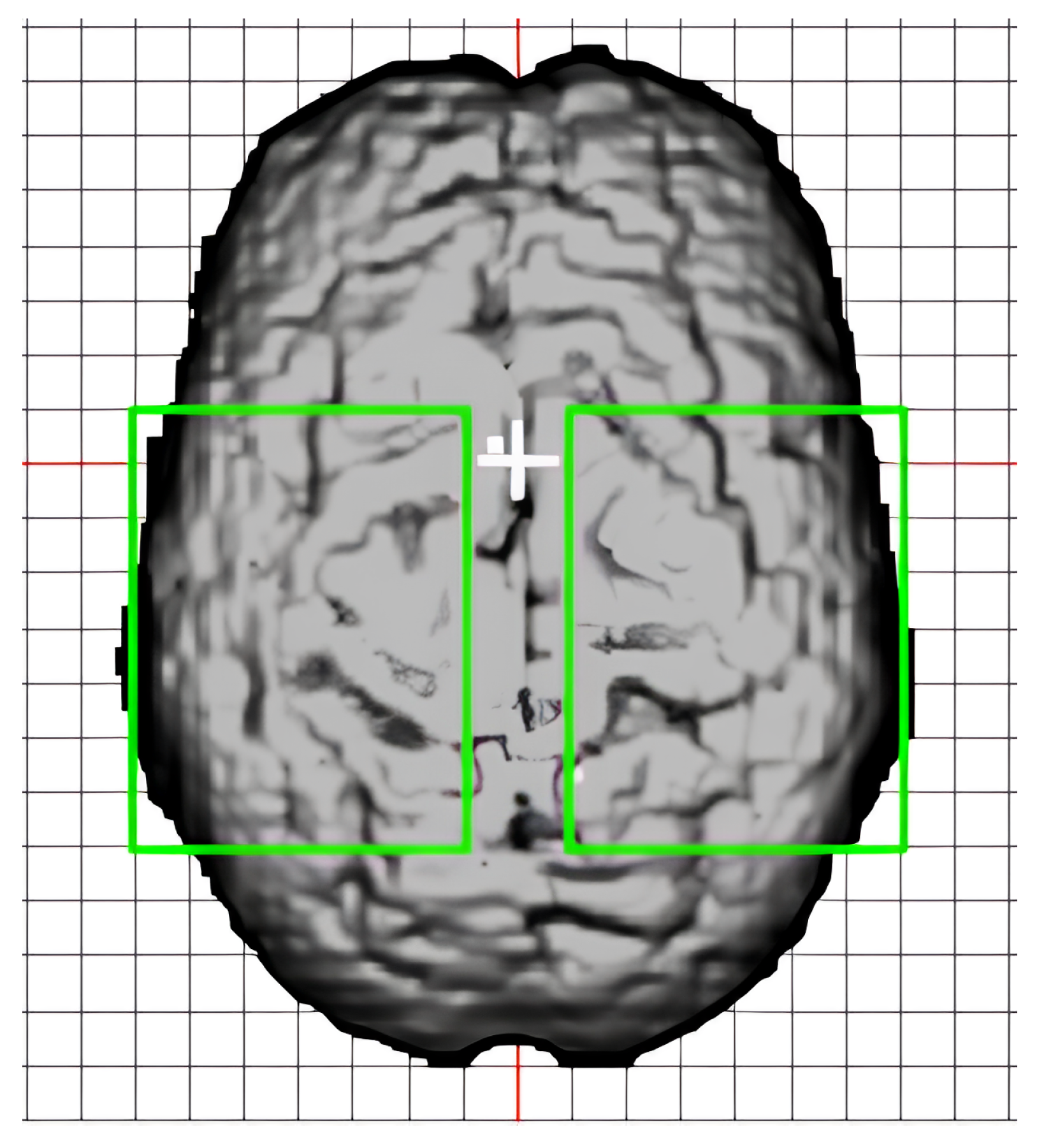
4.4. Neural Network Feature Relevance
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
- The following abbreviations are used in this manuscript:
| AC | alternating current light intensity |
| ANOVA | Analysis of Variance |
| AUROC | area under the curve of the Receiver Operating Characteristic |
| BCI | brain–computer interface |
| CI | confidence interval |
| CNN | convolutional neural network |
| CSP | common spatial patterns |
| CW | continuous-wave |
| DC | direct current light intensity |
| DT | decision tree |
| EEG | electroencephalography |
| EHR | electronic health records |
| ELU | exponential linear unit |
| EROS | event-related optical signals |
| FD | frequency-domain |
| FBCSP | filter bank common spatial patterns |
| fNIRS | functional near-infrared spectroscopy |
| FOS | fast optical signals |
| FPR | false positive rate |
| HSD | honestly significant difference |
| ICA | independent component analysis |
| KDA | kernel Fisher discriminant analysis |
| k-NN | k-nearest neighbors |
| LDA | linear discriminant analysis |
| LVQ | learning vector quantization |
| MI | motor imagery |
| MLP | multi-layer perceptron |
| MPRAGE | magnetization-prepared rapid gradient-echo |
| MR | magnetic resonance |
| NDA | nonlinear discriminant analysis |
| QDA | quadratic discriminant analysis |
| RFD | regularized Fischer discriminant |
| ROC | Receiver Operating Characteristic |
| ROI | region-of-interest |
| SAE | stacked autoencoder |
| SGD | stochastic gradient descent |
| SNR | signal-to-noise ratio |
| STFT | short-time Fourier transform |
| SVM | support vector machine |
| TPR | true positive rate |
Appendix A. Hyperparameter Tuning
| Hyperparameter | Default | Sampling Function | Value Range |
|---|---|---|---|
| # temporal filters () | 8 | for | [4, 16) |
| # spatial filters (D) | 2 | for | [4, 16) |
| # pointwise filters () | 16 | for | [4, 16) |
| Optimizer | Adam | Uniform | {Adam, SGD, RMSprop} |
| Learning rate | 0.001 | for | (0.0001, 0.01] |
| Weight decay | 0 | for | |
| Early stopping metric | Accuracy | Uniform | {Accuracy, AUROC} |
Appendix B. Baseline CNN Implementation Details
Appendix C. Additional Performance Metrics

References
- Coupé, C.; Oh, Y.M.; Dediu, D.; Pellegrino, F. Different languages, similar encoding efficiency: Comparable information rates across the human communicative niche. Sci. Adv. 2019, 5, eaaw2594. [Google Scholar] [CrossRef] [PubMed]
- Gratton, G.; Corballis, P.M.; Cho, E.; Fabiani, M.; Hood, D.C. Shades of gray matter: Noninvasive optical images of human brain reponses during visual stimulation. Psychophysiology 1995, 32, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Gratton, G.; Chiarelli, A.M.; Fabiani, M. From brain to blood vessels and back: A noninvasive optical imaging approach. Neurophotonics 2017, 4, 031208. [Google Scholar] [CrossRef]
- Radhakrishnan, H.; Vanduffel, W.; Deng, H.P.; Ekstrom, L.; Boas, D.A.; Franceschini, M.A. Fast optical signal not detected in awake behaving monkeys. NeuroImage 2009, 45, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Medvedev, A.V.; Kainerstorfer, J.; Borisov, S.V.; Barbour, R.L.; VanMeter, J. Event-related fast optical signal in a rapid object recognition task: Improving detection by the independent component analysis. Brain Res. 2008, 1236, 145–158. [Google Scholar] [CrossRef]
- Medvedev, A.V.; Kainerstorfer, J.M.; Borisov, S.V.; Gandjbakhche, A.H.; VanMeter, J.W. Seeing electroencephalogram through the skull: Imaging prefrontal cortex with fast optical signal. J. Biomed. Opt. 2010, 15, 061702. [Google Scholar] [CrossRef] [PubMed]
- Proulx, N.; Samadani, A.A.; Chau, T. Online classification of the near-infrared spectroscopy fast optical signal for brain-computer interfaces. Biomed. Phys. Eng. Express 2018, 4, 065010. [Google Scholar] [CrossRef]
- Baniqued, P.L.; Low, K.A.; Fabiani, M.; Gratton, G. Frontoparietal traffic signals: A fast optical imaging study of preparatory dynamics in response mode switching. J. Cogn. Neurosci. 2013, 25, 887–902. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.Y.; Bagul, A.; Langlotz, C.P.; Shpanskaya, K.S.; et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Faust, O.; Hagiwara, Y.; Hong, T.J.; Lih, O.S.; Acharya, U.R. Deep learning for healthcare applications based on physiological signals: A review. Comput. Methods Programs Biomed. 2018, 161, 1–13. [Google Scholar] [CrossRef]
- Shickel, B.; Tighe, P.J.; Bihorac, A.; Rashidi, P. Deep EHR: A Survey of Recent Advances on Deep Learning Techniques for Electronic Health Record (EHR) Analysis. arXiv 2017, arXiv:1706.03446. [Google Scholar] [CrossRef]
- Rajkomar, A.; Oren, E.; Chen, K.; Dai, A.M.; Hajaj, N.; Hardt, M.; Liu, P.J.; Liu, X.; Marcus, J.; Sun, M.; et al. Scalable and accurate deep learning with electronic health records. NPJ Digit. Med. 2018, 1, 1–10. [Google Scholar] [CrossRef]
- So, C.; Kim, J.U.; Luan, H.; Park, S.U.; Kim, H.; Han, S.; Kim, D.; Shin, C.; il Kim, T.; Lee, W.H.; et al. Epidermal piezoresistive structure with deep learning-assisted data translation. NPJ Flex. Electron. 2022, 6, 1–9. [Google Scholar] [CrossRef]
- Gong, S.; Zhang, X.; Nguyen, X.A.; Shi, Q.; Lin, F.; Chauhan, S.; Ge, Z.; Cheng, W. Hierarchically resistive skins as specific and multimetric on-throat wearable biosensors. Nat. Nanotechnol. 2023, 18, 889–897. [Google Scholar] [CrossRef]
- Guo, W.; Ma, Z.; Chen, Z.; Hua, H.; Wang, D.; Elhousseini Hilal, M.; Fu, Y.; Lu, P.; Lu, J.; Zhang, Y.; et al. Thin and soft Ti3C2Tx MXene sponge structure for highly sensitive pressure sensor assisted by deep learning. Chem. Eng. J. 2024, 485, 149659. [Google Scholar] [CrossRef]
- Herman, P.; Prasad, G.; McGinnity, T.M.; Coyle, D. Comparative Analysis of Spectral Approaches to Feature Extraction for EEG-Based Motor Imagery Classification. IEEE Trans. Neural Syst. Rehabil. Eng. 2008, 16, 317–326. [Google Scholar] [CrossRef]
- Wang, B.; Wong, C.M.; Wan, F.; Mak, P.U.; Mak, P.I.; Vai, M.I. Comparison of different classification methods for EEG-based brain computer interfaces: A case study. In Proceedings of the 2009 International Conference on Information and Automation, Zhuhai/Macau, China, 22–24 June 2009; pp. 1416–1421. [Google Scholar] [CrossRef]
- Zhang, R.; Xu, P.; Guo, L.; Zhang, Y.; Li, P.; Yao, D. Z-Score Linear Discriminant Analysis for EEG Based Brain-Computer Interfaces. PLoS ONE 2013, 8, e74433. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Tabar, Y.R.; Halici, U. A novel deep learning approach for classification of EEG motor imagery signals. J. Neural Eng. 2016, 14, 016003. [Google Scholar] [CrossRef]
- Olivas-Padilla, B.E.; Chacon-Murguia, M.I. Classification of multiple motor imagery using deep convolutional neural networks and spatial filters. Appl. Soft Comput. 2019, 75, 461–472. [Google Scholar] [CrossRef]
- Miao, M.; Hu, W.; Yin, H.; Zhang, K. Spatial-Frequency Feature Learning and Classification of Motor Imagery EEG Based on Deep Convolution Neural Network. Comput. Math. Methods Med. 2020, 2020, 1981728. [Google Scholar] [CrossRef]
- Zhang, C.; Kim, Y.K.; Eskandarian, A. EEG-inception: An accurate and robust end-to-end neural network for EEG-based motor imagery classification. J. Neural Eng. 2021, 18, 046014. [Google Scholar] [CrossRef]
- Schirrmeister, R.T.; Springenberg, J.T.; Fiederer, L.D.J.; Glasstetter, M.; Eggensperger, K.; Tangermann, M.; Hutter, F.; Burgard, W.; Ball, T. Deep learning with convolutional neural networks for EEG decoding and visualization. Hum. Brain Mapp. 2017, 38, 5391–5420. [Google Scholar] [CrossRef]
- Sakhavi, S.; Guan, C.; Yan, S. Parallel convolutional-linear neural network for motor imagery classification. In Proceedings of the 2015 23rd European Signal Processing Conference (EUSIPCO), Nice, France, 31 August–4 September 2015; pp. 2736–2740. [Google Scholar] [CrossRef]
- Cecotti, H.; Graser, A. Convolutional Neural Networks for P300 Detection with Application to Brain-Computer Interfaces. IEEE Trans. Pattern Anal. Mach. Intell. 2011, 33, 433–445. [Google Scholar] [CrossRef]
- Cecotti, H.; Eckstein, M.P.; Giesbrecht, B. Single-Trial Classification of Event-Related Potentials in Rapid Serial Visual Presentation Tasks Using Supervised Spatial Filtering. IEEE Trans. Neural Networks Learn. Syst. 2014, 25, 2030–2042. [Google Scholar] [CrossRef]
- Manor, R.; Geva, A.B. Convolutional Neural Network for Multi-Category Rapid Serial Visual Presentation BCI. Front. Comput. Neurosci. 2015, 9. [Google Scholar] [CrossRef]
- Shamwell, J.; Lee, H.; Kwon, H.; Marathe, A.R.; Lawhern, V.; Nothwang, W. Single-trial EEG RSVP classification using convolutional neural networks. In Proceedings of the Micro- and Nanotechnology Sensors, Systems, and Applications VIII; George, T., Dutta, A.K., Islam, M.S., Eds.; International Society for Optics and Photonics, SPIE: Bellingham, WA, USA, 2016; Volume 9836, p. 983622. [Google Scholar] [CrossRef]
- Antoniades, A.; Spyrou, L.; Took, C.C.; Sanei, S. Deep learning for epileptic intracranial EEG data. In Proceedings of the 2016 IEEE 26th International Workshop on Machine Learning for Signal Processing (MLSP), Salerno, Italy, 13–16 September 2016; pp. 1–6. [Google Scholar] [CrossRef]
- Liang, J.; Lu, R.; Zhang, C.; Wang, F. Predicting Seizures from Electroencephalography Recordings: A Knowledge Transfer Strategy. In Proceedings of the 2016 IEEE International Conference on Healthcare Informatics (ICHI), Chicago, IL, USA, 4–7 October 2016; pp. 184–191. [Google Scholar] [CrossRef]
- Mirowski, P.; Madhavan, D.; LeCun, Y.; Kuzniecky, R. Classification of patterns of EEG synchronization for seizure prediction. Clin. Neurophysiol. 2009, 120, 1927–1940. [Google Scholar] [CrossRef]
- Page, A.; Shea, C.; Mohsenin, T. Wearable seizure detection using convolutional neural networks with transfer learning. In Proceedings of the 2016 IEEE International Symposium on Circuits and Systems (ISCAS), Montreal, QC, Canada, 22–25 May 2016; pp. 1086–1089. [Google Scholar] [CrossRef]
- Thodoroff, P.; Pineau, J.; Lim, A. Learning Robust Features using Deep Learning for Automatic Seizure Detection. In Proceedings of the 1st Machine Learning for Healthcare Conference; Doshi-Velez, F., Fackler, J., Kale, D., Wallace, B., Wiens, J., Eds.; Proceedings of Machine Learning Research; Northeastern University: Boston, MA, USA, 2016; Volume 56, pp. 178–190. [Google Scholar]
- Lawhern, V.J.; Solon, A.J.; Waytowich, N.R.; Gordon, S.M.; Hung, C.P.; Lance, B.J. EEGNet: A Compact Convolutional Neural Network for EEG-based Brain-Computer Interfaces. J. Neural Eng. 2018, 15, 056013. [Google Scholar] [CrossRef]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. arXiv 2016, arXiv:1610.02357. [Google Scholar]
- Ogasawara, J.; Ikenoue, S.; Yamamoto, H.; Sato, M.; Kasuga, Y.; Mitsukura, Y.; Ikegaya, Y.; Yasui, M.; Tanaka, M.; Ochiai, D. Deep neural network-based classification of cardiotocograms outperformed conventional algorithms. Sci. Rep. 2021, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, Q.; Lu, H.; Won, D.; Yoon, S.W. 3D Medical Image Classification with Depthwise Separable Networks. Procedia Manuf. 2019, 39, 349–356. [Google Scholar] [CrossRef]
- Perpetuini, D.; Günal, M.; Chiou, N.; Koyejo, S.; Mathewson, K.; Low, K.A.; Fabiani, M.; Gratton, G.; Chiarelli, A.M. Fast Optical Signals for Real-Time Retinotopy and Brain Computer Interface. Bioengineering 2023, 10, 553. [Google Scholar] [CrossRef]
- Maclin, E.L.; Low, K.A.; Fabiani, M.; Gratton, G. Improving the signal-to-noise ratio of event-related optical signals. IEEE Eng. Med. Biol. Mag. 2007, 26, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Whalen, C.; Maclin, E.L.; Fabiani, M.; Gratton, G. Validation of a method for coregistering scalp recording locations with 3D structural MR images. Hum. Brain Mapp. 2008, 29, 1288–1301. [Google Scholar] [CrossRef] [PubMed]
- Gratton, G.; Corballis, P.M. Removing the heart from the brain: Compensation for the pulse artifact in the photon migration signal. Psychophysiology 1995, 32, 292–299. [Google Scholar] [CrossRef]
- Gratton, G.; Brumback, C.R.; Gordon, B.A.; Pearson, M.A.; Low, K.A.; Fabiani, M. Effects of measurement method, wavelength, and source-detector distance on the fast optical signal. Neuroimage 2006, 32, 1576–1590. [Google Scholar] [CrossRef]
- Gratton, G. “Opt-cont” and “Opt-3D”: A software suite for the analysis and 3D reconstruction of the event-related optical signal (EROS). Psychophysiology 2000, 37, S44. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. arXiv 2019, arXiv:1912.01703. [Google Scholar]
- Montavon, G.; Samek, W.; Müller, K.R. Methods for interpreting and understanding deep neural networks. Digit. Signal Process. 2018, 73, 1–15. [Google Scholar] [CrossRef]
- Ancona, M.; Ceolini, E.; Öztireli, C.; Gross, M. Towards better understanding of gradient-based attribution methods for Deep Neural Networks. arXiv 2017, arXiv:1711.06104. [Google Scholar]
- Shrikumar, A.; Greenside, P.; Kundaje, A. Learning Important Features Through Propagating Activation Differences. Int. Conf. Mach. Learn. (ICML) 2017, 7, 4844–4866. [Google Scholar]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should I trust you?” Explaining the predictions of any classifier. Proc. Acm Sigkdd Int. Conf. Knowl. Discov. Data Min. 2016, 13–17, 1135–1144. [Google Scholar] [CrossRef]
- Nguyen, A.; Yosinski, J.; Clune, J. Deep Neural Networks are Easily Fooled: High Confidence Predictions for Unrecognizable Images. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 427–436. [Google Scholar] [CrossRef]
- Zeiler, M.D.; Fergus, R. Visualizing and Understanding Convolutional Networks. Eur. Conf. Comput. Vis. (ECCV) 2013, 8689 LNCS, 818–833. [Google Scholar] [CrossRef]
- Baehrens, D.; Schroeter, T.; Harmeling, S.; Kawanabe, M.; Hansen, K.; Müller, K.R. How to Explain Individual Classification Decisions. J. Mach. Learn. Res. 2010, 11, 1803–1831. [Google Scholar]
- Kim, S.G.; Ashe, J.; Hendrich, K.; Ellermann, J.M.; Merkle, H.; Ugurbil, K.; Georgopoulos, A.P. Functional Magnetic Resonance Imaging of Motor Cortex: Hemispheric Asymmetry and Handedness. Science 1993, 261, 615–617. [Google Scholar] [CrossRef] [PubMed]
- Kübler, A.; Mushahwar, V.K.; Hochberg, L.R.; Donoghue, J.P. BCI Meeting 2005 - Workshop on clinical issues and applications. IEEE Trans. Neural Syst. Rehabil. Eng. 2006, 14, 131–134. [Google Scholar] [CrossRef]
- Chiarelli, A.M.; Romani, G.L.; Merla, A. Fast optical signals in the sensorimotor cortex: General Linear Convolution Model applied to multiple source–detector distance-based data. NeuroImage 2014, 85, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Gratton, G.; Fabiani, M.; Friedman, D.; Franceschini, M.A.; Fantini, S.; Corballis, P.; Gratton, E. Rapid changes of optical parameters in the human brain during a tapping task. J. Cogn. Neurosci. 1995, 7, 446–456. [Google Scholar] [CrossRef]
- Morren, G.; Wolf, M.; Lemmerling, P.; Wolf, U.; Choi, J.H.; Gratton, E.; De Lathauwer, L.; Van Huffel, S. Detection of fast neuronal signals in the motor cortex from functional near infrared spectroscopy measurements using independent component analysis. Med Biol. Eng. Comput. 2004, 42, 92–99. [Google Scholar] [CrossRef]
- Wolf, M.; Wolf, U.; Choi, J.H.; Toronov, V.; Adelina Paunescu, L.; Michalos, A.; Gratton, E. Fast cerebral functional signal in the 100-ms range detected in the visual cortex by frequency-domain near-infrared spectrophotometry. Psychophysiology 2003, 40, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Gratton, G.; Fabiani, M. The event-related optical signal (EROS) in visual cortex: Replicability, consistency, localization, and resolution. Psychophysiology 2003, 40, 561–571. [Google Scholar] [CrossRef] [PubMed]


| Author | BCI Application | ML Algorithm(s) | Data | Limitation |
|---|---|---|---|---|
| [17] | Motor imagery | LDA, RFD, SVM | EEG | Spectral density features limit applicability in online settings |
| [18] | Motor imagery, finger movement | LDA, QDA, KFD, SVM, MLP, LVQ, k-NN, DT | EEG | Reliance on task-specific feature extraction |
| [19] | Motor imagery | LDA, NDA, SVM | EEG | Reliance on task-specific feature extraction and selection |
| [22] | Motor imagery | CNN, SAE | EEG | Domain expertise required for frequency- band pre-processing, large trainable parameter count |
| [23] | Motor imagery | CNN | EEG | Feature extraction independently considered from classification algorithm, separate spatial filter for each pairwise combination of classes |
| [24] | Motor imagery | CNN | EEG | Discretized frequency subbands limit method’s capacity to identify trends across larger frequency ranges |
| [25] | Motor imagery | CNN | EEG | Large trainable parameter count |
| [26] | Motor imagery, hand and foot movement | CNN | EEG | Minimal performance gains over baselines in limited data regime |
| [37] | Various ERP- and oscillatory- based tasks | CNN | EEG | Subject-specific hyperparameter and architecture selection has yet to be explored |
| [5] | Object recognition | ICA | EROS | Event-related averaging across a large number of trials |
| [6] | Go-NoGo | ICA | EROS, EEG | Event-related averaging across a large number of trials |
| [7] | Visual oddball classification | LDA, SVM | EROS | Event-related averaging across a large number of trials, domain expertise required for feature extraction |
| [41] | Retinotopy classification | SVM | EROS | Domain expertise required for frequency-based feature extraction |
| Subject ID | Total Number of Trials | Viable Voxel Count | Channels Per Voxel | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | A | B | C | D | A | B | C | D | |
| 1 | 208 | 219 | 196 | 222 | 78 | 77 | 79 | 70 | 3.9 | 4.5 | 5.3 | 3.4 |
| 2 | 225 | 218 | 222 | 211 | 83 | 80 | 75 | 72 | 4.0 | 4.6 | 4.4 | 4.6 |
| 3 | 229 | 229 | 228 | 84 | 65 | 60 | 3.7 | 2.3 | 1.6 | |||
| 4 | 233 | 219 | 194 | 226 | 80 | 77 | 70 | 75 | 4.3 | 3.9 | 3.9 | 4.0 |
| 5 | 199 | 190 | 204 | 200 | 81 | 79 | 79 | 75 | 3.6 | 4.1 | 4.2 | 3.3 |
| 6 | 228 | 229 | 223 | 226 | 84 | 84 | 74 | 72 | 4.7 | 4.4 | 4.6 | 5.4 |
| 7 | 222 | 210 | 226 | 81 | 81 | 78 | 4.3 | 4.6 | 4.6 | |||
| 8 | 208 | 210 | 220 | 196 | 78 | 83 | 76 | 75 | 4.8 | 5.0 | 5.0 | 5.0 |
| 9 | 177 | 221 | 170 | 212 | 83 | 76 | 75 | 75 | 3.9 | 3.7 | 4.4 | 4.6 |
| 10 | 225 | 220 | 72 | 74 | 4.2 | 4.4 | ||||||
| 11 | 220 | 210 | 84 | 74 | 3.8 | 5.1 | ||||||
| 12 | 229 | 228 | 228 | 228 | 81 | 75 | 77 | 73 | 4.5 | 3.7 | 4.1 | 4.1 |
| Layer | # Filters | Size | # Params | Output | Activation | Options |
|---|---|---|---|---|---|---|
| Input | (C, T) | |||||
| Reshape | (1, C, T) | |||||
| Conv2D | (1, 20) | 20 ∗ | (, C, T) | Linear | mode = same | |
| BatchNorm | 2 ∗ | (, C, T) | ||||
| DepthwiseConv2D | D ∗ | (C, 1) | C ∗ D ∗ | (D ∗ , 1, T) | Linear | mode = valid, depth = D,max norm = 1 |
| BatchNorm | 2 ∗ D ∗ | (D ∗ , 1, T) | ||||
| Activation | (D ∗ , 1, T) | ELU | ||||
| Dropout | (D ∗ , 1, T) | |||||
| SeparableConv2D | (1, 20) | 20 ∗ D ∗ + ∗ (D ∗ ) | (, 1, T) | Linear | mode = same | |
| BatchNorm | 2 ∗ | (, 1, T) | ||||
| Activation | (, 1, T) | ELU | ||||
| AveragePool2D | (1, 8) | (, 1, T // 8) | ||||
| Dropout | (, 1, T // 8) | |||||
| Flatten | ( ∗ (T // 8)) | |||||
| Dense | ∗ (T // 8) | 1 | Sigmoid | max norm = 0.25 |
| Input Signal | Freq. (Hz) | Approach | Accuracy | >50% | AUROC | >0.5 |
|---|---|---|---|---|---|---|
| Phase | 0.1–12 | Best Montage | 0.628 ± 0.054 | 10/12 | 0.674 ± 0.059 | 12/12 |
| Phase | 0.1–12 | Cross-Montage | 0.559 ± 0.042 | 8/12 | 0.580 ± 0.054 | 8/12 |
| Phase | 0.1–12 | Pre-Train Best Montage | 0.597 ± 0.061 | 7/12 | 0.628 ± 0.074 | 8/12 |
| Phase | 4–7 | Best Montage | 0.519 ± 0.031 | 5/12 | 0.515 ± 0.037 | 3/12 |
| Phase | 8–13 | Best Montage | 0.506 ± 0.024 | 1/12 | 0.514 ± 0.031 | 1/12 |
| Phase | 13–20 | Best Montage | 0.477 ± 0.028 | 1/12 | 0.472 ± 0.040 | 0/12 |
| Intensity | 0.1–12 | Best Montage | 0.490 ± 0.025 | 1/12 | 0.484 ± 0.036 | 2/12 |
| Intensity | 0.1–12 | Cross-Montage | 0.502 ± 0.013 | 1/12 | 0.506 ± 0.019 | 2/12 |
| Intensity | 0.1–12 | Pre-Train Best Montage | 0.501 ± 0.036 | 4/12 | 0.501 ± 0.041 | 5/12 |
| Intensity | 4–7 | Best Montage | 0.507 ± 0.022 | 3/12 | 0.4950. ± 027 | 3/12 |
| Intensity | 8–13 | Best Montage | 0.522 ± 0.019 | 2/12 | 0.530 ± 0.029 | 3/12 |
| Intensity | 13–20 | Best Montage | 0.496 ± 0.025 | 2/12 | 0.496 ± 0.031 | 2/12 |
| Phase + Intensity | 0.1–12 + 0.1–12 | Dual-Input Best Montage | 0.580 ± 0.051 | 7/12 | 0.601 ± 0.067 | 7/12 |
| Phase + Intensity | 0.1–12 + 8–13 | Dual-Input Best Montage | 0.592 ± 0.051 | 7/12 | 0.608 ± 0.070 | 8/12 |
| Phase + Intensity | 0.1–12 + 13–20 | Dual-Input Best Montage | 0.586 ± 0.057 | 6/12 | 0.625 ± 0.069 | 7/12 |
| Input Signal | Freq. (Hz) | Approach | Accuracy | >50% | AUROC | >0.5 |
|---|---|---|---|---|---|---|
| Phase | 0.1–12 | Proposed CNN | 0.628 ± 0.054 | 10/12 | 0.674 ± 0.059 | 12/12 |
| Phase | 0.1–12 | DeepConvNet | 0.609 ± 0.094 | 7/12 | 0.635 ± 0.115 | 8/12 |
| Phase | 0.1–12 | EEGNet-8,2 | 0.604 ± 0.104 | 8/12 | 0.650 ± 0.118 | 9/12 |
| Phase | 0.1–12 | EEGNet-4,2 | 0.612 ± 0.093 | 7/12 | 0.643 ± 0.112 | 8/12 |
| Training Paradigm | Data Quality Measure | p-Value |
|---|---|---|
| Montage-specific | # viable voxels in L and R hemi. | 0.272 |
| Montage-specific | # viable voxels in R hemi. | 0.030 |
| Montage-specific | # viable voxels in L hemi. | 0.366 |
| Montage-specific | avg. # channels / viable voxel in L and R hemi. | 0.048 |
| Cross-montage | # training montages | <0.001 |
| Cross-montage | # montages w/ acc. >50% | <0.023 |
| Cross-montage | prop. montages w/ acc. >50% | 0.920 |
| Pre-train | # training montages | 0.514 |
| Pre-train | # montages w/ acc. >50% | <0.001 |
| Pre-train | prop. montages w/ acc. >50% | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiou, N.; Günal, M.; Koyejo, S.; Perpetuini, D.; Chiarelli, A.M.; Low, K.A.; Fabiani, M.; Gratton, G. Single-Trial Detection and Classification of Event-Related Optical Signals for a Brain–Computer Interface Application. Bioengineering 2024, 11, 781. https://doi.org/10.3390/bioengineering11080781
Chiou N, Günal M, Koyejo S, Perpetuini D, Chiarelli AM, Low KA, Fabiani M, Gratton G. Single-Trial Detection and Classification of Event-Related Optical Signals for a Brain–Computer Interface Application. Bioengineering. 2024; 11(8):781. https://doi.org/10.3390/bioengineering11080781
Chicago/Turabian StyleChiou, Nicole, Mehmet Günal, Sanmi Koyejo, David Perpetuini, Antonio Maria Chiarelli, Kathy A. Low, Monica Fabiani, and Gabriele Gratton. 2024. "Single-Trial Detection and Classification of Event-Related Optical Signals for a Brain–Computer Interface Application" Bioengineering 11, no. 8: 781. https://doi.org/10.3390/bioengineering11080781
APA StyleChiou, N., Günal, M., Koyejo, S., Perpetuini, D., Chiarelli, A. M., Low, K. A., Fabiani, M., & Gratton, G. (2024). Single-Trial Detection and Classification of Event-Related Optical Signals for a Brain–Computer Interface Application. Bioengineering, 11(8), 781. https://doi.org/10.3390/bioengineering11080781









