Automatic Transcranial Sonography-Based Classification of Parkinson’s Disease Using a Novel Dual-Channel CNXV2-DANet
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data
2.1.1. Data Acquisition
2.1.2. Image Pre-Processing
2.2. Deep Learning Neural Networks
2.2.1. ConvNext v2
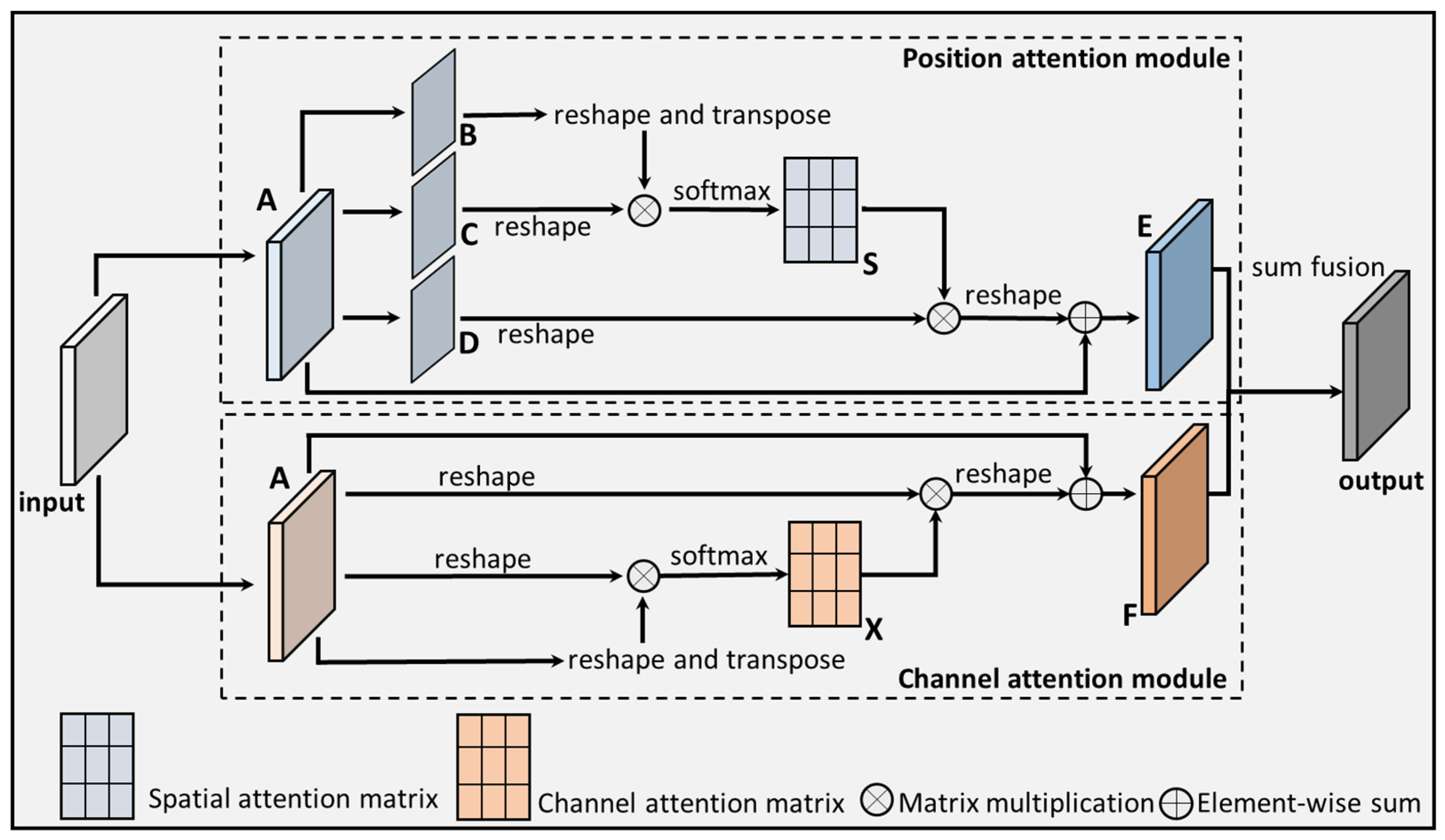
2.2.2. DANet
2.2.3. The Proposed CNXV2-DANet
2.3. Model Development and Comparison
2.3.1. Model Development
2.3.2. Model Comparison
2.4. Evaluation Metrics
3. Results
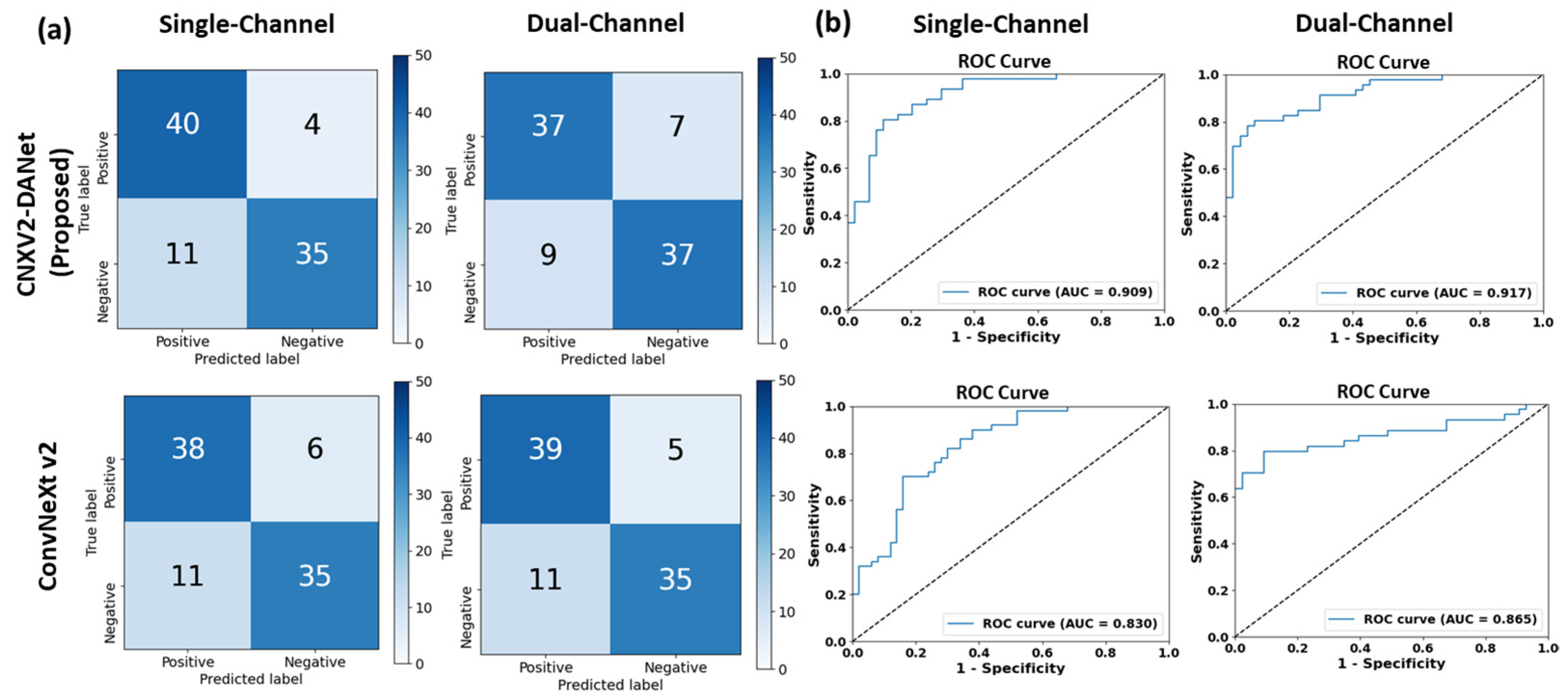
3.1. Single-Channel vs. Dual-Channel Methods
3.1.1. CNXV2-DANet
3.1.2. ConvNeXt V2
3.2. CNXV2-DANet vs. State-of-the-Art Networks on the Dual-Channel Setting
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maserejian, N.; Vinikoor-Imler, L.; Dilley, A. Estimation of the 2020 Global Population of Parkinson’s Disease (PD). Mov. Disord. 2020, 35, S79–S80. [Google Scholar]
- Feigin, V.L.; Nichols, E.; Alam, T.; Bannick, M.S.; Beghi, E.; Blake, N.; Culpepper, W.J.; Dorsey, E.R.; Elbaz, A.; Ellenbogen, R.G.; et al. Global, regional, and national burden of neurological disorders, 1990-2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 459–480. [Google Scholar] [CrossRef] [PubMed]
- Bloem, B.R.; Okun, M.S.; Klein, C. Parkinson’s disease. Lancet 2021, 397, 2284–2303. [Google Scholar] [CrossRef] [PubMed]
- Tolosa, E.; Garrido, A.; Scholz, S.W.; Poewe, W. Challenges in the diagnosis of Parkinson’s disease. Lancet Neurol. 2021, 20, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Poewe, W.; Seppi, K.; Tanner, C.M.; Halliday, G.M.; Brundin, P.; Volkmann, J.; Schrag, A.E.; Lang, A.E. Parkinson disease. Nat. Rev. Dis. Primers 2017, 3, 17013. [Google Scholar] [CrossRef]
- Kalia, L.V.; Lang, A.E. Parkinson’s disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef]
- Jankovic, J. Parkinson’s disease: Clinical features and diagnosis. J. Neurol. Neurosurg. Psychiatry 2008, 79, 368–376. [Google Scholar] [CrossRef]
- DeMaagd, G.; Philip, A. Parkinson’s Disease and Its Management: Part 1: Disease Entity, Risk Factors, Pathophysiology, Clinical Presentation, and Diagnosis. Pharm. Ther. 2015, 40, 504–532. [Google Scholar]
- Safi, K.; Aly, W.H.F.; AlAkkoumi, M.; Kanj, H.; Ghedira, M.; Hutin, E. EMD-Based Method for Supervised Classification of Parkinson’s Disease Patients Using Balance Control Data. Bioengineering 2022, 9, 283. [Google Scholar] [CrossRef]
- Mondol, S.I.M.M.R.; Kim, R.; Lee, S.M. Hybrid Machine Learning Framework for Multistage Parkinson’s Disease Classification Using Acoustic Features of Sustained Korean Vowels. Bioengineering 2023, 10, 984. [Google Scholar] [CrossRef]
- Ibarra, E.J.; Arias-Londoño, J.D.; Zañartu, M.; Godino-Llorente, J.I. Towards a Corpus (and Language)-Independent Screening of Parkinson’s Disease from Voice and Speech through Domain Adaptation. Bioengineering 2023, 10, 1316. [Google Scholar] [CrossRef] [PubMed]
- Connolly, B.S.; Lang, A.E. Pharmacological Treatment of Parkinson Disease A Review. JAMA J. Am. Med. Assoc. 2014, 311, 1670–1683. [Google Scholar] [CrossRef]
- Armstrong, M.J.; Okun, M.S. Diagnosis and Treatment of Parkinson Disease A Review. JAMA J. Am. Med. Assoc. 2020, 323, 548–560. [Google Scholar] [CrossRef]
- Hall, M.F.E.; Church, F.C. Integrative Medicine and Health Therapy for Parkinson Disease. Top. Geriatr. Rehabil. 2020, 36, 176–186. [Google Scholar] [CrossRef]
- Kim, S.N.; Wang, X.; Park, H.J. Editorial: Integrative Approach to Parkinson’s Disease. Front. Aging Neurosci. 2019, 11, 339. [Google Scholar] [CrossRef] [PubMed]
- Church, F.C. Treatment Options for Motor and Non-Motor Symptoms of Parkinson’s Disease. Biomolecules 2021, 11, 612. [Google Scholar] [CrossRef]
- Petzinger, G.M.; Fisher, B.E.; McEwen, S.; Beeler, J.A.; Walsh, J.P.; Jakowec, M.W. Exercise-enhanced neuroplasticity targeting motor and cognitive circuitry in Parkinson’s disease. Lancet Neurol. 2013, 12, 716–726. [Google Scholar] [CrossRef] [PubMed]
- Palasz, E.; Niewiadomski, W.; Gasiorowska, A.; Wysocka, A.; Stepniewska, A.; Niewiadomska, G. Exercise-Induced Neuroprotection and Recovery of Motor Function in Animal Models of Parkinson’s Disease. Front. Neurol. 2019, 10, 1143. [Google Scholar] [CrossRef] [PubMed]
- Crocker, A.D. The regulation of motor control: An evaluation of the role of dopamine receptors in the substantia nigra. Rev. Neurosci. 1997, 8, 55–76. [Google Scholar] [CrossRef]
- Brar, S.; Henderson, D.; Schenck, J.; Zimmerman, E.A. Iron accumulation in the substantia nigra of patients with Alzheimer disease and parkinsonism. Arch. Neurol. 2009, 66, 371–374. [Google Scholar] [CrossRef]
- de Bie, R.M.A.; Clarke, C.E.; Espay, A.J.; Fox, S.H.; Lang, A.E. Initiation of pharmacological therapy in Parkinson’s disease: When, why, and how. Lancet Neurol. 2020, 19, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Grosset, D.G.; Macphee, G.J.A.; Nairn, M.; Grp, G.D. Guidelines Diagnosis and pharmacological management of Parkinson’s disease: Summary of SIGN guidelines. BMJ Brit Med. J. 2010, 340, b5614. [Google Scholar] [CrossRef] [PubMed]
- Heim, B.; Krismer, F.; De Marzi, R.; Seppi, K. Magnetic resonance imaging for the diagnosis of Parkinson’s disease. J. Neural Transm. 2017, 124, 915–964. [Google Scholar] [CrossRef]
- Brooks, D.J. Imaging approaches to Parkinson disease. J. Nucl. Med. 2010, 51, 596–609. [Google Scholar] [CrossRef]
- Bae, Y.J.; Kim, J.M.; Sohn, C.H.; Choi, J.H.; Choi, B.S.; Song, Y.S.; Nam, Y.; Cho, S.J.; Jeon, B.; Kim, J.H. Imaging the Substantia Nigra in Parkinson Disease and Other Parkinsonian Syndromes. Radiology 2021, 300, 260–278. [Google Scholar] [CrossRef]
- Behnke, S.; Berg, D.; Naumann, M.; Becker, G. Differentiation of Parkinson’s disease and atypical parkinsonian syndromes by transcranial ultrasound. J. Neurol. Neurosur Ps. 2005, 76, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Tao, A.Y.; Chen, G.Z.; Deng, Y.B.; Xu, R.F. Accuracy of Transcranial Sonography of the Substantia Nigra for Detection of Parkinson’s Disease: A Systematic Review and Meta-Analysis. Ultrasound Med. Biol. 2019, 45, 628–641. [Google Scholar] [CrossRef]
- Mei, Y.L.; Yang, J.; Wu, Z.R.; Yang, Y.; Xu, Y.M. Transcranial Sonography of the Substantia Nigra for the Differential Diagnosis of Parkinson’s Disease and Other Movement Disorders: A Meta-Analysis. Park. Dis. 2021, 2021, 8891874. [Google Scholar] [CrossRef]
- Berg, D.; Seppi, K.; Behnke, S.; Liepelt, I.; Schweitzer, K.; Stockner, H.; Wollenweber, F.; Gaenslen, A.; Mahlknecht, P.; Spiegel, J.; et al. Enlarged substantia nigra hyperechogenicity and risk for Parkinson disease: A 37-month 3-center study of 1847 older persons. Arch. Neurol. 2011, 68, 932–937. [Google Scholar] [CrossRef]
- Skoloudík, D.; Fadrná, T.; Bártova, P.; Langová, K.; Ressner, P.; Zapletalová, O.; Hlustík, P.; Herzig, R.; Kannovsky, P. Reproducibility of sonographic measurement of the substantia nigra. Ultrasound Med. Biol. 2007, 33, 1347–1352. [Google Scholar] [CrossRef]
- Berg, D.; Godau, J.; Walter, U. Transcranial sonography in movement disorders. Lancet Neurol. 2008, 7, 1044–1055. [Google Scholar] [CrossRef]
- Berardelli, A.; Wenning, G.K.; Antonini, A.; Berg, D.; Bloem, B.R.; Bonifati, V.; Brooks, D.; Burn, D.J.; Colosimo, C.; Fanciulli, A.; et al. EFNS/MDS-ES recommendations for the diagnosis of Parkinson’s disease. Eur. J. Neurol. 2013, 20, 16–34. [Google Scholar] [CrossRef]
- Basukala, D.; Mukundan, R.; Lim, A.; Hurrell, M.A.; Keenan, R.J.; Dalrymple-Alford, J.C.; Anderson, T.J.; Myall, D.J.; Melzer, T.R. Automated segmentation of substantia nigra and red nucleus using quantitative susceptibility mapping images: Application to Parkinson’s disease. Comput. Electr. Eng. 2021, 91, 107091. [Google Scholar] [CrossRef]
- Choi, H.; Ha, S.; Im, H.J.; Paek, S.H.; Lee, D.S. Refining diagnosis of Parkinson’s disease with deep learning-based interpretation of dopamine transporter imaging. Neuroimage Clin. 2017, 16, 586–594. [Google Scholar] [CrossRef]
- Vásquez-Correa, J.C.; Arias-Vergara, T.; Orozco-Arroyave, J.R.; Eskofier, B.; Klucken, J.; Nöth, E. Multimodal Assessment of Parkinson’s Disease: A Deep Learning Approach. IEEE J. Biomed. Health 2019, 23, 1618–1630. [Google Scholar] [CrossRef]
- Piccardo, A.; Cappuccio, R.; Bottoni, G.; Cecchin, D.; Mazzella, L.; Cirone, A.; Righi, S.; Ugolini, M.; Bianchi, P.; Bertolaccini, P.; et al. The role of the deep convolutional neural network as an aid to interpreting brain [F]DOPA PET/CT in the diagnosis of Parkinson’s disease. Eur. Radiol. 2021, 31, 7003–7011. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Aich, S.; Kim, H.C. Detection of Parkinson’s Disease from 3T T1 Weighted MRI Scans Using 3D Convolutional Neural Network. Diagnostics 2020, 10, 402. [Google Scholar] [CrossRef] [PubMed]
- Sivaranjini, S.; Sujatha, C.M. Deep learning based diagnosis of Parkinson’s disease using convolutional neural network. Multimed. Tools Appl. 2020, 79, 15467–15479. [Google Scholar] [CrossRef]
- Manzanera, O.M.; Meles, S.K.; Leenders, K.L.; Renken, R.J.; Pagani, M.; Arnaldi, D.; Nobili, F.; Obeso, J.; Oroz, M.R.; Morbelli, S.; et al. Scaled Subprofile Modeling and Convolutional Neural Networks for the Identification of Parkinson’s Disease in 3D Nuclear Imaging Data. Int. J. Neural Syst. 2019, 29, 1950010. [Google Scholar] [CrossRef]
- Zhao, H.L.; Tsai, C.C.; Zhou, M.Y.; Liu, Y.P.; Chen, Y.L.; Huang, F.; Lin, Y.C.; Wang, J.J. Deep learning based diagnosis of Parkinson’s Disease using diffusion magnetic resonance imaging. Brain Imaging Behav. 2022, 16, 1749–1760. [Google Scholar] [CrossRef]
- Vyas, T.; Yadav, R.; Solanki, C.; Darji, R.; Desai, S.; Tanwar, S. Deep learning-based scheme to diagnose Parkinson’s disease. Expert. Syst. 2022, 39, e12739. [Google Scholar] [CrossRef]
- Shen, L.; Shi, J.; Dong, Y.; Ying, S.H.; Peng, Y.X.; Chen, L.; Zhang, Q.; An, H.D.; Zhang, Y.C. An Improved Deep Polynomial Network Algorithm for Transcranial Sonography-Based Diagnosis of Parkinson’s Disease. Cogn. Comput. 2020, 12, 553–562. [Google Scholar] [CrossRef]
- Woo, S.; Debnath, S.; Hu, R.; Chen, X.; Liu, Z.; Kweon, I.S.; Xie, S. Convnext v2: Co-designing and scaling convnets with masked autoencoders. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 16133–16142. [Google Scholar]
- Fu, J.; Liu, J.; Tian, H.; Li, Y.; Bao, Y.; Fang, Z.; Lu, H. Dual attention network for scene segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 3146–3154. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Zhang, J.; Lam, S.K.; Teng, X.Z.; Ma, Z.R.; Han, X.Y.; Zhang, Y.P.; Cheung, A.L.Y.; Chau, T.C.; Ng, S.C.Y.; Lee, F.K.H.; et al. Radiomic feature repeatability and its impact on prognostic model generalizability: A multi-institutional study on nasopharyngeal carcinoma patients. Radiother. Oncol. 2023, 183, 109578. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.K.; Zhang, Y.P.; Zhang, J.; Li, B.; Sun, J.C.; Liu, C.Y.T.; Chou, P.H.; Teng, X.Z.; Ma, Z.R.; Ni, R.Y.; et al. Multi-Organ Omics-Based Prediction for Adaptive Radiation Therapy Eligibility in Nasopharyngeal Carcinoma Patients Undergoing Concurrent Chemoradiotherapy. Front. Oncol. 2022, 11, 792024. [Google Scholar] [CrossRef]
- Liu, Z.; Mao, H.; Wu, C.-Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A convnet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 21–24 June 2022; pp. 11976–11986. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Vujovic, Z.D. Classification Model Evaluation Metrics. Int. J. Adv. Comput. Sc. 2021, 12, 599–606. [Google Scholar] [CrossRef]
- Delong, E.R.; Delong, D.M.; Clarkepearson, D.I. Comparing the Areas under 2 or More Correlated Receiver Operating Characteristic Curves—A Nonparametric Approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Baranauskas, M.; Jurkonis, R.; Lukosevicius, A.; Matijosaitis, V.; Gleizniene, R.; Rastenyte, D. Diagnostic Ability of Radiofrequency Ultrasound in Parkinson’s Disease Compared to Conventional Transcranial Sonography and Magnetic Resonance Imaging. Diagnostics 2020, 10, 778. [Google Scholar] [CrossRef]



| Method | Accuracy | Precision | Recall | F1-Score | AUC | |
|---|---|---|---|---|---|---|
| Single-channel | Seed1 | 0.767 | 0.871 | 0.614 | 0.720 | 0.884 |
| Seed2 | 0.789 | 0.772 | 0.791 | 0.782 | 0.868 | |
| Seed3 | 0.733 | 0.679 | 0.864 | 0.760 | 0.784 | |
| Seed4 | 0.800 | 0.865 | 0.711 | 0.780 | 0.861 | |
| Seed5 | 0.833 | 0.897 | 0.760 | 0.823 | 0.909 | |
| Average | 0.784 | 0.817 | 0.748 | 0.773 | 0.861 | |
| SD | 0.037 | 0.090 | 0.093 | 0.037 | 0.047 | |
| Dual-channel | Seed1 | 0.867 | 0.833 | 0.909 | 0.870 | 0.924 |
| Seed2 | 0.861 | 0.863 | 0.862 | 0.785 | 0.898 | |
| Seed3 | 0.844 | 0.841 | 0.841 | 0.841 | 0.898 | |
| Seed4 | 0.800 | 0.865 | 0.811 | 0.780 | 0.894 | |
| Seed5 | 0.823 | 0.841 | 0.804 | 0.822 | 0.917 | |
| Average | 0.839 | 0.849 | 0.845 | 0.820 | 0.906 | |
| SD | 0.028 | 0.014 | 0.043 | 0.038 | 0.013 |
| Method | Accuracy | Precision | Recall | F1-Score | AUC | |
|---|---|---|---|---|---|---|
| Single-channel | Seed1 | 0.744 | 0.763 | 0.674 | 0.716 | 0.852 |
| Seed2 | 0.844 | 0.787 | 0.902 | 0.841 | 0.884 | |
| Seed3 | 0.756 | 0.690 | 0.909 | 0.784 | 0.838 | |
| Seed4 | 0.800 | 0.783 | 0.818 | 0.800 | 0.876 | |
| Seed5 | 0.811 | 0.854 | 0.761 | 0.805 | 0.830 | |
| Average | 0.791 | 0.775 | 0.813 | 0.789 | 0.856 | |
| SD | 0.041 | 0.059 | 0.099 | 0.046 | 0.023 | |
| Dual-channel | Seed1 | 0.911 | 0.972 | 0.837 | 0.900 | 0.946 |
| Seed2 | 0.811 | 0.875 | 0.683 | 0.767 | 0.919 | |
| Seed3 | 0.856 | 0.878 | 0.818 | 0.847 | 0.903 | |
| Seed4 | 0.711 | 0.688 | 0. 750 | 0.717 | 0.847 | |
| Seed5 | 0.822 | 0.875 | 0.761 | 0.761 | 0.865 | |
| Average | 0.822 | 0.858 | 0.775 | 0.798 | 0.896 | |
| SD | 0.073 | 0.104 | 0.069 | 0.074 | 0.040 |
| Model | Accuracy (SD) | Precision (SD) | Recall (SD) | F1-Score (SD) | AUC (SD) |
|---|---|---|---|---|---|
| CNXV2-DANet (proposed) | 0.839 (0.028) | 0.849 (0.014) | 0.845 (0.043) | 0.820 (0.038) | 0.906 (0.013) |
| ConvNeXt V2 | 0.822 (0.073) | 0.858 (0.104) | 0.775(0.069) | 0.798 (0.074) | 0.896 (0.040) |
| ConvNeXt | 0.733 (0.045) | 0.710 (0.060) | 0.794 (0.117) | 0.744 (0.051) | 0.842 (0.016) |
| Swin Transformer | 0.753 (0.022) | 0.734 (0.057) | 0.799 (0.075) | 0.760 (0.028) | 0.834 (0.030) |
| Researcher | Year | Dataset | Modality | Method | Accuracy (%) | With an Independent Test Set |
|---|---|---|---|---|---|---|
| Sivaranjini et al. [38] | 2019 | 182 | MRI | AlexNet + transfer learning | 88.9 | No |
| Manzanera et al. [39] | 2019 | 310 | PET | CNN | 86.0 | Yes |
| Chakraborty et al. [37] | 2020 | 406 | MRI | CNN | 95.3 | No |
| Shen et al. [42] | 2020 | 153 | TCS | Deep polynomial network | 86.9 | No |
| Zhao et al. [40] | 2022 | 432 | MRI | 3D CNN | 80.7 | Yes |
| Our study | 2024 | 588 | TCS | CNXV2-DANet | 83.9 | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, H.; Wang, X.; Sun, Y.; Li, S.; Sun, X.; Li, F.; Hou, C.; Lam, S.-k.; Zhang, W.; Zheng, Y.-p. Automatic Transcranial Sonography-Based Classification of Parkinson’s Disease Using a Novel Dual-Channel CNXV2-DANet. Bioengineering 2024, 11, 889. https://doi.org/10.3390/bioengineering11090889
Kang H, Wang X, Sun Y, Li S, Sun X, Li F, Hou C, Lam S-k, Zhang W, Zheng Y-p. Automatic Transcranial Sonography-Based Classification of Parkinson’s Disease Using a Novel Dual-Channel CNXV2-DANet. Bioengineering. 2024; 11(9):889. https://doi.org/10.3390/bioengineering11090889
Chicago/Turabian StyleKang, Hongyu, Xinyi Wang, Yu Sun, Shuai Li, Xin Sun, Fangxian Li, Chao Hou, Sai-kit Lam, Wei Zhang, and Yong-ping Zheng. 2024. "Automatic Transcranial Sonography-Based Classification of Parkinson’s Disease Using a Novel Dual-Channel CNXV2-DANet" Bioengineering 11, no. 9: 889. https://doi.org/10.3390/bioengineering11090889
APA StyleKang, H., Wang, X., Sun, Y., Li, S., Sun, X., Li, F., Hou, C., Lam, S.-k., Zhang, W., & Zheng, Y.-p. (2024). Automatic Transcranial Sonography-Based Classification of Parkinson’s Disease Using a Novel Dual-Channel CNXV2-DANet. Bioengineering, 11(9), 889. https://doi.org/10.3390/bioengineering11090889








