Addition of ROCK Inhibitors Alleviates Prostaglandin-Induced Inhibition of Adipogenesis in 3T3L-1 Spheroids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Adipocyte Culture and Adipogenic Differentiation of 3T3-L1 Cells with or without PG and ROCK Inhibitors (ROCK-i)
2.2. Lipid Staining by Oil Red O (2D) or BODIPY (3D)
2.3. Measurement of Real-Time Cellular Metabolic Functions
2.4. Physical Properties, Size and Stiffness Analyses of the 3D 3T3-L1 Spheroids
2.5. Immunostaining of 3D 3T3-L1 Spheroids
2.6. Other Analytical Methods
3. Results
3.1. Additive Effects of Rock-Is on PGs on 2D Cultured 3T3-L1 Cells
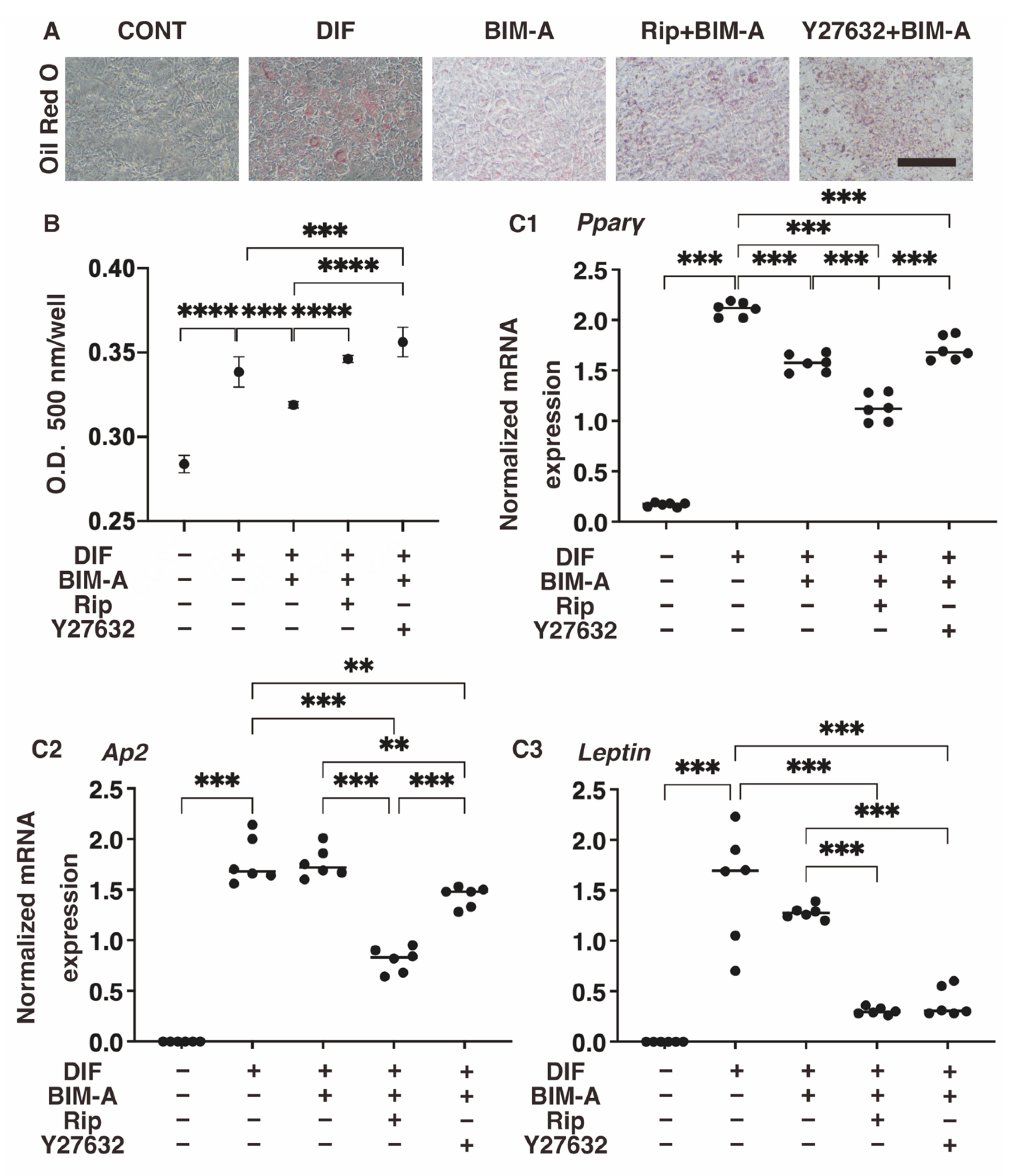
3.1.1. Lipid Staining with Oil Red O and the Quantitative PCR of Adipogenesis (DIF+)-Related Genes, including Pparγ, Ap2 and Leptin (Figure 2)
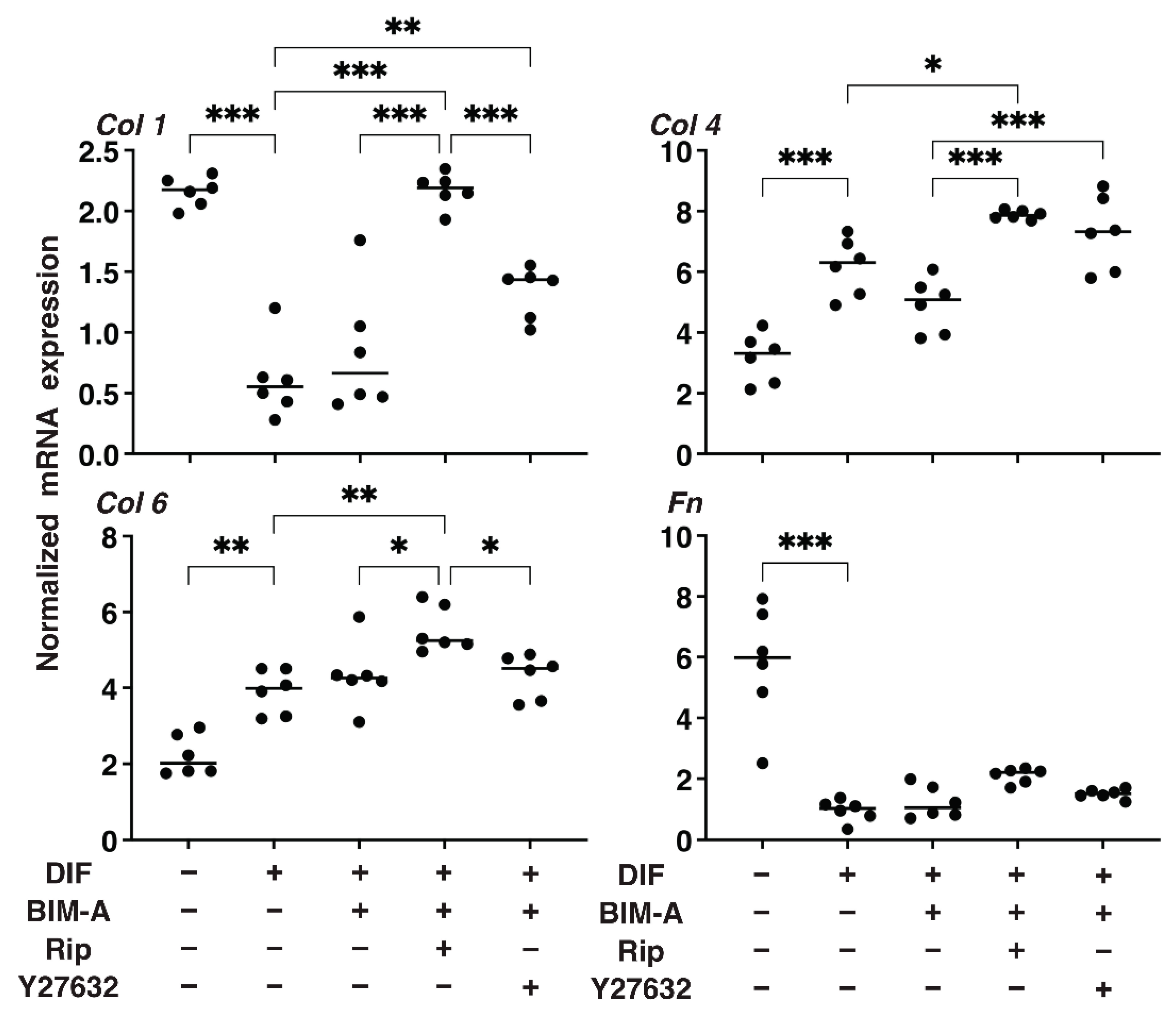
3.1.2. The Quantitative PCR Analysis of ECMs including Collagen (Col) 1, 4 and 6, and Fibronectin (Fn) (Figure 2), and Real-Time Cellular Metabolic Analysis (Figure 3)

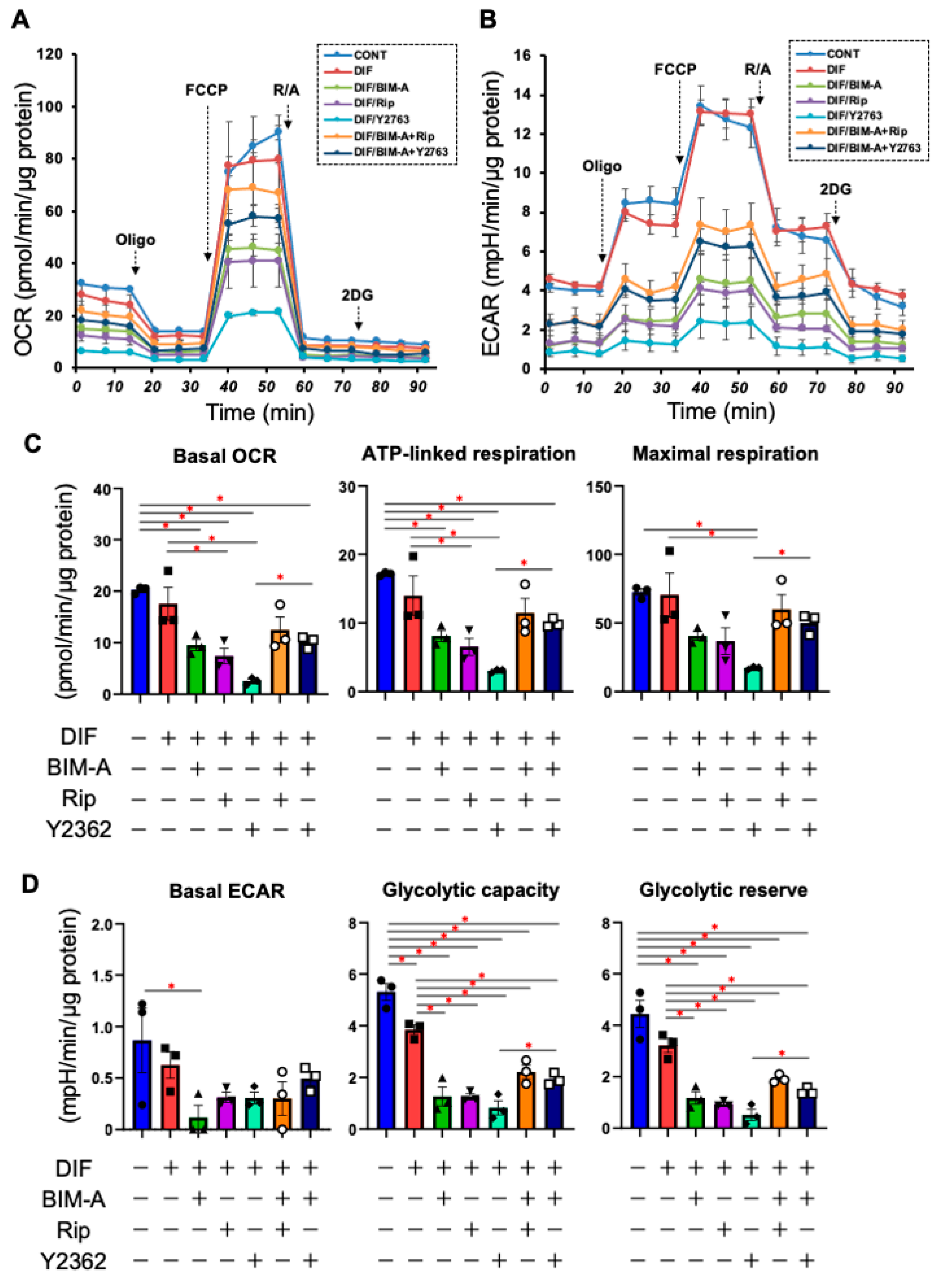
3.2. Additive Effects of ROCK-Is to PGs on 2D Cultured 3T3-L1 Cells
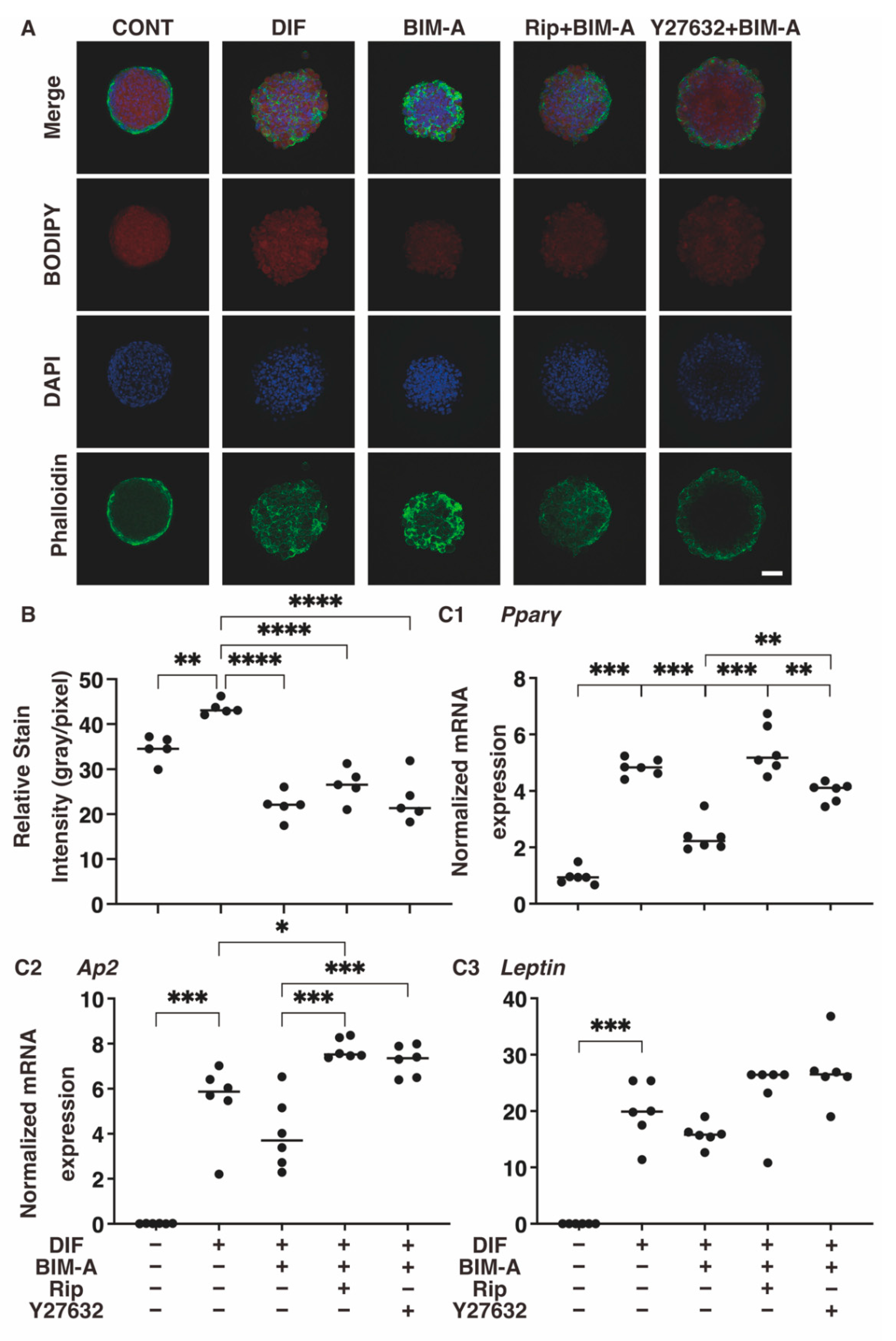
3.2.1. The 3D 3T3-L1 Spheroid Maturation and Their Physical Properties, Size (Figure 4) and Stiffness (Figure 5)
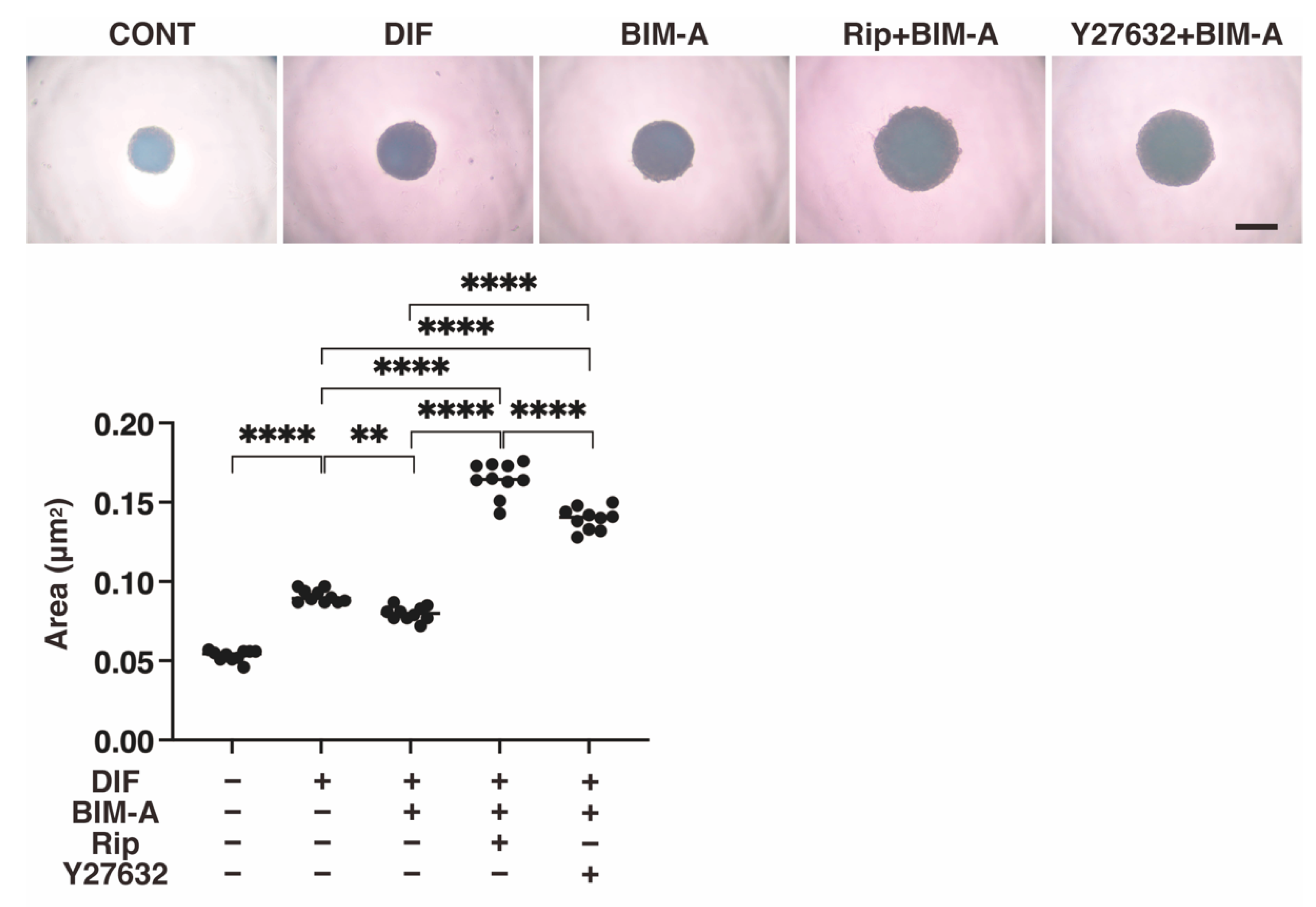

3.2.2. Lipid Staining with BODIPY and the Quantitative PCR of Adipogenesis (DIF+)-Related Genes, including Pparγ, Ap2 and Leptin (Figure 6)
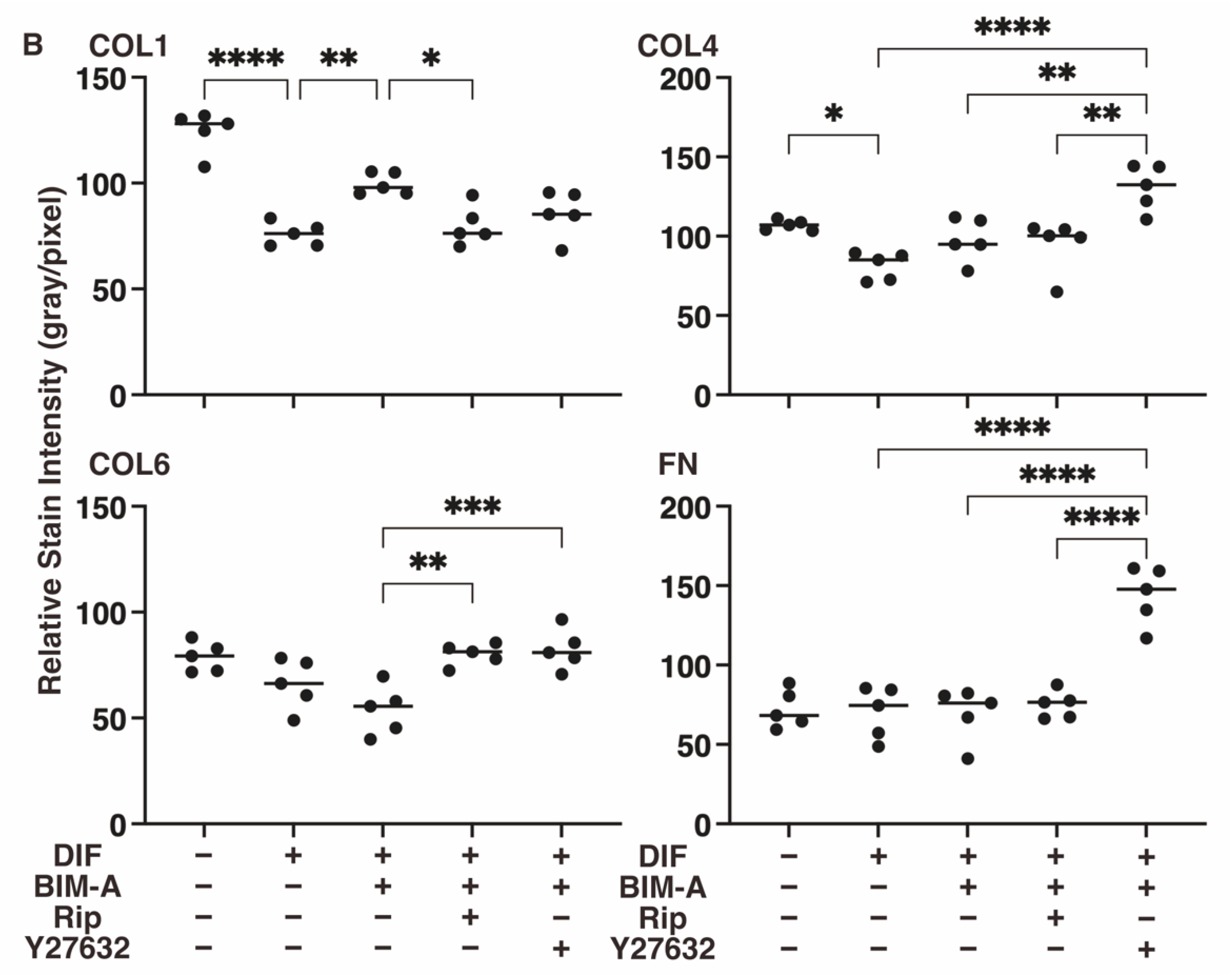
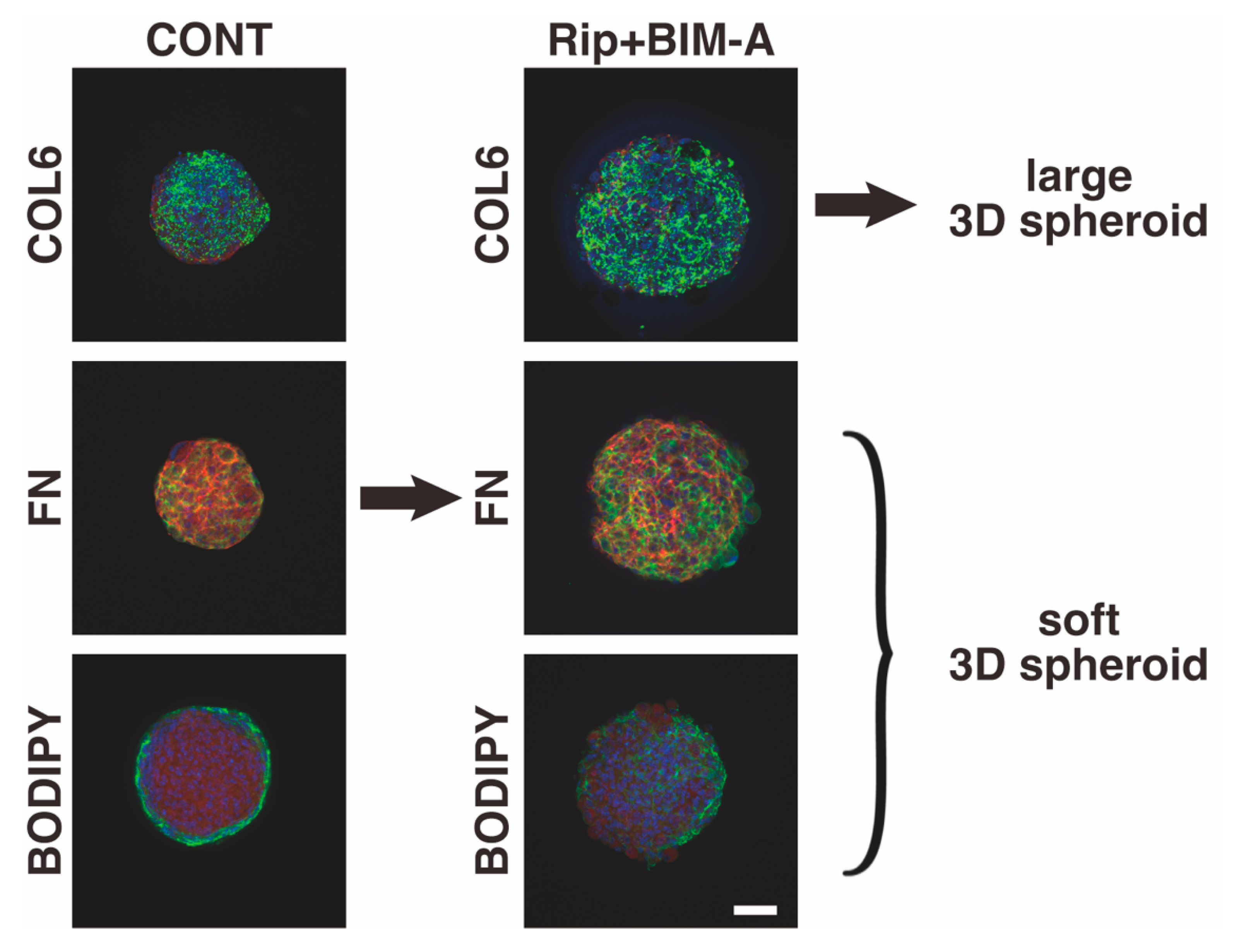
3.2.3. The Expression of ECMs including Collagen (Col) 1, 4 and 6, and Fibronectin (Fn) Analyzed by qPCR (Figure 8) and Immunocytochemistry (Figure 9)
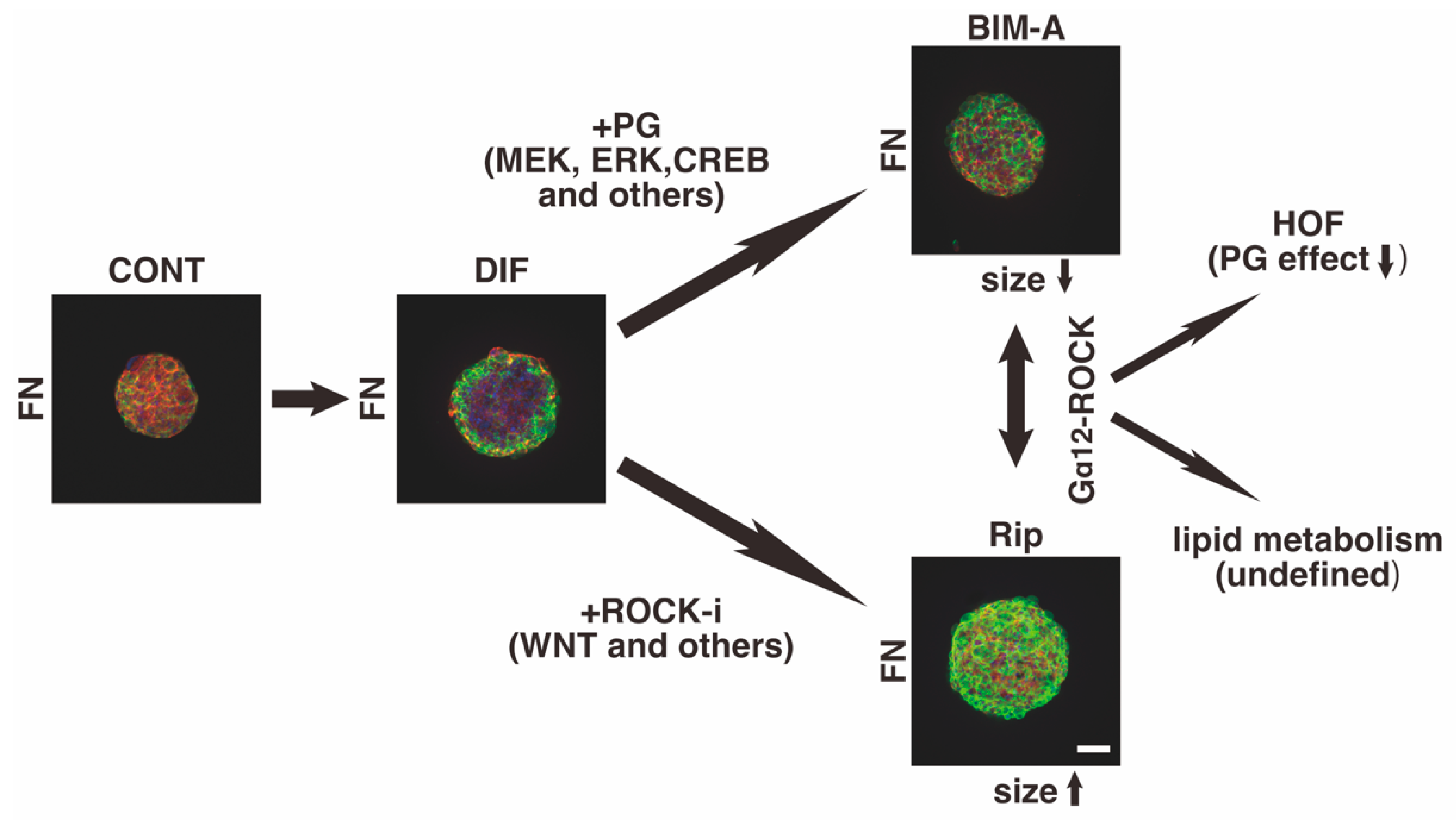
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cornier, M.A.; Dabelea, D.; Hernandez, T.L.; Lindstrom, R.C.; Steig, A.J.; Stob, N.R.; Van Pelt, R.E.; Wang, H.; Eckel, R.H. The metabolic syndrome. Endocr. Rev. 2008, 29, 777–822. [Google Scholar] [CrossRef] [PubMed]
- Pi-Sunyer, X. The medical risks of obesity. Postgrad. Med. 2009, 121, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Spiegelman, B.M.; Flier, J.S. Obesity and the regulation of energy balance. Cell 2001, 104, 531–543. [Google Scholar] [CrossRef] [Green Version]
- Fujimori, K.; Yano, M.; Ueno, T. Synergistic suppression of early phase of adipogenesis by microsomal PGE synthase-1 (PTGES1)-produced PGE2 and aldo-keto reductase 1B3-produced PGF2α. PLoS ONE 2012, 7, e44698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aubert, J.; Saint-Marc, P.; Belmonte, N.; Dani, C.; Négrel, R.; Ailhaud, G. Prostacyclin IP receptor up-regulates the early expression of C/EBPbeta and C/EBPdelta in preadipose cells. Mol. Cell. Endocrinol. 2000, 160, 149–156. [Google Scholar] [CrossRef]
- Vassaux, G.; Gaillard, D.; Ailhaud, G.; Négrel, R. Prostacyclin is a specific effector of adipose cell differentiation. Its dual role as a cAMP- and Ca(2+)-elevating agent. J. Biol. Chem. 1992, 267, 11092–11097. [Google Scholar] [CrossRef]
- Sasaki, Y.; Kuwata, H.; Akatsu, M.; Yamakawa, Y.; Ochiai, T.; Yoda, E.; Nakatani, Y.; Yokoyama, C.; Hara, S. Involvement of prostacyclin synthase in high-fat-diet-induced obesity. Prostaglandins Other Lipid Mediat. 2021, 153, 106523. [Google Scholar] [CrossRef]
- Fujimori, K.; Maruyama, T.; Kamauchi, S.; Urade, Y. Activation of adipogenesis by lipocalin-type prostaglandin D synthase-generated Δ12-PGJ₂ acting through PPARγ-dependent and independent pathways. Gene 2012, 505, 46–52. [Google Scholar] [CrossRef]
- Kliewer, S.A.; Lenhard, J.M.; Willson, T.M.; Patel, I.; Morris, D.C.; Lehmann, J.M. A prostaglandin J2 metabolite binds peroxisome proliferator-activated receptor gamma and promotes adipocyte differentiation. Cell 1995, 83, 813–819. [Google Scholar] [CrossRef] [Green Version]
- Wakai, E.; Aritake, K.; Urade, Y.; Fujimori, K. Prostaglandin D(2) enhances lipid accumulation through suppression of lipolysis via DP2 (CRTH2) receptors in adipocytes. Biochem. Biophys. Res. Commun. 2017, 490, 393–399. [Google Scholar] [CrossRef]
- Forman, B.M.; Tontonoz, P.; Chen, J.; Brun, R.P.; Spiegelman, B.M.; Evans, R.M. 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell 1995, 83, 803–812. [Google Scholar] [CrossRef] [Green Version]
- Matsui, T.; Amano, M.; Yamamoto, T.; Chihara, K.; Nakafuku, M.; Ito, M.; Nakano, T.; Okawa, K.; Iwamatsu, A.; Kaibuchi, K. Rho-associated kinase, a novel serine/threonine kinase, as a putative target for small GTP binding protein Rho. Embo J. 1996, 15, 2208–2216. [Google Scholar] [CrossRef]
- Ishizaki, T.; Maekawa, M.; Fujisawa, K.; Okawa, K.; Iwamatsu, A.; Fujita, A.; Watanabe, N.; Saito, Y.; Kakizuka, A.; Morii, N.; et al. The small GTP-binding protein Rho binds to and activates a 160 kDa Ser/Thr protein kinase homologous to myotonic dystrophy kinase. Embo J. 1996, 15, 1885–1893. [Google Scholar] [CrossRef]
- Nakagawa, O.; Fujisawa, K.; Ishizaki, T.; Saito, Y.; Nakao, K.; Narumiya, S. ROCK-I and ROCK-II, two isoforms of Rho-associated coiled-coil forming protein serine/threonine kinase in mice. FEBS Lett. 1996, 392, 189–193. [Google Scholar] [CrossRef] [Green Version]
- Leung, T.; Chen, X.Q.; Manser, E.; Lim, L. The p160 RhoA-binding kinase ROK alpha is a member of a kinase family and is involved in the reorganization of the cytoskeleton. Mol. Cell. Biol. 1996, 16, 5313–5327. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.; Shi, J. Insight Into Rho Kinase Isoforms in Obesity and Energy Homeostasis. Front. Endocrinol. 2022, 13, 886534. [Google Scholar] [CrossRef]
- van der Valk, R.; Webers, C.A.; Schouten, J.S.; Zeegers, M.P.; Hendrikse, F.; Prins, M.H. Intraocular pressure-lowering effects of all commonly used glaucoma drugs: A meta-analysis of randomized clinical trials. Ophthalmology 2005, 112, 1177–1185. [Google Scholar] [CrossRef] [PubMed]
- Weinreb, R.N.; Khaw, P.T. Primary open-angle glaucoma. Lancet 2004, 363, 1711–1720. [Google Scholar] [CrossRef]
- Quigley, H.A.; Broman, A.T. The number of people with glaucoma worldwide in 2010 and 2020. Br. J. Ophthalmol. 2006, 90, 262–267. [Google Scholar] [CrossRef] [Green Version]
- Caprioli, J.; Coleman, A.L. Blood pressure, perfusion pressure, and glaucoma. Am. J. Ophthalmol. 2010, 149, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Lindsley, K.; Rouse, B.; Hong, H.; Shi, Q.; Friedman, D.S.; Wormald, R.; Dickersin, K. Comparative Effectiveness of First-Line Medications for Primary Open-Angle Glaucoma: A Systematic Review and Network Meta-analysis. Ophthalmology 2016, 123, 129–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, K.; Shiokawa, M.; Wakakura, M.; Tomita, G. Deepening of the upper eyelid sulcus caused by 5 types of prostaglandin analogs. J. Glaucoma 2013, 22, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Filippopoulos, T.; Paula, J.S.; Torun, N.; Hatton, M.P.; Pasquale, L.R.; Grosskreutz, C.L. Periorbital changes associated with topical bimatoprost. Ophthalmic Plast. Reconstr. Surg. 2008, 24, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Jayaprakasam, A.; Ghazi-Nouri, S. Periorbital fat atrophy—An unfamiliar side effect of prostaglandin analogues. Orbit 2010, 29, 357–359. [Google Scholar] [CrossRef]
- Hikage, F.; Atkins, S.; Kahana, A.; Smith, T.J.; Chun, T.H. HIF2A-LOX Pathway Promotes Fibrotic Tissue Remodeling in Thyroid-Associated Orbitopathy. Endocrinology 2019, 160, 20–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hikage, F.; Ichioka, H.; Watanabe, M.; Umetsu, A.; Ohguro, H.; Ida, Y. Addition of ROCK inhibitors to prostaglandin derivative (PG) synergistically affects adipogenesis of the 3D spheroids of human orbital fibroblasts (HOFs). Hum. Cell 2022, 35, 125–132. [Google Scholar] [CrossRef]
- Itoh, K.; Hikage, F.; Ida, Y.; Ohguro, H. Prostaglandin F2alpha Agonists Negatively Modulate the Size of 3D Organoids from Primary Human Orbital Fibroblasts. Investig. Ophthalmol. Vis. Sci. 2020, 61, 13. [Google Scholar] [CrossRef]
- Itoh, K.; Ida, Y.; Ohguro, H.; Hikage, F. Prostaglandin F2α agonists induced enhancement in collagen1 expression is involved in the pathogenesis of the deepening of upper eyelid sulcus. Sci. Rep. 2021, 11, 9002. [Google Scholar] [CrossRef] [PubMed]
- Hikage, F.; Ichioka, H.; Watanabe, M.; Umetsu, A.; Ohguro, H.; Ida, Y. ROCK inhibitors modulate the physical properties and adipogenesis of 3D spheroids of human orbital fibroblasts in different manners. FASEB BioAdvances 2021, 3, 866–872. [Google Scholar] [CrossRef]
- Shen, J.X.; Couchet, M.; Dufau, J.; de Castro Barbosa, T.; Ulbrich, M.H.; Helmstädter, M.; Kemas, A.M.; Zandi Shafagh, R.; Marques, M.A.; Hansen, J.B.; et al. 3D Adipose Tissue Culture Links the Organotypic Microenvironment to Improved Adipogenesis. Adv. Sci. 2021, 8, e2100106. [Google Scholar] [CrossRef]
- Ohguro, H.; Ida, Y.; Hikage, F.; Umetsu, A.; Ichioka, H.; Watanabe, M.; Furuhashi, M. STAT3 Is the Master Regulator for the Forming of 3D Spheroids of 3T3-L1 Preadipocytes. Cells 2022, 11, 300. [Google Scholar] [CrossRef] [PubMed]
- Ida, Y.; Hikage, F.; Ohguro, H. ROCK inhibitors enhance the production of large lipid-enriched 3D organoids of 3T3-L1 cells. Sci. Rep. 2021, 11, 5479. [Google Scholar] [CrossRef] [PubMed]
- Ida, Y.; Hikage, F.; Itoh, K.; Ida, H.; Ohguro, H. Prostaglandin F2alpha agonist-induced suppression of 3T3-L1 cell adipogenesis affects spatial formation of extra-cellular matrix. Sci. Rep. 2020, 10, 7958. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Chang, H.C.; Bayeva, M.; Shapiro, J.S.; Ramos-Alonso, L.; Kouzu, H.; Jiang, X.; Liu, T.; Yar, S.; Sawicki, K.T.; et al. mRNA-binding protein tristetraprolin is essential for cardiac response to iron deficiency by regulating mitochondrial function. Proc. Natl. Acad. Sci. USA 2018, 115, E6291–E6300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, T.; Ichise, N.; Kobayashi, T.; Fusagawa, H.; Yamazaki, H.; Kudo, T.; Tohse, N. Enhanced glucose metabolism through activation of HIF-1α covers the energy demand in a rat embryonic heart primordium after heartbeat initiation. Sci. Rep. 2022, 12, 74. [Google Scholar] [CrossRef]
- Peplinski, L.S.; Albiani Smith, K. Deepening of lid sulcus from topical bimatoprost therapy. Optom. Vis. Sci. 2004, 81, 574–577. [Google Scholar] [CrossRef]
- Aihara, M.; Shirato, S.; Sakata, R. Incidence of deepening of the upper eyelid sulcus after switching from latanoprost to bimatoprost. Jpn. J. Ophthalmol. 2011, 55, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Guru, A.; Issac, P.K.; Velayutham, M.; Saraswathi, N.T.; Arshad, A.; Arockiaraj, J. Molecular mechanism of down-regulating adipogenic transcription factors in 3T3-L1 adipocyte cells by bioactive anti-adipogenic compounds. Mol. Biol. Rep. 2021, 48, 743–761. [Google Scholar] [CrossRef]
- Ida, Y.; Hikage, F.; Umetsu, A.; Ida, H.; Ohguro, H. Omidenepag, a non-prostanoid EP2 receptor agonist, induces enlargement of the 3D organoid of 3T3-L1 cells. Sci. Rep. 2020, 10, 16018. [Google Scholar] [CrossRef]
- Matsuda, H.; Nakamura, S.; Yoshikawa, M. Search for new type of PPARγ agonist-like anti-diabetic compounds from medicinal plants. Biol. Pharm. Bull. 2014, 37, 884–891. [Google Scholar] [CrossRef]
- Härmä, V.; Schukov, H.P.; Happonen, A.; Ahonen, I.; Virtanen, J.; Siitari, H.; Åkerfelt, M.; Lötjönen, J.; Nees, M. Quantification of dynamic morphological drug responses in 3D organotypic cell cultures by automated image analysis. PLoS ONE 2014, 9, e96426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosen, E.D. The transcriptional basis of adipocyte development. Prostaglandins Leukot. Essent. Fat. Acids 2005, 73, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Dai, F.; Liu, Y.; Yu, X.; Huang, C.; Wang, Y.; Yao, W. RhoA/ROCK signaling regulates smooth muscle phenotypic modulation and vascular remodeling via the JNK pathway and vimentin cytoskeleton. Pharmacol. Res. 2018, 133, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; He, J.T.; Zhang, K.K.; Chen, L.J.; Wang, Q.; Xie, X.L. Methamphetamine reduces expressions of tight junction proteins, rearranges F-actin cytoskeleton and increases the blood brain barrier permeability via the RhoA/ROCK-dependent pathway. Biochem. Biophys. Res. Commun. 2019, 509, 395–401. [Google Scholar] [CrossRef]
- Mack, N.A.; Georgiou, M. The interdependence of the Rho GTPases and apicobasal cell polarity. Small GTPases 2014, 5, 10. [Google Scholar] [CrossRef]
- Julian, L.; Olson, M.F. Rho-associated coiled-coil containing kinases (ROCK): Structure, regulation, and functions. Small GTPases 2014, 5, e29846. [Google Scholar] [CrossRef] [PubMed]
- Chun, K.H.; Araki, K.; Jee, Y.; Lee, D.H.; Oh, B.C.; Huang, H.; Park, K.S.; Lee, S.W.; Zabolotny, J.M.; Kim, Y.B. Regulation of glucose transport by ROCK1 differs from that of ROCK2 and is controlled by actin polymerization. Endocrinology 2012, 153, 1649–1662. [Google Scholar] [CrossRef] [Green Version]
- Street, C.A.; Bryan, B.A. Rho kinase proteins--pleiotropic modulators of cell survival and apoptosis. Anticancer Res. 2011, 31, 3645–3657. [Google Scholar]
- Noguchi, M.; Hosoda, K.; Fujikura, J.; Fujimoto, M.; Iwakura, H.; Tomita, T.; Ishii, T.; Arai, N.; Hirata, M.; Ebihara, K.; et al. Genetic and pharmacological inhibition of Rho-associated kinase II enhances adipogenesis. J. Biol. Chem. 2007, 282, 29574–29583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diep, D.T.V.; Hong, K.; Khun, T.; Zheng, M.; Ul-Haq, A.; Jun, H.S.; Kim, Y.B.; Chun, K.H. Anti-adipogenic effects of KD025 (SLx-2119), a ROCK2-specific inhibitor, in 3T3-L1 cells. Sci. Rep. 2018, 8, 2477. [Google Scholar] [CrossRef] [Green Version]
- Inoue, T.; Tanihara, H. Rho-associated kinase inhibitors: A novel glaucoma therapy. Prog. Retin. Eye Res. 2013, 37, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yoneda, A.; Ushakov, D.; Multhaupt, H.A.; Couchman, J.R. Fibronectin matrix assembly requires distinct contributions from Rho kinases I and -II. Mol. Biol. Cell 2007, 18, 66–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, C.; Chrzanowska-Wodnicka, M.; Brown, J.; Shaub, A.; Belkin, A.M.; Burridge, K. Rho-mediated contractility exposes a cryptic site in fibronectin and induces fibronectin matrix assembly. J. Cell Biol. 1998, 141, 539–551. [Google Scholar] [CrossRef]
- Honjo, M.; Tanihara, H.; Inatani, M.; Kido, N.; Sawamura, T.; Yue, B.Y.J.T.; Narumiya, S.; Honda, Y. Effects of Rho-Associated Protein Kinase Inhibitor Y-27632 on Intraocular Pressure and Outflow Facility. Investig. Ophthalmol. Vis. Sci. 2001, 42, 137–144. [Google Scholar]
- Rao, P.V.; Deng, P.-F.; Kumar, J.; Epstein, D.L. Modulation of Aqueous Humor Outflow Facility by the Rho Kinase–Specific Inhibitor Y-27632. Investig. Ophthalmol. Vis. Sci. 2001, 42, 1029–1037. [Google Scholar]

| BIM-A | Rip | Y27632 | BIM-A+Rip | BIM-A+Y27632 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L1 | HOF ** | L1 * | HOF ** | L1 * | L1 | HOF ** | L1 | |||||
| size 3D | ↓ | ↓↓ | ↑ | → | ↑ | ↑ | (B < R) | ↓↓ | (B > R) | ↑ | (B < Y) | |
| stiffness 3D | ↑ | ↑↑ | ↓ | ↓ | ↓ | ↓ | (B < R) | ↓ | (B < R) | ↓ | (B < Y) | |
| lipid stain | 2D | ↓ | ↑ | ↑ | → | (B = R) | ↑ | (B < Y) | ||||
| 3D | ↓ | ↓↓ | → | ↑ | ↑ | ↓ | (B > R) | → | (B = R) | ↓ | (B > Y) | |
| Pparγ | 2D | ↓ | → | ↑ | ↓↓ | N. I. | ↓ | (B > Y) | ||||
| 3D | ↓↓↓ | ↓ | → | → | ↑↑ | → | N. I. | → | N.I. | ↓↓ | (B > Y) | |
| Ap2 | 2D | → | → | → | ↓↓↓ | N. I. | ↓ | N. I. | ||||
| 3D | → | ↓ | ↑↑ | ↑↑ | ↑↑↑↑ | ↑ | N. I. | ↑↑ | (B < R) | → | N. I. | |
| Leptin | 2D | → | ↓↓↓ | → | ↓↓↓ | (B < R) | ↓↓↓ | N. I. | ||||
| 3D | → | ↑ | ↑↑↑ | → | N. I. | → | N. I. | |||||
| Col1 | 2D | → | ↓ | → | ↓ | (B < R) | ↓ | N. I. | ||||
| 3D | → | ↑↑ | ↑ | → | → | → | N. I. | ↑↑ | (B > R) | → | N. I. | |
| Col4 | 2D | → | ↓ | → | ↓ | (B < R) | ↓ | N. I. | ||||
| 3D | → | → | ↑ | ↑↑ | ↑ | → | N. I. | → | N. I. | ↓ | N. I. | |
| Col6 | 2D | → | → | ↑ | ↓ | N. I. | ↓ | N. I. | ||||
| 3D | → | → | ↑ | → | ↑ | ↑ | (B < R) | ↑ | N. I. | → | N. I. | |
| Fn | 2D | → | → | → | ↓ | N. I. | ↓ | N. I. | ||||
| 3D | → | → | → | → | → | ↑ | N. I. | → | B=R | ↑ | N. I. | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ida, Y.; Sato, T.; Umetsu, A.; Watanabe, M.; Furuhashi, M.; Hikage, F.; Ohguro, H. Addition of ROCK Inhibitors Alleviates Prostaglandin-Induced Inhibition of Adipogenesis in 3T3L-1 Spheroids. Bioengineering 2022, 9, 702. https://doi.org/10.3390/bioengineering9110702
Ida Y, Sato T, Umetsu A, Watanabe M, Furuhashi M, Hikage F, Ohguro H. Addition of ROCK Inhibitors Alleviates Prostaglandin-Induced Inhibition of Adipogenesis in 3T3L-1 Spheroids. Bioengineering. 2022; 9(11):702. https://doi.org/10.3390/bioengineering9110702
Chicago/Turabian StyleIda, Yosuke, Tatsuya Sato, Araya Umetsu, Megumi Watanabe, Masato Furuhashi, Fumihito Hikage, and Hiroshi Ohguro. 2022. "Addition of ROCK Inhibitors Alleviates Prostaglandin-Induced Inhibition of Adipogenesis in 3T3L-1 Spheroids" Bioengineering 9, no. 11: 702. https://doi.org/10.3390/bioengineering9110702
APA StyleIda, Y., Sato, T., Umetsu, A., Watanabe, M., Furuhashi, M., Hikage, F., & Ohguro, H. (2022). Addition of ROCK Inhibitors Alleviates Prostaglandin-Induced Inhibition of Adipogenesis in 3T3L-1 Spheroids. Bioengineering, 9(11), 702. https://doi.org/10.3390/bioengineering9110702









