Emotion Detection from EEG Signals Using Machine Deep Learning Models
Abstract
1. Introduction
2. Related Works
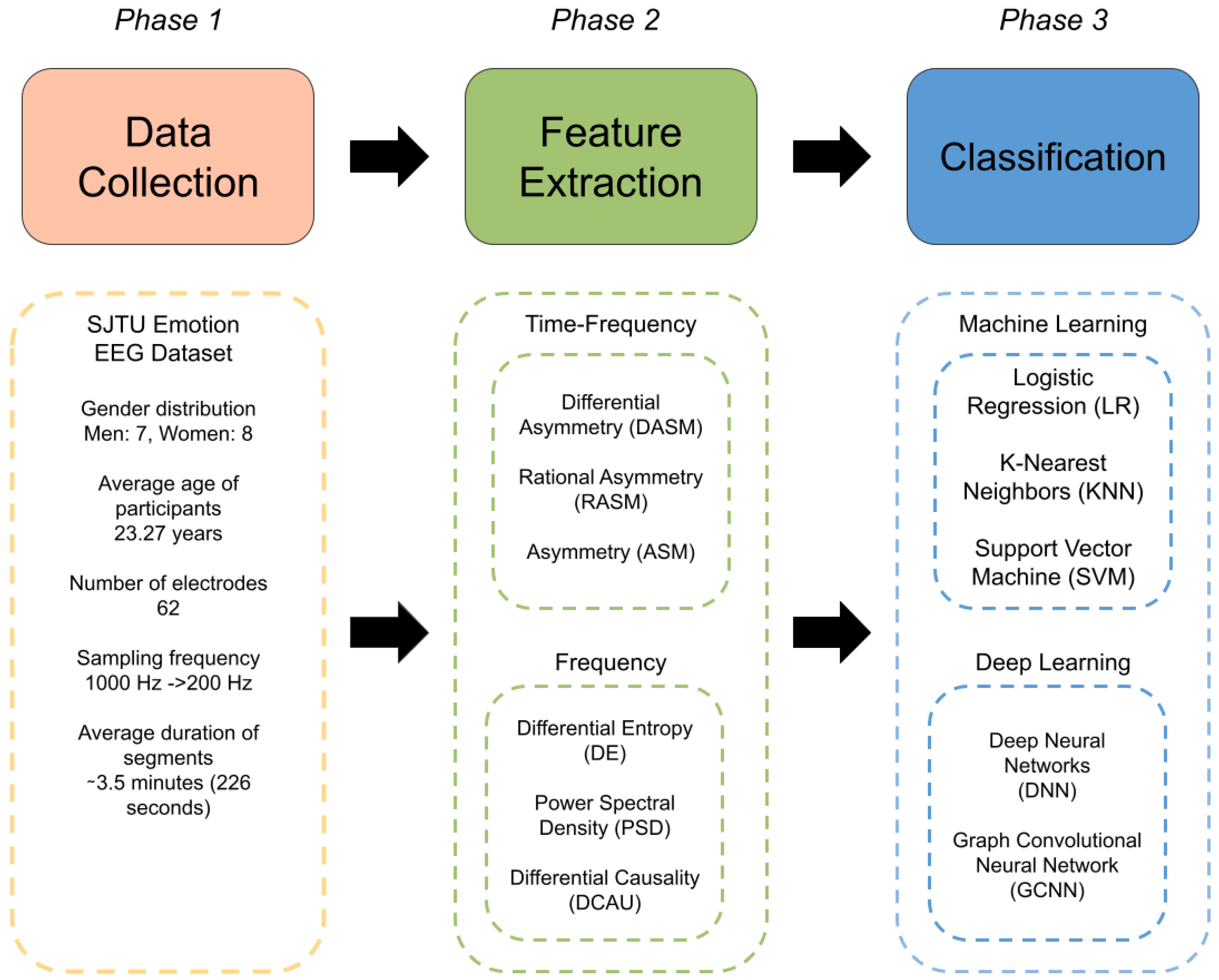
3. Materials and Methods
3.1. EEG Datasets
3.2. SEED
3.3. EEG Pre-Processing
3.3.1. Feature Extraction
- Differential entropy (DE);
- Power spectral density (PSD);
- Differential asymmetry (DASM);
- Rational asymmetry (RASM);
- Asymmetry (ASM);
- Differential causality (DCAU).
3.3.2. Feature Smoothing
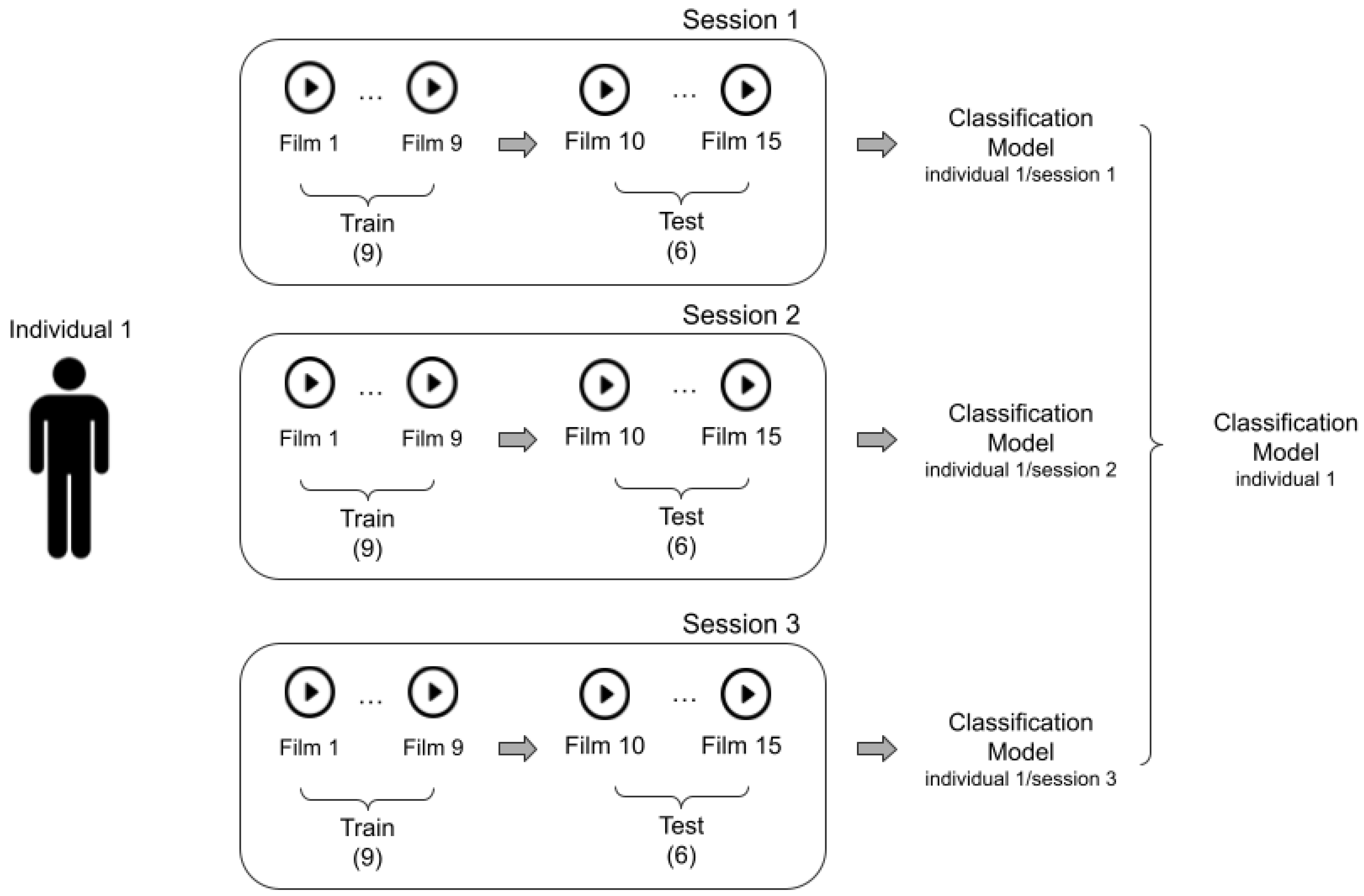
3.4. Experimental Design
Results Analysis
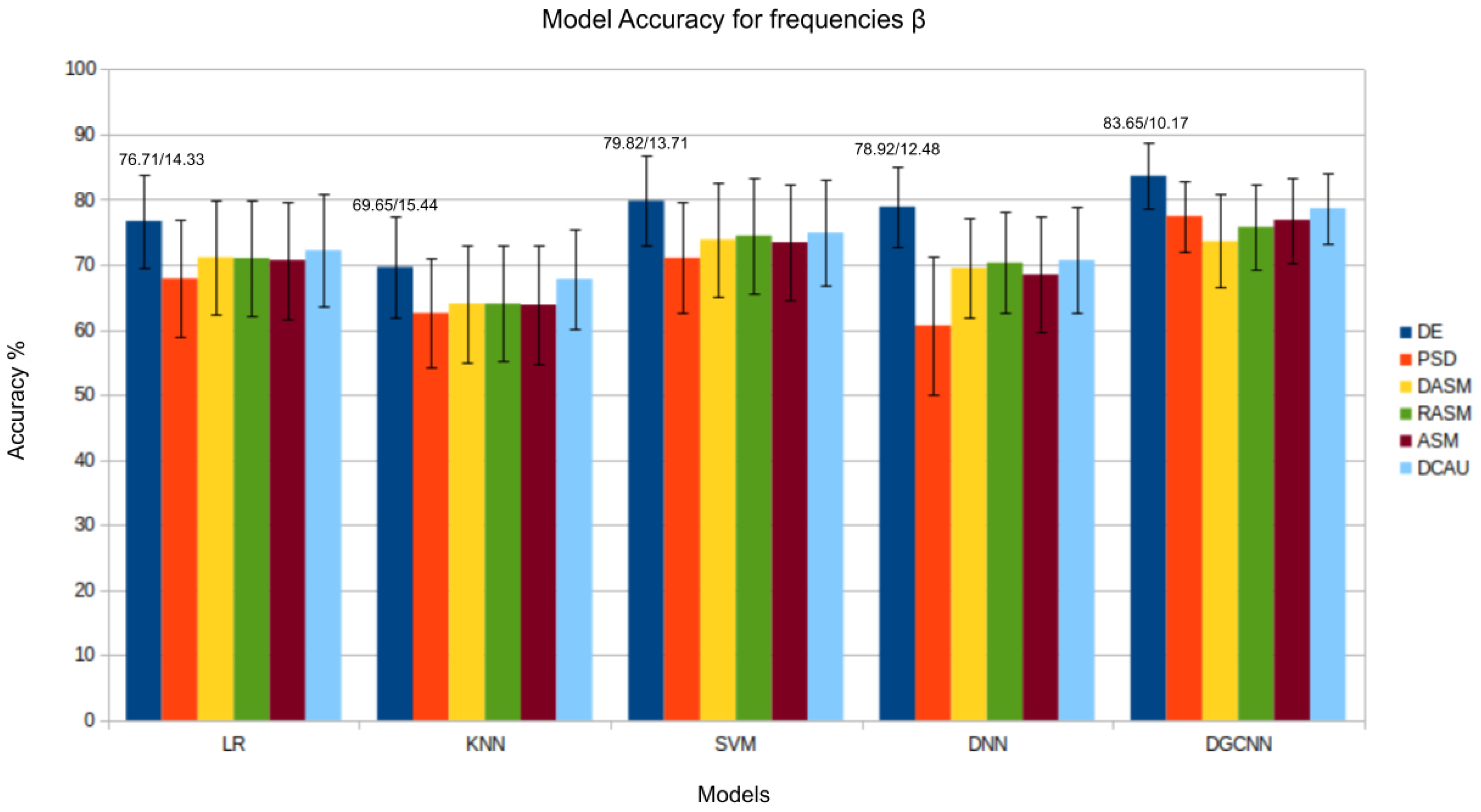
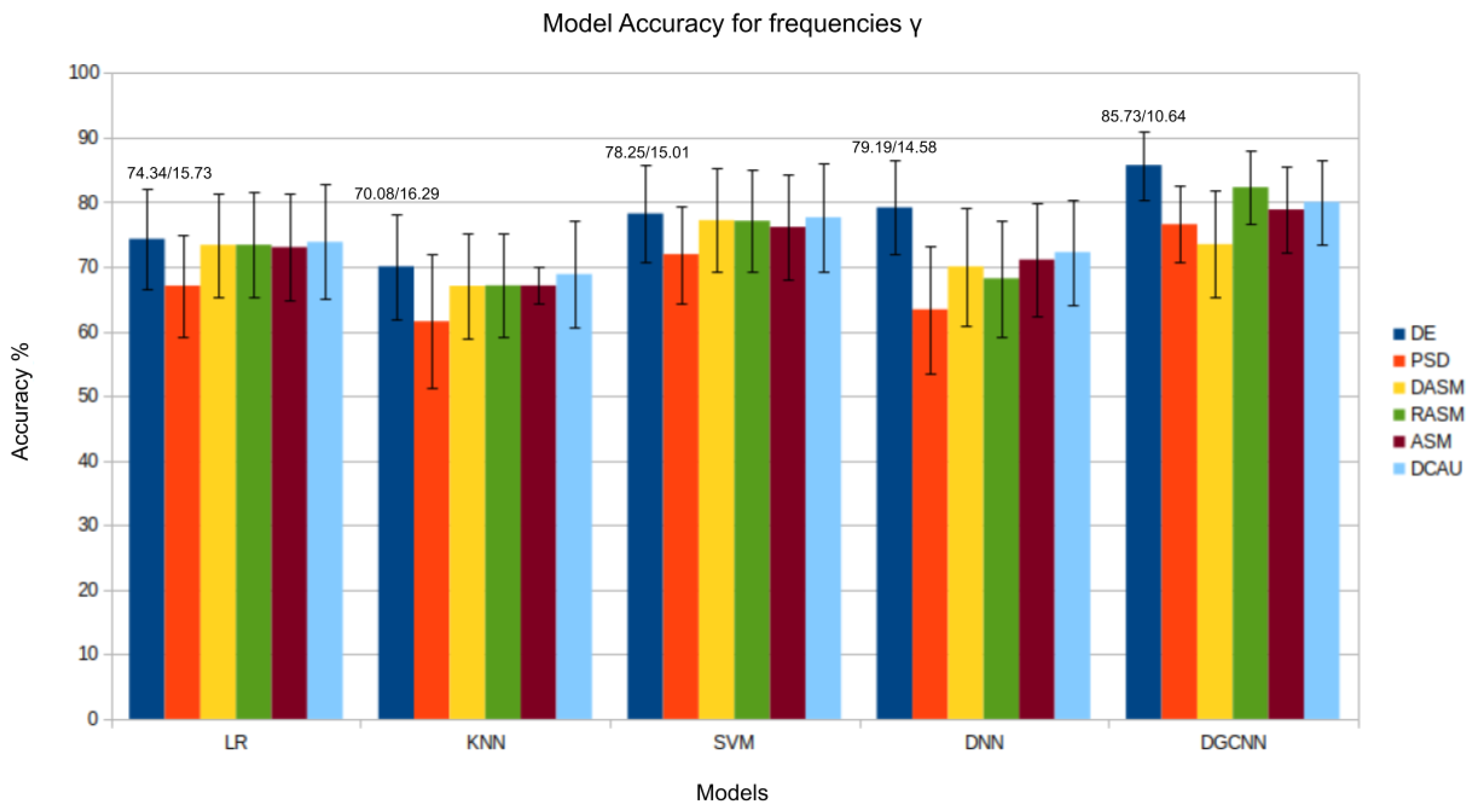
4. Results and Discussion
4.1. Traditional Machine Learning Models
4.2. Deep Learning Networks
- The use of a non-linear neural network like DGCNN makes it more effective in learning non-linear discriminative features.
- The graph representation of DGCNN provides a useful way to characterize the intrinsic relationships between various EEG channels, which is advantageous for extracting the most discriminative features for the emotion recognition task.
- The DGCNN model adaptively learns the intrinsic relationships of EEG channels by optimizing the adjacency matrix W.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, X.; Zhang, Y.; Tiwari, P.; Song, D.; Hu, B.; Yang, M.; Zhao, Z.; Kumar, N.; Marttinen, P. EEG Based Emotion Recognition: A Tutorial and Review. ACM Comput. Surv. 2022, 55, 1–57. [Google Scholar] [CrossRef]
- Roy, Y.; Banville, H.; Albuquerque, I.; Gramfort, A.; Falk, T.H.; Faubert, J. Deep learning-based electroencephalography analysis: A systematic review. J. Neural Eng. 2019, 16, 51001. [Google Scholar] [CrossRef]
- Wang, X.; Ren, Y.; Luo, Z.; He, W.; Hong, J.; Huang, Y. Deep learning-based EEG emotion recognition: Current trends and future perspectives. Front. Psychol. 2023, 14, 1126994. [Google Scholar] [CrossRef]
- Zhong, P.; Wang, D.; Miao, C. EEG-Based Emotion Recognition Using Regularized Graph Neural Networks. IEEE Trans. Affect. Comput. 2022, 13, 1290–1301. [Google Scholar] [CrossRef]
- Alarcao, S.M.; Fonseca, M.J. Emotions Recognition Using EEG Signals: A Survey. IEEE Trans. Affect. Comput. 2019, 10, 374–393. [Google Scholar] [CrossRef]
- Liu, W.; Zheng, W.L.; Li, Z.; Wu, S.Y.; Gan, L.; Lu, B.L. Identifying similarities and differences in emotion recognition with EEG and eye movements among Chinese, German, and French People. J. Neural Eng. 2022, 19, 026012. [Google Scholar] [CrossRef]
- Shu, L.; Xie, J.; Yang, M.; Li, Z.; Li, Z.; Liao, D.; Xu, X.; Yang, X. A Review of Emotion Recognition Using Physiological Signals. Sensors 2018, 18, 2074. [Google Scholar] [CrossRef]
- Bota, P.J.; Wang, C.; Fred, A.L.N.; Placido Da Silva, H. A Review, Current Challenges, and Future Possibilities on Emotion Recognition Using Machine Learning and Physiological Signals. IEEE Access 2019, 7, 140990–141020. [Google Scholar] [CrossRef]
- Gan, L.; Liu, W.; Luo, Y.; Wu, X.; Lu, B.L. A Cross-Culture Study on Multimodal Emotion Recognition Using Deep Learning. In Proceedings of the Neural Information Processing; Gedeon, T., Wong, K.W., Lee, M., Eds.; Communications in Computer and Information Science; Springer: Cham, Switzerland, 2019; pp. 670–680. [Google Scholar] [CrossRef]
- Zhang, G.; Yu, M.; Liu, Y.J.; Zhao, G.; Zhang, D.; Zheng, W. SparseDGCNN: Recognizing Emotion From Multichannel EEG Signals. IEEE Trans. Affect. Comput. 2023, 14, 537–548. [Google Scholar] [CrossRef]
- Gheisari, M.; Ebrahimzadeh, F.; Rahimi, M.; Moazzamigodarzi, M.; Liu, Y.; Dutta Pramanik, P.K.; Heravi, M.A.; Mehbodniya, A.; Ghaderzadeh, M.; Feylizadeh, M.R.; et al. Deep learning: Applications, architectures, models, tools, and frameworks: A comprehensive survey. CAAI Trans. Intell. Technol. 2023, 8, 581–606. [Google Scholar] [CrossRef]
- Zheng, W.L.; Zhu, J.Y.; Lu, B.L. Identifying Stable Patterns over Time for Emotion Recognition from EEG. IEEE Trans. Affect. Comput. 2019, 10, 417–429. [Google Scholar] [CrossRef]
- Li, Y.; Wang, L.; Zheng, W.; Zong, Y.; Qi, L.; Cui, Z.; Zhang, T.; Song, T. A Novel Bi-Hemispheric Discrepancy Model for EEG Emotion Recognition. IEEE Trans. Cogn. Dev. Syst. 2021, 13, 354–367. [Google Scholar] [CrossRef]
- Yongqiang, Y. EEG emotion recognition using fusion model of graph convolutional neural networks and LSTM. Appl. Soft Comput. 2021, 100, 106954. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, X.; Xu, X.; Chen, C.L.P. GCB-Net: Graph Convolutional Broad Network and Its Application in Emotion Recognition. IEEE Trans. Affect. Comput. 2022, 13, 379–388. [Google Scholar] [CrossRef]
- Jeong, D.K.; Kim, H.G.; Kim, J.Y. Emotion Recognition Using Hierarchical Spatiotemporal Electroencephalogram Information from Local to Global Brain Regions. Bioengineering 2023, 10, 1040. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, D.; Li, H.; Zhang, Y.; Xia, Y.; Liu, J. Self-training maximum classifier discrepancy for EEG emotion recognition. CAAI Trans. Intell. Technol. 2023, 8, 1480–1491. [Google Scholar] [CrossRef]
- Dwivedi, A.K.; Verma, O.P.; Taran, S. EEG-Based Emotion Recognition Using Optimized Deep-Learning Techniques. In Proceedings of the 2024 11th International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 21–22 March 2024; pp. 372–377. [Google Scholar] [CrossRef]
- Fan, C.; Xie, H.; Tao, J.; Li, Y.; Pei, G.; Li, T.; Lv, Z. ICaps-ResLSTM: Improved capsule network and residual LSTM for EEG emotion recognition. Biomed. Signal Process. Control 2024, 87, 105422. [Google Scholar] [CrossRef]
- Roshdy, A.; Karar, A.; Kork, S.A.; Beyrouthy, T.; Nait-ali, A. Advancements in EEG Emotion Recognition: Leveraging Multi-Modal Database Integration. Appl. Sci. 2024, 14, 2487. [Google Scholar] [CrossRef]
- Rajwal, S.; Aggarwal, S. Convolutional Neural Network-Based EEG Signal Analysis: A Systematic Review. Arch. Comput. Methods Eng. 2023, 30, 3585–3615. [Google Scholar] [CrossRef]
- Abhang, P.A.; Gawali, B.W.; Mehrotra, S.C. Chapter 2—Technological Basics of EEG Recording and Operation of Apparatus. In Introduction to EEG- and Speech-Based Emotion Recognition; Abhang, P.A., Gawali, B.W., Mehrotra, S.C., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 19–50. [Google Scholar] [CrossRef]
- Lotte, F.; Bougrain, L.; Cichocki, A.; Clerc, M.; Congedo, M.; Rakotomamonjy, A.; Yger, F. A review of classification algorithms for EEG-based brain–computer interfaces: A 10 year update. J. Neural Eng. 2018, 15, 31005. [Google Scholar] [CrossRef] [PubMed]
- Koelstra, S.; Muhl, C.; Soleymani, M.; Lee, J.S.; Yazdani, A.; Ebrahimi, T.; Pun, T.; Nijholt, A.; Patras, I. DEAP: A Database for Emotion Analysis; Using Physiological Signals. IEEE Trans. Affect. Comput. 2012, 3, 18–31. [Google Scholar] [CrossRef]
- Katsigiannis, S.; Ramzan, N. DREAMER: A Database for Emotion Recognition through EEG and ECG Signals from Wireless Low-cost Off-the-Shelf Devices. IEEE J. Biomed. Health Inform. 2018, 22, 98–107. [Google Scholar] [CrossRef]
- Lee, K.; Jeong, H.; Kim, S.; Yang, D.; Kang, H.C.; Choi, E. Real-Time Seizure Detection using EEG: A Comprehensive Comparison of Recent Approaches under a Realistic Setting. Proc. Conf. Health Inference Learn. 2022, 174, 311–337. [Google Scholar]
- Babayan, A.; Erbey, M.; Kumral, D.; Reinelt, J.D.; Reiter, A.M.F.; Röbbig, J.; Schaare, H.L.; Uhlig, M.; Anwander, A.; Bazin, P.L.; et al. A mind-brain-body dataset of MRI, EEG, cognition, emotion, and peripheral physiology in young and old adults. Sci. Data 2019, 6, 180308. [Google Scholar] [CrossRef]
- Soleymani, M.; Lichtenauer, J.; Pun, T.; Pantic, M. A Multimodal Database for Affect Recognition and Implicit Tagging. IEEE Trans. Affect. Comput. 2012, 3, 42–55. [Google Scholar] [CrossRef]
- Correa, A.G.; Laciar, E.; Patiño, H.D.; Valentinuzzi, M.E. Artifact removal from EEG signals using adaptive filters in cascade. J. Phys. Conf. Ser. 2007, 90, 012081. [Google Scholar] [CrossRef]
- Jenke, R.; Peer, A.; Buss, M. Feature Extraction and Selection for Emotion Recognition from EEG. IEEE Trans. Affect. Comput. 2014, 5, 327–339. [Google Scholar] [CrossRef]
- Wang, J.; Wang, M. Review of the emotional feature extraction and classification using EEG signals. Cogn. Robot. 2021, 1, 29–40. [Google Scholar] [CrossRef]
- Sanei, S.; Chambers, J. EEG Signal Processing; Wiley: Hoboken, NJ, USA, 2013. [Google Scholar]
- Du, X.; Ma, C.; Zhang, G.; Li, J.; Lai, Y.K.; Zhao, G.; Deng, X.; Liu, Y.J.; Wang, H. An Efficient LSTM Network for Emotion Recognition From Multichannel EEG Signals. IEEE Trans. Affect. Comput. 2022, 13, 1528–1540. [Google Scholar] [CrossRef]
- Ahmed, M.Z.I.; Sinha, N.; Phadikar, S.; Ghaderpour, E. Automated Feature Extraction on AsMap for Emotion Classification Using EEG. Sensors 2022, 22, 2346. [Google Scholar] [CrossRef]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information: Criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Lotte, F.; Congedo, M.; Lécuyer, A.; Lamarche, F.; Arnaldi, B. A review of classification algorithms for EEG-based brain-computer interfaces. J. Neural Eng. 2007, 4, R1–R13. [Google Scholar] [CrossRef]
- Song, T.; Zheng, W.; Liu, S.; Zong, Y.; Cui, Z.; Li, Y. Graph-Embedded Convolutional Neural Network for Image-Based EEG Emotion Recognition. IEEE Trans. Emerg. Top. Comput. 2022, 10, 1399–1413. [Google Scholar] [CrossRef]
- Song, T.; Zheng, W.; Song, P.; Cui, Z. EEG Emotion Recognition Using Dynamical Graph Convolutional Neural Networks. IEEE Trans. Affect. Comput. 2018, 11, 532–541. [Google Scholar] [CrossRef]



| Year | Authors | Model | DATASET | Features | Highest Accuracy |
|---|---|---|---|---|---|
| 2019 | ZHENG, ZHU, AND LU [12] | KNN, LR, SVM, GELM | SEED | DE, PSD, DASM, RASM, DCAU | 91.07% |
| 2021 | LI et al. [13] | Bi-Hemispheric Discrepancy Model -4 RNNS in 2 spatial orientations | SEED, SEED IV, MPED | DE—SEED e SEED IV STFT—MPED | SEED: 93.12% SEED IV: 74.35% MPED: 40.34% |
| 2021 | YONGQIANG [14] | LSTM + GCNN | DEAP | DE | 90.45% |
| 2022 | ZHANG et al. [15] | GCB-net | SEED, DREAMER | DE, PSD, DASM, RASM, DCAU—SEED PSD—DREAMER | SEED: 92.30% DREAMER: 86.99% |
| 2022 | ZHONG, WANG, AND MIAO [4] | RGCNN | SEED, SEED IV | DE | SEED: 94.24% SEED IV: 79.37% |
| 2023 | ZHANG et al. [10] | sparse DGCNN | SEED, DEAP, DREAMER e CMEED | DE, PSD, DASM, RASM e DCAU—SEED e DEAP PSD—DREAMER, CMEED | SEED: 98.53% DEAP: 95.72% DREAMER: 92.11% CMED: 91.72% |
| 2023 | JEONG, KIN, AND KIN [16] | HSCFLM | DEAP, MAHNOB-HCI, and SEED | local and global features | DEAP: 92.10% MAHNOB-HCI: 93.3% SEED: 90.9% |
| 2023 | Zhang, X., et al. [16] | Self-training Maximum ClassifierDiscrepancy (SMCD | SEED, SEED IV | DE as 3D cube | SEED: 96.36% SEED IV: 78.49% |
| 2023 | DWIVEDI VERMA AND TARAM [18] | SPWVD | GAMEEMO | GoogleNet | 84.2% |
| 2024 | FAN et al.[19] | ICaps-ResLSTM | DEAP, DREAMER | ICapsNet, ResLSTM | DEAP: 97.94% DREAMER: 94.71% |
| 2024 | ROSHDY et al. [20] | DeepFace and CNN | Local | CNN | 91.21% |
| Feature | Number of EEG Features per Experiment | ||||
|---|---|---|---|---|---|
| (0.5–4 Hz) | (4–8 Hz) | (8–13 Hz) | (13–30 Hz) | (30–100 Hz) | |
| PSD | 62 | 62 | 62 | 62 | 62 |
| DE | 62 | 62 | 62 | 62 | 62 |
| DASM | 27 | 27 | 27 | 27 | 27 |
| RASM | 27 | 27 | 27 | 27 | 27 |
| ASM | 54 | 54 | 54 | 54 | 54 |
| DCAU | 23 | 23 | 23 | 23 | 23 |
| Model | Hyperparameter | Values/Description |
|---|---|---|
| Logistic Regression (LR) | Penalty | “L2” |
| Solver | “lbfgs” | |
| K-Nearest Neighbors (KNN) | K | 3 to 10 |
| Distance Metric | p = 2 (Euclidean Distance) | |
| Support Vector Machine (SVM) | Kernel | Linear |
| Penalty | “L2” | |
| Loss | “squared hinge” | |
| C | Grid Search: and | |
| Deep Neural Network (DNN) | Hidden Layers | 128 × 64 × 32 |
| Activation Function | ReLU | |
| Optimization | RMSProp | |
| Epochs | 5000 | |
| Learning Rate | 0.007 | |
| Graph Convolutional Neural Network (GCNN) | Chebyshev Order | 2 |
| Convolution Layer | 32 | |
| Activation Function | ReLU | |
| Optimization | Adam | |
| Epochs | 20 | |
| Learning Rate | 0.01 | |
| Weight of Decay | 0.005 |
| Model | Feature | All Frequencies | |||||
|---|---|---|---|---|---|---|---|
| LR | DE | 61.18/13.71 | 65.32/14.16 | 67.58/13.74 | 76.71/14.33 | 74.34/15.73 | 82.46/11.11 |
| PSD | 58.66/15.66 | 66.02/12.92 | 62.81/17.23 | 67.87/18.09 | 67.09/15.92 | 75.44/15.02 | |
| DASM | 45.31/12.83 | 49.85/15.55 | 56.42/14.7 | 71.02/17.92 | 73.41/16.25 | 75.34/15.56 | |
| RASM | 45.72/12.8 | 49.93/15.67 | 56.57/14.54 | 71.12/17.71 | 73.39/16.1 | 75.41/15.53 | |
| ASM | 45.36/12.95 | 50.05/15.56 | 56.92/14.27 | 70.71/17.96 | 73.05/16.59 | 74.73/15.48 | |
| DCAU | 53.77/15.15 | 55.22/15.72 | 60.98/14.92 | 72.19/17.31 | 73.86/17.74 | 80.69/11.72 |
| Feature | Subject | Session | All Frequencies | |||||
|---|---|---|---|---|---|---|---|---|
| DE | 1 | 1 | 56.58 | 85.19 | 82.15 | 88.22 | 82.15 | 95.3 |
| 2 | 60.84 | 71.97 | 51.73 | 63.08 | 73.55 | 79.55 | ||
| 3 | 45.66 | 69.44 | 53.61 | 61.2 | 56 | 70.09 | ||
| 2 | 1 | 79.99 | 19.58 | 35.84 | 45.74 | 55.42 | 79.48 | |
| 2 | 73.99 | 57.73 | 68.64 | 65.03 | 65.61 | 91.84 | ||
| 3 | 59.47 | 65.97 | 81.65 | 52.89 | 51.23 | 61.13 | ||
| 3 | 1 | 67.92 | 52.96 | 63.37 | 78.83 | 68.28 | 89.88 | |
| 2 | 46.68 | 70.45 | 64.96 | 64.02 | 73.84 | 80.27 | ||
| 3 | 63.58 | 74.49 | 90.39 | 91.76 | 87.43 | 97.18 | ||
| 4 | 1 | 57.37 | 57.88 | 75.43 | 71.97 | 65.1 | 71.46 | |
| 2 | 59.03 | 56.58 | 59.03 | 58.38 | 63.73 | 68.5 | ||
| 3 | 61.71 | 78.11 | 71.68 | 55.64 | 67.05 | 74.06 | ||
| 5 | 1 | 67.27 | 88.15 | 61.56 | 55.06 | 37.14 | 70.74 | |
| 2 | 58.89 | 74.35 | 71.6 | 90.68 | 77.02 | 90.39 | ||
| 3 | 67.12 | 71.97 | 61.78 | 72.83 | 55.71 | 75.14 | ||
| 6 | 1 | 82.88 | 86.27 | 88.51 | 91.11 | 84.75 | 99.64 | |
| 2 | 46.32 | 71.6 | 75.29 | 82.73 | 84.1 | 78.03 | ||
| 3 | 51.81 | 69.58 | 70.95 | 80.27 | 77.38 | 80.78 | ||
| 7 | 1 | 52.1 | 80.71 | 77.67 | 86.85 | 64.31 | 84.03 | |
| 2 | 26.81 | 65.68 | 64.02 | 100 | 99.21 | 99.78 | ||
| 3 | 51.95 | 52.17 | 72.04 | 65.82 | 45.88 | 69.87 | ||
| 8 | 1 | 60.55 | 48.55 | 65.17 | 79.41 | 77.24 | 77.31 | |
| 2 | 42.27 | 64.02 | 69.58 | 87.21 | 57.44 | 89.02 | ||
| 3 | 69.65 | 50.87 | 84.03 | 97.54 | 68.28 | 96.46 | ||
| 9 | 1 | 78.83 | 65.46 | 54.91 | 77.96 | 97.4 | 92.77 | |
| 2 | 58.96 | 71.39 | 84.25 | 95.95 | 83.16 | 87.86 | ||
| 3 | 55.92 | 53.83 | 62.5 | 61.56 | 71.97 | 67.99 | ||
| 10 | 1 | 57.3 | 56.36 | 60.77 | 69.15 | 77.46 | 66.62 | |
| 2 | 69.94 | 34.9 | 41.55 | 66.69 | 68.35 | 70.16 | ||
| 3 | 52.17 | 48.27 | 54.7 | 76.59 | 78.11 | 81.65 | ||
| 11 | 1 | 48.99 | 65.75 | 79.62 | 64.52 | 76.23 | 70.59 | |
| 2 | 54.84 | 58.16 | 60.26 | 72.47 | 74.78 | 72.47 | ||
| 3 | 77.82 | 48.63 | 84.68 | 85.69 | 86.2 | 96.03 | ||
| 12 | 1 | 85.98 | 94.44 | 61.42 | 74.57 | 86.2 | 92.2 | |
| 2 | 29.91 | 49.78 | 54.48 | 62.07 | 56.07 | 73.27 | ||
| 3 | 64.16 | 62.21 | 77.17 | 89.81 | 96.17 | 85.91 | ||
| 13 | 1 | 59.75 | 63.87 | 43.06 | 73.92 | 86.05 | 84.39 | |
| 2 | 43.28 | 71.89 | 57.88 | 93.71 | 93.21 | 88.08 | ||
| 3 | 69.65 | 86.49 | 55.49 | 87.93 | 83.74 | 80.85 | ||
| 14 | 1 | 85.19 | 72.47 | 60.48 | 86.78 | 81.21 | 83.67 | |
| 2 | 64.52 | 70.59 | 62.36 | 72.47 | 68.42 | 79.84 | ||
| 3 | 52.1 | 64.23 | 61.27 | 63.22 | 44.15 | 66.33 | ||
| 15 | 1 | 70.23 | 72.83 | 91.33 | 93.06 | 98.7 | 100 | |
| 2 | 80.85 | 70.66 | 72.11 | 100 | 100 | 100 | ||
| 3 | 82.44 | 73.05 | 100 | 97.62 | 100 | 100 |
| Feature | Subject | Session | All Frequencies | |||||
|---|---|---|---|---|---|---|---|---|
| PSD | 1 | 1 | 46.24 | 73.7 | 88.8 | 70.09 | 74.71 | 90.17 |
| 2 | 47.11 | 78.76 | 55.13 | 56.21 | 62.93 | 76.3 | ||
| 3 | 35.4 | 52.53 | 58.24 | 71.68 | 60.84 | 54.99 | ||
| 2 | 1 | 82.44 | 55.78 | 21.17 | 38.44 | 49.71 | 65.03 | |
| 2 | 72.9 | 55.42 | 60.62 | 49.21 | 60.12 | 81.58 | ||
| 3 | 64.23 | 63.44 | 81.07 | 48.34 | 47.18 | 64.16 | ||
| 3 | 1 | 61.92 | 58.67 | 70.23 | 61.92 | 62.43 | 78.61 | |
| 2 | 69.44 | 70.16 | 56.43 | 60.55 | 74.35 | 76.16 | ||
| 3 | 54.26 | 65.82 | 92.34 | 93.93 | 78.83 | 93.42 | ||
| 4 | 1 | 39.31 | 67.7 | 63.8 | 54.12 | 65.03 | 78.47 | |
| 2 | 72.9 | 73.05 | 66.69 | 68.21 | 69.73 | 75.51 | ||
| 3 | 50.29 | 71.68 | 73.84 | 45.3 | 54.7 | 72.25 | ||
| 5 | 1 | 63.29 | 82.01 | 67.63 | 52.75 | 43.79 | 66.11 | |
| 2 | 57.51 | 71.97 | 62.72 | 84.68 | 74.42 | 82.95 | ||
| 3 | 69.15 | 63.87 | 49.57 | 70.45 | 59.03 | 73.99 | ||
| 6 | 1 | 70.45 | 81.36 | 57.51 | 73.63 | 67.05 | 85.04 | |
| 2 | 40.17 | 48.48 | 44.22 | 45.66 | 45.88 | 37.64 | ||
| 3 | 52.96 | 63.01 | 42.92 | 50.65 | 75.14 | 55.78 | ||
| 7 | 1 | 51.23 | 76.01 | 71.39 | 60.19 | 44.58 | 72.98 | |
| 2 | 27.6 | 85.77 | 56.72 | 96.24 | 88.29 | 95.38 | ||
| 3 | 66.26 | 53.83 | 62.36 | 35.4 | 34.47 | 64.09 | ||
| 8 | 1 | 65.25 | 33.67 | 56.5 | 82.51 | 66.33 | 65.82 | |
| 2 | 64.96 | 62.21 | 60.77 | 79.77 | 62.43 | 75.94 | ||
| 3 | 69.94 | 69.08 | 60.48 | 94.58 | 69.8 | 85.69 | ||
| 9 | 1 | 71.75 | 66.55 | 60.04 | 71.53 | 71.1 | 84.32 | |
| 2 | 64.81 | 58.24 | 69.65 | 83.96 | 64.67 | 71.46 | ||
| 3 | 59.75 | 48.63 | 79.19 | 61.49 | 61.34 | 69.94 | ||
| 10 | 1 | 50.43 | 45.74 | 56.65 | 60.69 | 39.96 | 59.68 | |
| 2 | 45.3 | 58.16 | 39.88 | 57.88 | 52.75 | 43.86 | ||
| 3 | 63.73 | 55.49 | 26.81 | 45.95 | 53.25 | 62.21 | ||
| 11 | 1 | 28.97 | 65.75 | 83.89 | 54.34 | 82.44 | 86.71 | |
| 2 | 48.41 | 78.4 | 65.1 | 77.96 | 75.51 | 67.77 | ||
| 3 | 63.44 | 65.61 | 94.08 | 91.26 | 86.2 | 98.77 | ||
| 12 | 1 | 86.78 | 94.44 | 54.55 | 75.72 | 71.24 | 97.54 | |
| 2 | 25.79 | 35.04 | 40.68 | 28.47 | 46.6 | 42.2 | ||
| 3 | 38.73 | 69.94 | 41.47 | 75.29 | 88.44 | 70.52 | ||
| 13 | 1 | 70.81 | 54.12 | 53.83 | 64.02 | 86.27 | 87.21 | |
| 2 | 41.47 | 66.11 | 52.75 | 81.07 | 84.1 | 80.06 | ||
| 3 | 59.83 | 78.83 | 57.08 | 84.47 | 67.12 | 77.96 | ||
| 14 | 1 | 60.33 | 68.93 | 51.01 | 73.27 | 79.55 | 77.89 | |
| 2 | 54.19 | 83.45 | 67.05 | 79.55 | 62.64 | 74.57 | ||
| 3 | 53.97 | 70.09 | 70.38 | 52.46 | 57.66 | 74.28 | ||
| 15 | 1 | 86.78 | 71.89 | 91.69 | 91.76 | 97.62 | 100 | |
| 2 | 89.45 | 86.71 | 89.74 | 100 | 100 | 100 | ||
| 3 | 79.84 | 70.66 | 100 | 98.48 | 98.63 | 100 |
| Feature | Subject | Session | All Frequencies | |||||
|---|---|---|---|---|---|---|---|---|
| DASM | 1 | 1 | 59.97 | 49.71 | 68.06 | 86.42 | 96.1 | 96.82 |
| 2 | 40.25 | 73.34 | 32.23 | 63.66 | 57.66 | 73.48 | ||
| 3 | 46.82 | 52.46 | 36.13 | 47.47 | 56.43 | 72.04 | ||
| 2 | 1 | 34.32 | 27.24 | 47.04 | 65.46 | 68.35 | 75.29 | |
| 2 | 46.82 | 24.86 | 42.85 | 76.16 | 59.54 | 74.78 | ||
| 3 | 52.89 | 23.55 | 61.92 | 41.91 | 47.33 | 62.57 | ||
| 3 | 1 | 50.72 | 45.74 | 54.34 | 52.82 | 58.82 | 66.76 | |
| 2 | 75.94 | 61.34 | 32.37 | 47.62 | 64.67 | 64.88 | ||
| 3 | 56.5 | 51.3 | 56.5 | 78.03 | 75.07 | 96.32 | ||
| 4 | 1 | 41.4 | 50.94 | 65.75 | 69 | 69.29 | 68.71 | |
| 2 | 46.24 | 25.72 | 43.57 | 57.37 | 59.61 | 46.6 | ||
| 3 | 68.35 | 32.95 | 63.29 | 42.2 | 57.95 | 56.58 | ||
| 5 | 1 | 32.59 | 63.87 | 35.69 | 41.62 | 29.7 | 45.01 | |
| 2 | 51.73 | 64.74 | 56.21 | 50.87 | 74.13 | 69.94 | ||
| 3 | 49.42 | 32.51 | 35.62 | 40.03 | 52.96 | 57.01 | ||
| 6 | 1 | 28.76 | 39.52 | 64.38 | 97.54 | 88.15 | 97.4 | |
| 2 | 17.77 | 55.78 | 59.39 | 65.39 | 72.04 | 61.71 | ||
| 3 | 32.8 | 71.6 | 66.26 | 84.68 | 80.13 | 78.32 | ||
| 7 | 1 | 45.38 | 70.81 | 72.69 | 74.64 | 78.68 | 84.32 | |
| 2 | 31.65 | 63.58 | 75.22 | 94.15 | 96.6 | 96.89 | ||
| 3 | 40.46 | 37.36 | 66.26 | 50.79 | 50.94 | 71.03 | ||
| 8 | 1 | 59.25 | 17.2 | 32.51 | 77.6 | 84.83 | 63.95 | |
| 2 | 48.41 | 55.64 | 48.34 | 100 | 72.83 | 91.33 | ||
| 3 | 35.4 | 52.96 | 56.65 | 65.39 | 93.64 | 77.17 | ||
| 9 | 1 | 62.43 | 66.62 | 53.25 | 75.29 | 68.5 | 74.13 | |
| 2 | 25.14 | 62.86 | 71.89 | 85.69 | 87.79 | 87.14 | ||
| 3 | 53.32 | 46.68 | 72.98 | 48.55 | 68.64 | 68.06 | ||
| 10 | 1 | 53.03 | 48.41 | 78.47 | 71.6 | 66.62 | 59.1 | |
| 2 | 56.21 | 35.19 | 49.93 | 61.71 | 78.68 | 75.94 | ||
| 3 | 52.82 | 38.58 | 39.81 | 71.97 | 76.81 | 73.41 | ||
| 11 | 1 | 47.9 | 35.91 | 82.37 | 77.53 | 85.12 | 90.61 | |
| 2 | 15.97 | 42.34 | 52.38 | 73.84 | 86.49 | 60.62 | ||
| 3 | 58.02 | 32.15 | 84.47 | 97.76 | 89.81 | 93.42 | ||
| 12 | 1 | 39.6 | 59.47 | 65.46 | 76.23 | 85.98 | 89.88 | |
| 2 | 40.17 | 27.6 | 50.94 | 48.7 | 39.23 | 40.03 | ||
| 3 | 63.73 | 53.25 | 45.66 | 92.34 | 99.35 | 99.06 | ||
| 13 | 1 | 38.58 | 74.78 | 50.51 | 79.48 | 91.98 | 92.63 | |
| 2 | 32.01 | 52.1 | 67.92 | 92.92 | 72.83 | 69.73 | ||
| 3 | 53.97 | 57.73 | 52.1 | 86.85 | 91.47 | 77.38 | ||
| 14 | 1 | 44 | 61.05 | 49.13 | 69.29 | 75.72 | 72.83 | |
| 2 | 59.68 | 77.38 | 50.14 | 77.38 | 75.43 | 74.78 | ||
| 3 | 41.62 | 57.08 | 56.65 | 62.07 | 58.82 | 57.88 | ||
| 15 | 1 | 29.48 | 44.44 | 40.46 | 88.29 | 86.99 | 91.4 | |
| 2 | 51.3 | 72.54 | 75.36 | 92.2 | 74.64 | 96.32 | ||
| 3 | 44.36 | 56.14 | 82.44 | 100 | 96.32 | 100 |
| Feature | Subject | Session | All Frequencies | |||||
|---|---|---|---|---|---|---|---|---|
| RASM | 1 | 1 | 65.46 | 52.75 | 67.34 | 85.4 | 95.95 | 97.25 |
| 2 | 38.15 | 73.92 | 32.37 | 63.8 | 57.88 | 73.41 | ||
| 3 | 46.24 | 51.66 | 35.84 | 46.1 | 57.51 | 72.69 | ||
| 2 | 1 | 32.37 | 26.45 | 46.1 | 65.03 | 68.57 | 76.73 | |
| 2 | 47.62 | 26.66 | 40.97 | 76.01 | 57.95 | 73.05 | ||
| 3 | 54.26 | 26.08 | 61.56 | 39.09 | 46.17 | 62.57 | ||
| 3 | 1 | 51.37 | 40.17 | 51.73 | 50.07 | 56.43 | 63.22 | |
| 2 | 74.78 | 60.4 | 33.53 | 49.21 | 68.86 | 66.84 | ||
| 3 | 58.96 | 49.42 | 55.71 | 78.68 | 73.84 | 96.32 | ||
| 4 | 1 | 39.38 | 59.75 | 63.01 | 67.41 | 64.81 | 67.49 | |
| 2 | 44.94 | 24.78 | 51.45 | 56.94 | 62.21 | 50.29 | ||
| 3 | 62.79 | 30.49 | 63.08 | 40.1 | 55.27 | 55.2 | ||
| 5 | 1 | 36.05 | 62.5 | 34.18 | 40.75 | 26.16 | 44.08 | |
| 2 | 52.1 | 58.31 | 55.92 | 52.46 | 73.7 | 69.73 | ||
| 3 | 53.03 | 30.85 | 31.79 | 41.62 | 56.79 | 55.56 | ||
| 6 | 1 | 25.72 | 37.14 | 69.65 | 98.05 | 86.63 | 98.12 | |
| 2 | 19.15 | 57.08 | 59.32 | 64.31 | 64.88 | 60.77 | ||
| 3 | 27.6 | 75.07 | 67.27 | 84.97 | 82.37 | 77.89 | ||
| 7 | 1 | 43.5 | 69.51 | 75.29 | 77.24 | 77.67 | 83.89 | |
| 2 | 38.73 | 65.53 | 75.43 | 94.15 | 96.53 | 96.82 | ||
| 3 | 38.87 | 35.69 | 64.09 | 51.95 | 50.94 | 72.47 | ||
| 8 | 1 | 57.95 | 19.51 | 30.56 | 80.27 | 85.91 | 64.81 | |
| 2 | 50.29 | 52.67 | 48.7 | 99.49 | 77.1 | 91.69 | ||
| 3 | 35.98 | 48.41 | 57.01 | 65.53 | 93.57 | 77.1 | ||
| 9 | 1 | 61.34 | 67.99 | 53.18 | 73.63 | 70.01 | 74.57 | |
| 2 | 24.57 | 57.95 | 70.81 | 84.61 | 87.64 | 86.49 | ||
| 3 | 48.05 | 51.01 | 58.02 | 49.28 | 68.14 | 66.04 | ||
| 10 | 1 | 51.81 | 48.55 | 82.66 | 69.94 | 64.52 | 58.82 | |
| 2 | 59.03 | 38.95 | 52.53 | 63.22 | 79.12 | 77.1 | ||
| 3 | 51.01 | 38.95 | 41.47 | 69.44 | 75.58 | 72.11 | ||
| 11 | 1 | 51.59 | 36.71 | 81 | 77.24 | 86.49 | 90.46 | |
| 2 | 14.67 | 43.06 | 54.19 | 73.48 | 86.34 | 61.99 | ||
| 3 | 55.06 | 32.08 | 83.67 | 97.62 | 90.39 | 92.56 | ||
| 12 | 1 | 35.69 | 59.54 | 64.88 | 76.95 | 85.84 | 89.45 | |
| 2 | 39.31 | 27.67 | 48.63 | 48.84 | 43.64 | 40.03 | ||
| 3 | 57.51 | 51.3 | 43.86 | 92.34 | 100 | 99.13 | ||
| 13 | 1 | 42.12 | 72.76 | 48.92 | 79.77 | 92.05 | 91.76 | |
| 2 | 34.18 | 50.87 | 68.86 | 92.99 | 73.84 | 70.66 | ||
| 3 | 50.36 | 58.45 | 45.09 | 87.07 | 92.49 | 77.89 | ||
| 14 | 1 | 45.45 | 60.48 | 50.79 | 68.93 | 75.58 | 71.32 | |
| 2 | 62.07 | 78.03 | 49.71 | 78.25 | 72.18 | 77.17 | ||
| 3 | 40.82 | 62.79 | 55.71 | 61.71 | 62.86 | 57.15 | ||
| 15 | 1 | 25.58 | 45.81 | 51.16 | 88.37 | 89.31 | 91.18 | |
| 2 | 47.76 | 69.73 | 79.41 | 93.64 | 74.28 | 96.53 | ||
| 3 | 45.52 | 55.85 | 82.59 | 100 | 95.59 | 100 |
| Feature | Subject | Session | All Frequencies | |||||
|---|---|---|---|---|---|---|---|---|
| ASM | 1 | 1 | 66.18 | 49.35 | 67.12 | 86.99 | 97.04 | 97.4 |
| 2 | 42.63 | 73.12 | 32.8 | 66.26 | 52.67 | 70.09 | ||
| 3 | 44.29 | 53.47 | 35.19 | 53.11 | 56.58 | 77.53 | ||
| 2 | 1 | 37.21 | 26.81 | 44.65 | 66.18 | 67.56 | 76.01 | |
| 2 | 44 | 30.13 | 45.16 | 74.57 | 58.45 | 71.82 | ||
| 3 | 54.91 | 25.14 | 62.21 | 38.01 | 46.97 | 59.9 | ||
| 3 | 1 | 52.02 | 41.84 | 53.25 | 51.66 | 58.96 | 62.57 | |
| 2 | 74.35 | 59.03 | 31.86 | 44.58 | 66.91 | 59.39 | ||
| 3 | 58.53 | 50.65 | 57.66 | 77.96 | 77.02 | 91.47 | ||
| 4 | 1 | 40.68 | 63.08 | 65.68 | 67.63 | 68.21 | 65.9 | |
| 2 | 46.53 | 27.67 | 45.81 | 57.3 | 62.28 | 51.16 | ||
| 3 | 60.19 | 29.12 | 63.08 | 39.45 | 56.86 | 55.71 | ||
| 5 | 1 | 33.74 | 64.09 | 37.07 | 38.01 | 27.67 | 42.77 | |
| 2 | 52.02 | 64.38 | 52.82 | 49.42 | 65.17 | 68.57 | ||
| 3 | 50.72 | 30.27 | 33.82 | 43.14 | 52.67 | 54.62 | ||
| 6 | 1 | 28.25 | 36.78 | 62.86 | 99.21 | 86.27 | 98.41 | |
| 2 | 19.08 | 55.49 | 59.61 | 69.08 | 70.16 | 57.51 | ||
| 3 | 33.09 | 75.87 | 66.33 | 83.67 | 81.21 | 79.05 | ||
| 7 | 1 | 44.65 | 70.23 | 71.97 | 74.78 | 77.67 | 86.13 | |
| 2 | 32.08 | 60.26 | 74.42 | 95.45 | 94.65 | 95.95 | ||
| 3 | 36.05 | 33.53 | 68.71 | 50.43 | 50.94 | 74.57 | ||
| 8 | 1 | 60.84 | 19.65 | 35.77 | 81.07 | 80.71 | 62.79 | |
| 2 | 50.87 | 57.51 | 50.72 | 98.63 | 83.67 | 91.84 | ||
| 3 | 40.25 | 51.88 | 62.72 | 66.18 | 94.29 | 79.84 | ||
| 9 | 1 | 62.43 | 67.7 | 53.68 | 76.81 | 69.8 | 74.57 | |
| 2 | 22.11 | 62.93 | 74.28 | 84.68 | 87.86 | 86.13 | ||
| 3 | 50.65 | 56.72 | 65.68 | 49.86 | 65.32 | 70.52 | ||
| 10 | 1 | 51.95 | 43.93 | 76.81 | 73.63 | 67.63 | 58.09 | |
| 2 | 59.25 | 38.01 | 52.6 | 61.2 | 80.85 | 79.19 | ||
| 3 | 51.88 | 39.81 | 41.33 | 63.44 | 75.29 | 72.76 | ||
| 11 | 1 | 49.28 | 36.78 | 81.58 | 74.06 | 86.49 | 91.62 | |
| 2 | 15.25 | 42.05 | 53.68 | 74.49 | 86.49 | 61.13 | ||
| 3 | 57.51 | 32.95 | 83.45 | 98.7 | 90.25 | 93.79 | ||
| 12 | 1 | 37.93 | 58.53 | 64.74 | 76.23 | 84.68 | 85.77 | |
| 2 | 39.45 | 26.01 | 52.53 | 44.08 | 30.85 | 41.55 | ||
| 3 | 55.85 | 52.46 | 43.57 | 88.29 | 95.16 | 92.7 | ||
| 13 | 1 | 40.61 | 67.27 | 48.19 | 79.12 | 91.69 | 90.32 | |
| 2 | 32.23 | 49.13 | 71.53 | 93.5 | 77.24 | 70.52 | ||
| 3 | 54.05 | 58.74 | 53.47 | 84.47 | 88.44 | 75.43 | ||
| 14 | 1 | 38.22 | 59.97 | 51.73 | 68.79 | 75.79 | 70.66 | |
| 2 | 60.98 | 79.34 | 50.65 | 79.48 | 73.41 | 75.87 | ||
| 3 | 41.76 | 62.93 | 58.02 | 63.22 | 66.26 | 56.65 | ||
| 15 | 1 | 22.4 | 45.38 | 41.76 | 86.92 | 87.28 | 88.22 | |
| 2 | 52.1 | 68.35 | 80.42 | 88.15 | 74.78 | 96.46 | ||
| 3 | 41.98 | 53.83 | 80.56 | 100 | 97.04 | 100 |
| Feature | Subject | Session | All Frequencies | |||||
|---|---|---|---|---|---|---|---|---|
| DCAU | 1 | 1 | 29.19 | 60.91 | 81.65 | 74.21 | 86.27 | 88.01 |
| 2 | 47.83 | 80.42 | 45.16 | 70.3 | 65.17 | 74.49 | ||
| 3 | 49.64 | 69.44 | 71.68 | 68.42 | 32.08 | 80.35 | ||
| 2 | 1 | 58.16 | 28.18 | 35.4 | 58.82 | 71.39 | 75.29 | |
| 2 | 64.16 | 42.05 | 57.95 | 56.72 | 69.08 | 75.79 | ||
| 3 | 47.47 | 60.26 | 65.75 | 57.08 | 44.08 | 68.57 | ||
| 3 | 1 | 52.31 | 49.35 | 55.13 | 55.92 | 61.85 | 81.43 | |
| 2 | 38.37 | 78.97 | 56.79 | 68.21 | 56.58 | 79.48 | ||
| 3 | 71.03 | 32.73 | 48.7 | 86.34 | 71.82 | 93.79 | ||
| 4 | 1 | 72.04 | 20.01 | 54.05 | 48.41 | 75.29 | 66.33 | |
| 2 | 65.39 | 40.75 | 49.78 | 53.11 | 72.11 | 72.18 | ||
| 3 | 60.26 | 54.05 | 60.84 | 66.26 | 70.66 | 73.84 | ||
| 5 | 1 | 58.45 | 57.08 | 41.04 | 35.48 | 39.96 | 56.79 | |
| 2 | 57.51 | 56.07 | 49.49 | 41.33 | 81.5 | 88.51 | ||
| 3 | 66.55 | 50 | 24.71 | 52.96 | 26.08 | 50.36 | ||
| 6 | 1 | 63.95 | 82.59 | 91.11 | 77.89 | 72.4 | 97.98 | |
| 2 | 41.98 | 57.23 | 57.73 | 85.98 | 89.16 | 82.66 | ||
| 3 | 49.28 | 67.63 | 62.64 | 71.75 | 71.53 | 84.9 | ||
| 7 | 1 | 42.77 | 56.65 | 56.94 | 46.03 | 60.84 | 61.78 | |
| 2 | 29.77 | 70.01 | 56.94 | 98.84 | 95.66 | 98.19 | ||
| 3 | 25.43 | 34.68 | 61.49 | 52.31 | 55.13 | 68.57 | ||
| 8 | 1 | 44.8 | 56.14 | 69.29 | 96.97 | 75.22 | 79.48 | |
| 2 | 35.77 | 70.45 | 71.53 | 100 | 88.51 | 93.57 | ||
| 3 | 52.75 | 65.9 | 43.35 | 96.39 | 85.91 | 88.37 | ||
| 9 | 1 | 75.72 | 45.01 | 40.97 | 73.48 | 92.56 | 90.61 | |
| 2 | 47.9 | 55.64 | 73.7 | 90.25 | 95.3 | 74.93 | ||
| 3 | 66.26 | 44.36 | 69.58 | 51.08 | 51.01 | 65.53 | ||
| 10 | 1 | 31.07 | 82.3 | 56.14 | 72.47 | 71.24 | 69.36 | |
| 2 | 77.89 | 35.33 | 58.09 | 68.06 | 82.01 | 80.64 | ||
| 3 | 27.6 | 43.93 | 54.12 | 56.5 | 67.63 | 78.4 | ||
| 11 | 1 | 62.79 | 65.17 | 84.9 | 71.75 | 78.18 | 77.53 | |
| 2 | 36.34 | 45.52 | 79.41 | 89.38 | 88.73 | 84.32 | ||
| 3 | 71.39 | 34.1 | 92.34 | 93.42 | 86.49 | 97.18 | ||
| 12 | 1 | 74.13 | 75.36 | 76.52 | 80.78 | 80.49 | 88.51 | |
| 2 | 35.26 | 45.59 | 51.3 | 59.68 | 65.61 | 71.39 | ||
| 3 | 44.94 | 76.81 | 77.38 | 91.76 | 92.41 | 89.6 | ||
| 13 | 1 | 64.02 | 82.08 | 36.27 | 79.91 | 90.9 | 81.94 | |
| 2 | 42.92 | 59.83 | 64.81 | 79.99 | 86.34 | 85.62 | ||
| 3 | 68.71 | 59.32 | 60.98 | 88.08 | 93.57 | 96.46 | ||
| 14 | 1 | 84.32 | 42.27 | 51.3 | 80.64 | 94.36 | 81.94 | |
| 2 | 51.66 | 56.36 | 59.03 | 65.53 | 50.29 | 90.03 | ||
| 3 | 42.41 | 48.19 | 74.49 | 65.46 | 59.75 | 64.31 | ||
| 15 | 1 | 55.56 | 32.3 | 59.47 | 71.53 | 90.61 | 83.74 | |
| 2 | 64.67 | 55.78 | 79.99 | 100 | 92.05 | 98.48 | ||
| 3 | 69.44 | 58.31 | 73.99 | 98.92 | 95.95 | 100 |
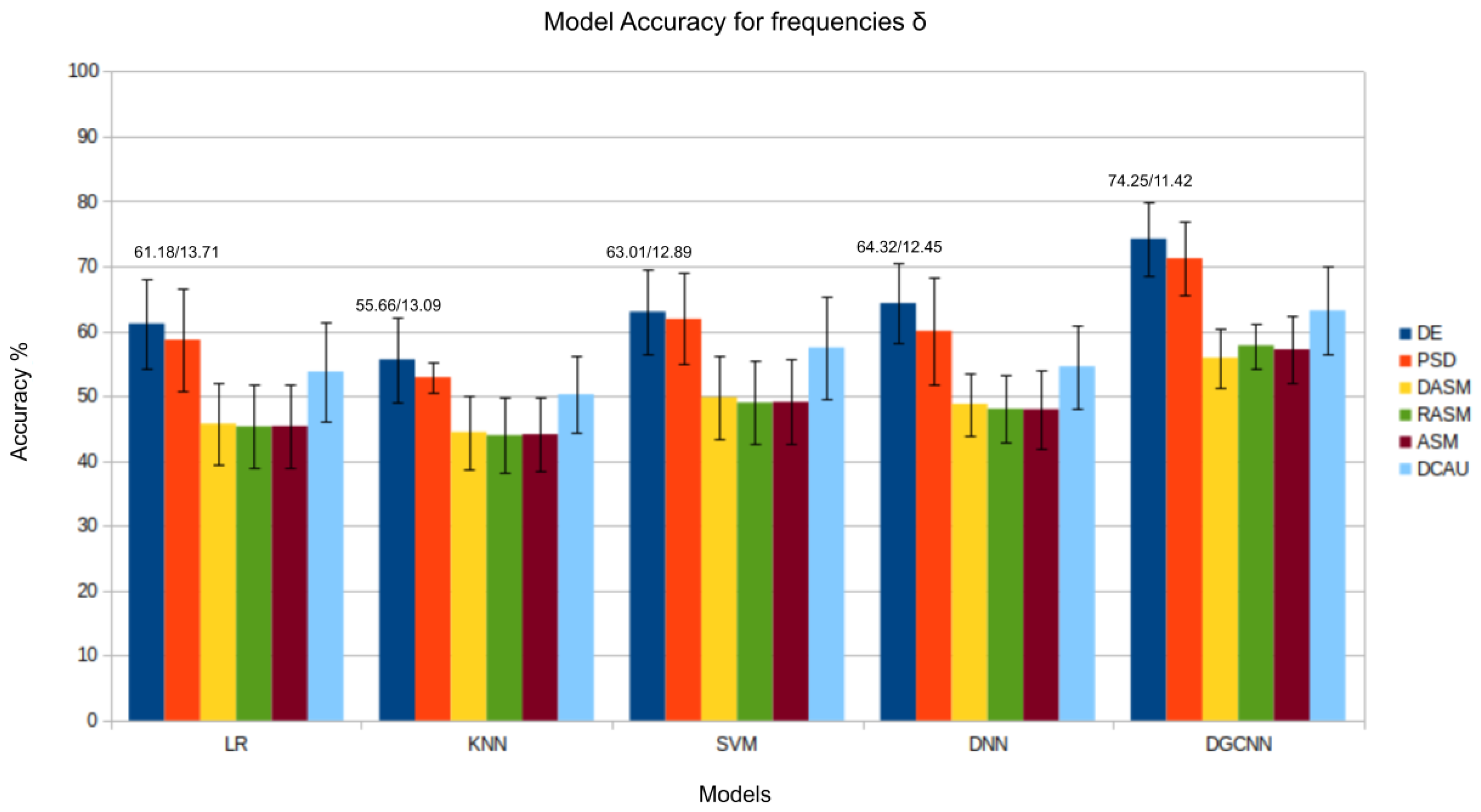
| Model | Feature | All Frequencies | |||||
|---|---|---|---|---|---|---|---|
| LR | DE | 61.18/13.71 | 65.32/14.16 | 67.58/13.74 | 76.71/14.33 | 74.34/15.73 | 82.46/11.11 |
| PSD | 58.66/15.66 | 66.02/12.92 | 62.81/17.23 | 67.87/18.09 | 67.09/15.92 | 75.44/15.02 | |
| DASM | 45.31/12.83 | 49.85/15.55 | 56.42/14.7 | 71.02/17.92 | 73.41/16.25 | 75.34/15.56 | |
| RASM | 45.72/12.8 | 49.93/15.67 | 56.57/14.54 | 71.12/17.71 | 73.39/16.1 | 75.41/15.53 | |
| ASM | 45.36/12.95 | 50.05/15.56 | 56.92/14.27 | 70.71/17.96 | 73.05/16.59 | 74.73/15.48 | |
| DCAU | 53.77/15.15 | 55.22/15.72 | 60.98/14.92 | 72.19/17.31 | 73.86/17.74 | 80.69/11.72 | |
| KNN | DE | 55.66/13.09 | 58.41/13.19 | 60.24/16.26 | 69.65/15.44 | 70.08/16.29 | 75.23/13.37 |
| PSD | 52.9/14.64 | 60.84/14.77 | 56.63/16.06 | 62.57/16.92 | 61.56/20.8 | 69.14/16.86 | |
| DASM | 43.95/11.7 | 43.09/13.37 | 49.42/15.38 | 64.03/17.66 | 67.11/16.14 | 66.24/17.53 | |
| RASM | 44.43/11.41 | 43.41/13.27 | 49.42/15.8 | 64.04/17.96 | 67.05/16.25 | 65.94/17.35 | |
| ASM | 44.09/11.43 | 43.1/12.99 | 49.26/15.33 | 63.84/18.09 | 67.1/15.57 | 66.16/17.09 | |
| DCAU | 50.27/11.77 | 49.5/10.38 | 53.6/11.18 | 67.8/15.09 | 68.88/16.44 | 72.84/12.75 | |
| SVM | DE | 63.01/12.89 | 69.61/13.78 | 71.53/14.22 | 79.82/13.71 | 78.25/15.01 | 84.44/11.58 |
| PSD | 61.91/14.1 | 69.43/12.41 | 66.33/16.42 | 71.04/17.01 | 71.96/15.02 | 77.99/13.91 | |
| DASM | 48.99/12.89 | 53.49/14.54 | 61.21/14.96 | 74.48/17.64 | 77.11/15.83 | 76.88/14.46 | |
| RASM | 49.83/12.83 | 53.53/14.9 | 61.95/15.02 | 73.87/17.67 | 77.22/15.9 | 76.8/14.44 | |
| ASM | 49.09/13.04 | 53.76/15.03 | 61.48/14.58 | 73.44/17.71 | 76.17/16.07 | 76.45/14.37 | |
| DCAU | 57.49/15.75 | 60.16/14.62 | 64.77/14.3 | 74.9/16.34 | 77.67/16.85 | 81.43/11.62 |
| Model | Feature | All Frequencies | |||||
|---|---|---|---|---|---|---|---|
| DNN | DE | 73.35/10.94 | 74.52/8.38 | 75.32/12.61 | 80.47/13.51 | 83.34/10.13 | 85.22/9.51 |
| PSD | 66.97/12.13 | 75.00/9.45 | 71.05/12.43 | 76.99/13.41 | 78.94/12.4 | 80.56/12.35 | |
| DASM | 57.01/7.23 | 61.91/10.56 | 67.82/11.67 | 79.01/13.57 | 81.85/12.17 | 79.59/12.15 | |
| RASM | 56.68/9.05 | 60.81/11.02 | 66.89/13.58 | 79.2/13.62 | 81.73/12.16 | 79.08/12.25 | |
| ASM | 55.33/8.25 | 59.97/10.46 | 66.96/11.82 | 78.35/13 | 80.14/12.38 | 81.19/11.5 | |
| DCAU | 70.07/6.89 | 68.39/11.6 | 72.42/9.54 | 80.94/11.63 | 83.06/10.74 | 84.71/9.82 | |
| DGCNN | DE | 74.81/8.41 | 69.81/11.6 | 71.62/13.71 | 83.83/9.4 | 83.62/10.76 | 89.97/5.57 |
| PSD | 70.25/13.41 | 69.63/8.63 | 65.26/10.99 | 71.67/15.5 | 75.87/18.33 | 79.39/15.08 | |
| DASM | 60.47/9.53 | 63.23/8.72 | 63.15/11.68 | 81.81/11.91 | 84.73/8.72 | 85.86/7.41 | |
| RASM | 60.14/10.36 | 62.17/10.09 | 63.01/11.11 | 81.92/10.26 | 84.53/9.29 | 84.82/7.79 | |
| ASM | 62.69/8.18 | 61.00/9.29 | 62.20/11.82 | 82.19/11.74 | 83.68/10.48 | 85.46/9.43 | |
| DCAU | 71.18/10.85 | 67.69/10.12 | 67.08/8.96 | 81.98/9.72 | 82.20/9.22 | 89.36/8.63 |
| Feature | Model | All Frequencies | |||||
|---|---|---|---|---|---|---|---|
| DE | LR | 61.18/13.71 | 65.32/14.16 | 67.58/13.74 | 76.71/14.33 | 74.34/15.73 | 82.46/11.11 |
| KNN | 55.66/13.09 | 58.41/13.19 | 60.24/16.26 | 69.65/15.44 | 70.08/16.29 | 75.23/13.37 | |
| SVM | 63.01/12.89 | 69.61/13.78 | 71.53/14.22 | 79.82/13.71 | 78.25/15.01 | 84.44/11.58 | |
| DNN | 73.35/10.94 | 74.52/8.38 | 75.32/12.61 | 80.47/13.51 | 83.34/10.13 | 85.22/9.51 | |
| DGCNN | 74.81/8.41 | 69.81/11.60 | 71.62/13.71 | 83.83/9.40 | 83.62/10.76 | 89.97/5.57 | |
| PSD | LR | 58.66/15.66 | 66.02/12.92 | 62.81/17.23 | 67.87/18.09 | 67.09/15.92 | 75.44/15.02 |
| KNN | 52.9/14.64 | 60.84/14.77 | 56.63/16.06 | 62.57/16.92 | 61.56/20.8 | 69.14/16.86 | |
| SVM | 61.91/14.1 | 69.43/12.41 | 66.33/16.42 | 71.04/17.01 | 71.96/15.02 | 77.99/13.91 | |
| DNN | 66.97/12.13 | 75.00/9.45 | 71.05/12.43 | 76.99/13.41 | 78.94/12.40 | 80.56/12.35 | |
| DGCNN | 70.25/13.41 | 69.63/8.63 | 65.26/10.99 | 71.67/15.50 | 75.87/18.33 | 79.39/15.08 | |
| DASM | LR | 45.72/12.8 | 49.93/15.67 | 56.57/14.54 | 71.12/17.71 | 73.39/16.1 | 75.41/15.53 |
| KNN | 44.43/11.41 | 43.41/13.27 | 49.42/15.8 | 64.04/17.96 | 67.05/16.25 | 65.94/17.35 | |
| SVM | 49.83/12.83 | 53.53/14.9 | 61.95/15.02 | 73.87/17.67 | 77.22/15.9 | 76.8/14.44 | |
| DNN | 57.01/7.23 | 61.91/10.56 | 67.82/11.67 | 79.01/13.57 | 81.85/12.17 | 79.59/12.15 | |
| DGCNN | 60.47/9.53 | 63.23/8.72 | 63.15/11.68 | 81.81/11.91 | 84.73/8.72 | 85.86/7.41 | |
| RASM | LR | 45.31/12.83 | 49.85/15.55 | 56.42/14.7 | 71.02/17.92 | 73.41/16.25 | 75.34/15.56 |
| KNN | 43.95/11.7 | 43.09/13.37 | 49.42/15.38 | 64.03/17.66 | 67.11/16.14 | 66.24/17.53 | |
| SVM | 48.99/12.89 | 53.49/14.54 | 61.21/14.96 | 74.48/17.64 | 77.11/15.83 | 76.88/14.46 | |
| DNN | 56.68/9.05 | 60.81/11.02 | 66.89/13.58 | 79.2/13.62 | 81.73/12.16 | 79.08/12.25 | |
| DGCNN | 60.14/10.36 | 62.17/10.09 | 63.01/11.11 | 81.92/10.26 | 84.53/9.29 | 84.82/7.79 | |
| ASM | LR | 45.36/12.95 | 50.05/15.56 | 56.92/14.27 | 70.71/17.96 | 73.05/16.59 | 74.73/15.48 |
| KNN | 44.09/11.43 | 43.1/12.99 | 49.26/15.33 | 63.84/18.09 | 67.1/15.57 | 66.16/17.09 | |
| SVM | 49.09/13.04 | 53.76/15.03 | 61.48/14.58 | 73.44/17.71 | 76.17/16.07 | 76.45/14.37 | |
| DNN | 55.33/8.25 | 59.97/10.46 | 66.96/11.82 | 78.35/13.00 | 80.14/12.38 | 81.19/11.50 | |
| DGCNN | 62.69/8.18 | 61.00/9.29 | 62.2/11.82 | 82.19/11.74 | 83.68/10.48 | 85.46/9.43 | |
| DCAU | LR | 53.77/15.15 | 55.22/15.72 | 60.98/14.92 | 72.19/17.31 | 73.86/17.74 | 80.69/11.72 |
| KNN | 50.27/11.77 | 49.5/10.38 | 53.6/11.18 | 67.8/15.09 | 68.88/16.44 | 72.84/12.75 | |
| SVM | 57.49/15.75 | 60.16/14.62 | 64.77/14.3 | 74.9/16.34 | 77.67/16.85 | 81.43/11.62 | |
| DNN | 70.07/6.89 | 68.39/11.60 | 72.42/9.54 | 80.94/11.63 | 83.06/10.74 | 84.71/9.82 | |
| DGCNN | 71.18/10.85 | 67.69/10.12 | 67.08/8.96 | 81.98/9.72 | 82.2/9.22 | 89.36/8.63 |
| Feature | Model | All Frequencies | |||||
|---|---|---|---|---|---|---|---|
| DE | LR | 61.18/13.71 | 65.32/14.16 | 67.58/13.74 | 76.71/14.33 | 74.34/15.73 | 82.46/11.11 |
| KNN | 55.66/13.09 | 58.41/13.19 | 60.24/16.26 | 69.65/15.44 | 70.08/16.29 | 75.23/13.37 | |
| SVM | 63.01/12.89 | 69.61/13.78 | 71.53/14.22 | 79.82/13.71 | 78.25/15.01 | 84.44/11.58 | |
| SVM [12] | 60.50/14.14 | 60.95/10.20 | 66.64/14.41 | 80.76/11.56 | 79.56/11.38 | 83.99/9.72 | |
| DNN | 73.35/10.94 | 74.52/8.38 | 75.32/12.61 | 80.47/13.51 | 83.34/10.13 | 85.22/9.51 | |
| DBN [12] | 64.32/12.45 | 60.77/10.42 | 64.01/15.97 | 78.92/12.48 | 79.19/14.58 | 86.08/8.34 | |
| DGCNN | 74.81/8.41 | 69.81/11.60 | 71.62/13.71 | 83.83/9.40 | 83.62/10.76 | 89.97/5.57 | |
| DGCNN [38] | 74.25/11.42 | 71.52/5.99 | 74.43/12.16 | 83.65/10.17 | 85.73/10.64 | 90.40/8.49 | |
| PSD | LR | 58.66/15.66 | 66.02/12.92 | 62.81/17.23 | 67.87/18.09 | 67.09/15.92 | 75.44/15.02 |
| KNN | 52.9/14.64 | 60.84/14.77 | 56.63/16.06 | 62.57/16.92 | 61.56/20.8 | 69.14/16.86 | |
| SVM | 61.91/14.1 | 69.43/12.41 | 66.33/16.42 | 71.04/17.01 | 71.96/15.02 | 77.99/13.91 | |
| SVM [12] | 58.03/15.39 | 57.26/15.09 | 59.04/15.75 | 73.34/15.20 | 71.24/16.38 | 59.60/15.93 | |
| DNN | 66.97/12.13 | 75.00/9.45 | 71.05/12.43 | 76.99/13.41 | 78.94/12.40 | 80.56/12.35 | |
| DBN [12] | 60.05/16.66 | 55.03/13.88 | 52.79/15.38 | 60.68/21.31 | 63.42/19.66 | 61.90/16.65 | |
| DGCNN | 70.25/13.41 | 69.63/8.63 | 65.26/10.99 | 71.67/15.50 | 75.87/18.33 | 79.39/15.08 | |
| DGCNN [38] | 71.23/11.42 | 71.20/8.99 | 73.45/12.25 | 77.45/10.81 | 76.60/11.83 | 81.73/9.94 | |
| DASM | LR | 45.72/12.8 | 49.93/15.67 | 56.57/14.54 | 71.12/17.71 | 73.39/16.1 | 75.41/15.53 |
| KNN | 44.43/11.41 | 43.41/13.27 | 49.42/15.8 | 64.04/17.96 | 67.05/16.25 | 65.94/17.35 | |
| SVM | 49.83/12.83 | 53.53/14.9 | 61.95/15.02 | 73.87/17.67 | 77.22/15.9 | 76.8/14.44 | |
| SVM [12] | 48.87/10.49 | 53.02/12.76 | 59.81/14.67 | 75.03/15.72 | 73.59/16.57 | 72.81/16.57 | |
| DNN | 57.01/7.23 | 61.91/10.56 | 67.82/11.67 | 79.01/13.57 | 81.85/12.17 | 79.59/12.15 | |
| DBN [12] | 48.79/9.62 | 51.59/13.98 | 54.03/17.05 | 69.51/15.22 | 70.06/18.14 | 72.73/15.93 | |
| DGCNN | 60.47/9.53 | 63.23/8.72 | 63.15/11.68 | 81.81/11.91 | 84.73/8.72 | 85.86/7.41 | |
| DGCNN [38] | 55.93/9.14 | 56.12/7.86 | 64.27/12.72 | 73.61/14.35 | 73.50/16.6 | 78.45/11.84 | |
| RASM | LR | 45.31/12.83 | 49.85/15.55 | 56.42/14.7 | 71.02/17.92 | 73.41/16.25 | 75.34/15.56 |
| KNN | 43.95/11.7 | 43.09/13.37 | 49.42/15.38 | 64.03/17.66 | 67.11/16.14 | 66.24/17.53 | |
| SVM | 48.99/12.89 | 53.49/14.54 | 61.21/14.96 | 74.48/17.64 | 77.11/15.83 | 76.88/14.46 | |
| SVM [12] | 47.75/10.59 | 51.40/12.53 | 60.71/14.57 | 74.59/16.18 | 74.61/15.57 | 74.74/14.79 | |
| DNN | 56.68/9.05 | 60.81/11.02 | 66.89/13.58 | 79.2/13.62 | 81.73/12.16 | 79.08/12.25 | |
| DBN [12] | 48.05/10.37 | 50.62/14.02 | 56.15/15.28 | 70.31/15.62 | 68.22/18.09 | 71.30/16.16 | |
| DGCNN | 60.14/10.36 | 62.17/10.09 | 63.01/11.11 | 81.92/10.26 | 84.53/9.29 | 84.82/7.79 | |
| DGCNN [38] | 57.79/6.90 | 55.79/8.10 | 61.58/12.63 | 75.79/13.07 | 82.32/11.54 | 85.00/12.47 | |
| ASM | LR | 45.36/12.95 | 50.05/15.56 | 56.92/14.27 | 70.71/17.96 | 73.05/16.59 | 74.73/15.48 |
| KNN | 44.09/11.43 | 43.1/12.99 | 49.26/15.33 | 63.84/18.09 | 67.1/15.57 | 66.16/17.09 | |
| SVM | 49.09/13.04 | 53.76/15.03 | 61.48/14.58 | 73.44/17.71 | 76.17/16.07 | 76.45/14.37 | |
| SVM [12] | - | - | - | - | - | - | |
| DNN | 55.33/8.25 | 59.97/10.46 | 66.96/11.82 | 78.35/13.00 | 80.14/12.38 | 81.19/11.50 | |
| DBN [12] | - | - | - | - | - | - | |
| DGCNN | 62.69/8.18 | 61.00/9.29 | 62.2/11.82 | 82.19/11.74 | 83.68/10.48 | 85.46/9.43 | |
| DGCNN [38] | - | - | - | - | - | - | |
| DCAU | LR | 53.77/15.15 | 55.22/15.72 | 60.98/14.92 | 72.19/17.31 | 73.86/17.74 | 80.69/11.72 |
| KNN | 50.27/11.77 | 49.5/10.38 | 53.6/11.18 | 67.8/15.09 | 68.88/16.44 | 72.84/12.75 | |
| SVM | 57.49/15.75 | 60.16/14.62 | 64.77/14.3 | 74.9/16.34 | 77.67/16.85 | 81.43/11.62 | |
| SVM [12] | 55.92/14.62 | 57.16/10.77 | 61.37/15.97 | 75.17/15.58 | 76.44/15.41 | 77.38/11.98 | |
| DNN | 70.07/6.89 | 68.39/11.60 | 72.42/9.54 | 80.94/11.63 | 83.06/10.74 | 84.71/9.82 | |
| DBN [12] | 54.58/12.81 | 56.94/12.54 | 57.62/13.58 | 70.70/16.33 | 72.27/16.12 | 77.20/14.24 | |
| DGCNN | 71.18/10.85 | 67.69/10.12 | 67.08/8.96 | 81.98/9.72 | 82.2/9.22 | 89.36/8.63 | |
| DGCNN [38] | 63.18/13.48 | 62.55/7.96 | 67.71/10.74 | 78.68/10.81 | 80.05/13.03 | 81.91/10.06 |
| Feature | Model | All Frequencies | |||||
|---|---|---|---|---|---|---|---|
| DE | DNN | 73.35/10.94 | 74.52/8.38 | 75.32/12.61 | 80.47/13.51 | 83.34/10.13 | 85.22/9.51 |
| DBN [12] | 64.32/12.45 | 60.77/10.42 | 64.01/15.97 | 78.92/12.48 | 79.19/14.58 | 86.08/8.34 | |
| PSD | DNN | 66.97/12.13 | 75.00/9.45 | 71.05/12.43 | 76.99/13.41 | 78.94/12.40 | 80.56/12.35 |
| DBN [12] | 60.05/16.66 | 55.03/13.88 | 52.79/15.38 | 60.68/21.31 | 63.42/19.66 | 61.90/16.65 | |
| DASM | DNN | 57.01/7.23 | 61.91/10.56 | 67.82/11.67 | 79.01/13.57 | 81.85/12.17 | 79.59/12.15 |
| DBN [12] | 48.79/9.62 | 51.59/13.98 | 54.03/17.05 | 69.51/15.22 | 70.06/18.14 | 72.73/15.93 | |
| RASM | DNN | 56.68/9.05 | 60.81/11.02 | 66.89/13.58 | 79.2/13.62 | 81.73/12.16 | 79.08/12.25 |
| DBN [12] | 48.05/10.37 | 50.62/14.02 | 56.15/15.28 | 70.31/15.62 | 68.22/18.09 | 71.30/16.16 | |
| ASM | DNN | 55.33/8.25 | 59.97/10.46 | 66.96/11.82 | 78.35/13.00 | 80.14/12.38 | 81.19/11.50 |
| DBN [12] | - | - | - | - | - | - | |
| DCAU | DNN | 70.07/6.89 | 68.39/11.60 | 72.42/9.54 | 80.94/11.63 | 83.06/10.74 | 84.71/9.82 |
| DBN [12] | 54.58/12.81 | 56.94/12.54 | 57.62/13.58 | 70.70/16.33 | 72.27/16.12 | 77.20/14.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes, J.V.M.R.; Alexandria, A.R.d.; Marques, J.A.L.; Assis, D.F.d.; Motta, P.C.; Silva, B.R.d.S. Emotion Detection from EEG Signals Using Machine Deep Learning Models. Bioengineering 2024, 11, 782. https://doi.org/10.3390/bioengineering11080782
Fernandes JVMR, Alexandria ARd, Marques JAL, Assis DFd, Motta PC, Silva BRdS. Emotion Detection from EEG Signals Using Machine Deep Learning Models. Bioengineering. 2024; 11(8):782. https://doi.org/10.3390/bioengineering11080782
Chicago/Turabian StyleFernandes, João Vitor Marques Rabelo, Auzuir Ripardo de Alexandria, João Alexandre Lobo Marques, Débora Ferreira de Assis, Pedro Crosara Motta, and Bruno Riccelli dos Santos Silva. 2024. "Emotion Detection from EEG Signals Using Machine Deep Learning Models" Bioengineering 11, no. 8: 782. https://doi.org/10.3390/bioengineering11080782
APA StyleFernandes, J. V. M. R., Alexandria, A. R. d., Marques, J. A. L., Assis, D. F. d., Motta, P. C., & Silva, B. R. d. S. (2024). Emotion Detection from EEG Signals Using Machine Deep Learning Models. Bioengineering, 11(8), 782. https://doi.org/10.3390/bioengineering11080782







