Bioengineering Strategies to Create 3D Cardiac Constructs from Human Induced Pluripotent Stem Cells
Abstract
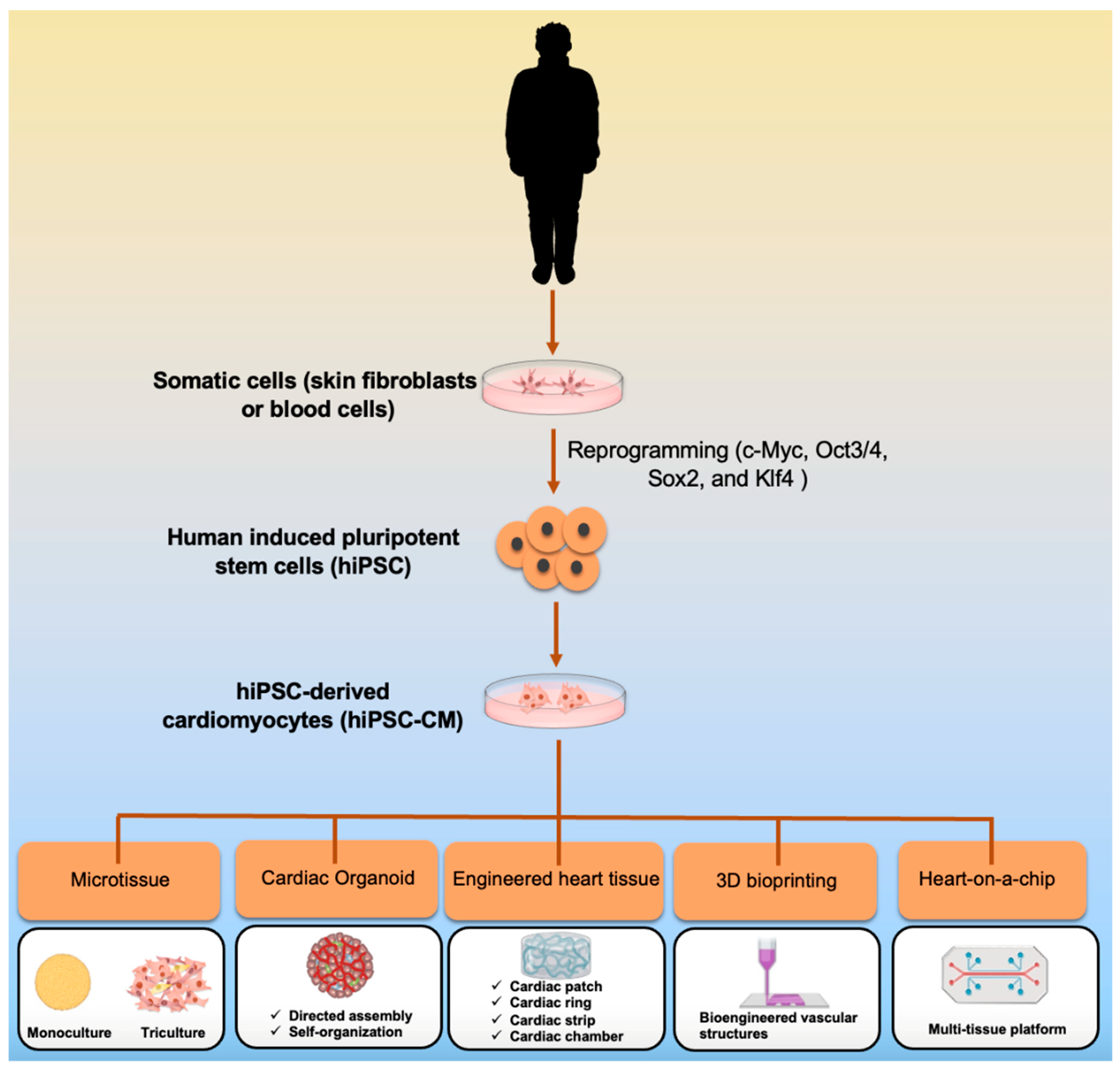
:1. Introduction
2. 3D Culture Using Scaffold-Free and Scaffold-Based Approaches
3. Cardiac Organoids
4. 3D Bioprinting
5. Heart-on-a-Chip
6. Cardiac Disease Modeling by 3D Engineering Tissue
7. Quality Control of hiPSC-CMs by Artificial Intelligence and Machine Learning
8. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update From the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef]
- Mozaffarian, D.; Benjamin, E.J.; Go, A.S.; Arnett, D.K.; Blaha, M.J.; Cushman, M.; de Ferranti, S.; Despres, J.P.; Fullerton, H.J.; Howard, V.J.; et al. Heart disease and stroke statistics—2015 update: A report from the American Heart Association. Circulation 2015, 131, e29–e322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Q.; Cai, H.; Zhan, Y.; Li, B.; Hu, S.; Sun, N. Applications of human-induced pluripotent stem cells in the investigation of inherited cardiomyopathy. Int. J. Cardiol. 2014, 177, 604–606. [Google Scholar] [CrossRef] [PubMed]
- Suh, T.C.; Amanah, A.Y.; Gluck, J.M. Electrospun Scaffolds and Induced Pluripotent Stem Cell-Derived Cardiomyocytes for Cardiac Tissue Engineering Applications. Bioengineering 2020, 7, 105. [Google Scholar] [CrossRef] [PubMed]
- Karakikes, I.; Ameen, M.; Termglinchan, V.; Wu, J.C. Human induced pluripotent stem cell-derived cardiomyocytes: Insights into molecular, cellular, and functional phenotypes. Circ. Res. 2015, 117, 80–88. [Google Scholar] [CrossRef] [Green Version]
- Pimpaneau, V.; Voisin, E. Innovative medicine development. Technol. Health Care 2022, 30, 499–503. [Google Scholar] [CrossRef]
- Sardella, M.; Belcher, G.; Lungu, C.; Ignoni, T.; Camisa, M.; Stenver, D.I.; Porcelli, P.; D’Antuono, M.; Castiglione, N.G.; Adams, A.; et al. Monitoring the manufacturing and quality of medicines: A fundamental task of pharmacovigilance. Ther. Adv. Drug Saf. 2021, 12, 20420986211038436. [Google Scholar] [CrossRef]
- Buono, M.F.; von Boehmer, L.; Strang, J.; Hoerstrup, S.P.; Emmert, M.Y.; Nugraha, B. Human Cardiac Organoids for Modeling Genetic Cardiomyopathy. Cells 2020, 9, 1733. [Google Scholar] [CrossRef]
- Abulaiti, M.; Yalikun, Y.; Murata, K.; Sato, A.; Sami, M.M.; Sasaki, Y.; Fujiwara, Y.; Minatoya, K.; Shiba, Y.; Tanaka, Y.; et al. Establishment of a heart-on-a-chip microdevice based on human iPS cells for the evaluation of human heart tissue function. Sci. Rep. 2020, 10, 19201. [Google Scholar] [CrossRef]
- Hnatiuk, A.P.; Briganti, F.; Staudt, D.W.; Mercola, M. Human iPSC modeling of heart disease for drug development. Cell Chem. Biol. 2021, 28, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Huch, M.; Knoblich, J.A.; Lutolf, M.P.; Martinez-Arias, A. The hope and the hype of organoid research. Development 2017, 144, 938–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kansakar, U.; Varzideh, F.; Jankauskas, S.S.; Gambardella, J.; Trimarco, B.; Santulli, G. Advances in the understanding of excitation-contraction coupling: The pulsing quest for drugs against heart failure and arrhythmias. Eur. Heart J. Cardiovasc. Pharmacother. 2021, 7, e91–e93. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Pu, W.T. Cardiomyocyte Maturation: New Phase in Development. Circ. Res. 2020, 126, 1086–1106. [Google Scholar] [CrossRef] [PubMed]
- Veerman, C.C.; Kosmidis, G.; Mummery, C.L.; Casini, S.; Verkerk, A.O.; Bellin, M. Immaturity of human stem-cell-derived cardiomyocytes in culture: Fatal flaw or soluble problem? Stem Cells Dev. 2015, 24, 1035–1052. [Google Scholar] [CrossRef] [Green Version]
- Scuderi, G.J.; Butcher, J. Naturally Engineered Maturation of Cardiomyocytes. Front. Cell Dev. Biol. 2017, 5, 50. [Google Scholar] [CrossRef]
- Tenreiro, M.F.; Louro, A.F.; Alves, P.M.; Serra, M. Next generation of heart regenerative therapies: Progress and promise of cardiac tissue engineering. NPJ Regen. Med. 2021, 6, 30. [Google Scholar] [CrossRef]
- Li, J.; Hua, Y.; Miyagawa, S.; Zhang, J.; Li, L.; Liu, L.; Sawa, Y. hiPSC-Derived Cardiac Tissue for Disease Modeling and Drug Discovery. Int. J. Mol. Sci. 2020, 21, 8893. [Google Scholar] [CrossRef]
- Deng, Y.; Lao, Y.; Ruan, Q.; Zhang, J.; Luo, C.; Shi, D.; Lu, F. Activation of Wnt/beta-Catenin Signaling Pathway Enhances the Derivation of Buffalo (Bubalus bubalis) Embryonic Stem Cell-Like Cells. Cell Reprogram. 2020, 22, 217–225. [Google Scholar] [CrossRef]
- Laco, F.; Woo, T.L.; Zhong, Q.; Szmyd, R.; Ting, S.; Khan, F.J.; Chai, C.L.L.; Reuveny, S.; Chen, A.; Oh, S. Unraveling the Inconsistencies of Cardiac Differentiation Efficiency Induced by the GSK3beta Inhibitor CHIR99021 in Human Pluripotent Stem Cells. Stem Cell Rep. 2018, 10, 1851–1866. [Google Scholar] [CrossRef] [Green Version]
- Halloin, C.; Schwanke, K.; Lobel, W.; Franke, A.; Szepes, M.; Biswanath, S.; Wunderlich, S.; Merkert, S.; Weber, N.; Osten, F.; et al. Continuous WNT Control Enables Advanced hPSC Cardiac Processing and Prognostic Surface Marker Identification in Chemically Defined Suspension Culture. Stem Cell Rep. 2019, 13, 775. [Google Scholar] [CrossRef] [PubMed]
- Mazzotta, S.; Neves, C.; Bonner, R.J.; Bernardo, A.S.; Docherty, K.; Hoppler, S. Distinctive Roles of Canonical and Noncanonical Wnt Signaling in Human Embryonic Cardiomyocyte Development. Stem Cell Rep. 2016, 7, 764–776. [Google Scholar] [CrossRef] [Green Version]
- Branco, M.A.; Cotovio, J.P.; Rodrigues, C.A.V.; Vaz, S.H.; Fernandes, T.G.; Moreira, L.M.; Cabral, J.M.S.; Diogo, M.M. Transcriptomic analysis of 3D Cardiac Differentiation of Human Induced Pluripotent Stem Cells Reveals Faster Cardiomyocyte Maturation Compared to 2D Culture. Sci. Rep. 2019, 9, 9229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correia, C.; Koshkin, A.; Duarte, P.; Hu, D.; Carido, M.; Sebastiao, M.J.; Gomes-Alves, P.; Elliott, D.A.; Domian, I.J.; Teixeira, A.P.; et al. 3D aggregate culture improves metabolic maturation of human pluripotent stem cell derived cardiomyocytes. Biotechnol. Bioeng. 2018, 115, 630–644. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, L.; Yu, L.; Minami, I.; Miyagawa, S.; Horning, M.; Dong, J.; Qiao, J.; Qu, X.; Hua, Y.; et al. Circulating re-entrant waves promote maturation of hiPSC-derived cardiomyocytes in self-organized tissue ring. Commun. Biol. 2020, 3, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Shadrin, I.Y.; Lam, J.; Xian, H.Q.; Snodgrass, H.R.; Bursac, N. Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials 2013, 34, 5813–5820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, P.; Deng, G.; Sai, X.; Guo, H.; Huang, H.; Zhu, P. Maturation strategies and limitations of induced pluripotent stem cell-derived cardiomyocytes. Biosci. Rep. 2021, 41, BSR20200833. [Google Scholar] [CrossRef] [PubMed]
- Beauchamp, P.; Jackson, C.B.; Ozhathil, L.C.; Agarkova, I.; Galindo, C.L.; Sawyer, D.B.; Suter, T.M.; Zuppinger, C. 3D Co-culture of hiPSC-Derived Cardiomyocytes With Cardiac Fibroblasts Improves Tissue-Like Features of Cardiac Spheroids. Front. Mol. Biosci. 2020, 7, 14. [Google Scholar] [CrossRef]
- Laco, F.; Lam, A.T.; Woo, T.L.; Tong, G.; Ho, V.; Soong, P.L.; Grishina, E.; Lin, K.H.; Reuveny, S.; Oh, S.K. Selection of human induced pluripotent stem cells lines optimization of cardiomyocytes differentiation in an integrated suspension microcarrier bioreactor. Stem Cell Res. Ther. 2020, 11, 118. [Google Scholar] [CrossRef] [Green Version]
- Varzideh, F.; Mahmoudi, E.; Pahlavan, S. Coculture with noncardiac cells promoted maturation of human stem cell-derived cardiomyocyte microtissues. J. Cell Biochem. 2019, 120, 16681–16691. [Google Scholar] [CrossRef]
- Archer, C.R.; Sargeant, R.; Basak, J.; Pilling, J.; Barnes, J.R.; Pointon, A. Characterization and Validation of a Human 3D Cardiac Microtissue for the Assessment of Changes in Cardiac Pathology. Sci. Rep. 2018, 8, 10160. [Google Scholar] [CrossRef] [PubMed]
- Mattapally, S.; Zhu, W.; Fast, V.G.; Gao, L.; Worley, C.; Kannappan, R.; Borovjagin, A.V.; Zhang, J. Spheroids of cardiomyocytes derived from human-induced pluripotent stem cells improve recovery from myocardial injury in mice. Am. J. Physiol. Heart Circ. Physiol. 2018, 315, H327–H339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Serpooshan, V.; Tong, X.; Venkatraman, S.; Lee, M.; Lee, J.; Chirikian, O.; Wu, J.C.; Wu, S.M.; Yang, F. Contractile force generation by 3D hiPSC-derived cardiac tissues is enhanced by rapid establishment of cellular interconnection in matrix with muscle-mimicking stiffness. Biomaterials 2017, 131, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Khanna, A.; Zamani, M.; Huang, N.F. Extracellular Matrix-Based Biomaterials for Cardiovascular Tissue Engineering. J. Cardiovasc. Dev. Dis. 2021, 8, 137. [Google Scholar] [CrossRef] [PubMed]
- Almeida, H.V.; Tenreiro, M.F.; Louro, A.F.; Abecasis, B.; Santinha, D.; Calmeiro, T.; Fortunato, E.; Ferreira, L.; Alves, P.M.; Serra, M. Human Extracellular-Matrix Functionalization of 3D hiPSC-Based Cardiac Tissues Improves Cardiomyocyte Maturation. ACS Appl. Bio. Mater. 2021, 4, 1888–1899. [Google Scholar] [CrossRef]
- Andrysiak, K.; Stepniewski, J.; Dulak, J. Human-induced pluripotent stem cell-derived cardiomyocytes, 3D cardiac structures, and heart-on-a-chip as tools for drug research. Pflug. Arch. 2021, 473, 1061–1085. [Google Scholar] [CrossRef]
- Bayomy, A.F.; Bauer, M.; Qiu, Y.; Liao, R. Regeneration in heart disease-Is ECM the key? Life Sci. 2012, 91, 823–827. [Google Scholar] [CrossRef] [Green Version]
- Bejleri, D.; Davis, M.E. Decellularized Extracellular Matrix Materials for Cardiac Repair and Regeneration. Adv. Healthc. Mater. 2019, 8, e1801217. [Google Scholar] [CrossRef]
- Kim, Y.S.; Majid, M.; Melchiorri, A.J.; Mikos, A.G. Applications of decellularized extracellular matrix in bone and cartilage tissue engineering. Bioeng. Transl. Med. 2019, 4, 83–95. [Google Scholar] [CrossRef] [Green Version]
- Sackett, S.D.; Tremmel, D.M.; Ma, F.; Feeney, A.K.; Maguire, R.M.; Brown, M.E.; Zhou, Y.; Li, X.; O’Brien, C.; Li, L.; et al. Extracellular matrix scaffold and hydrogel derived from decellularized and delipidized human pancreas. Sci. Rep. 2018, 8, 10452. [Google Scholar] [CrossRef]
- Giobbe, G.G.; Crowley, C.; Luni, C.; Campinoti, S.; Khedr, M.; Kretzschmar, K.; De Santis, M.M.; Zambaiti, E.; Michielin, F.; Meran, L.; et al. Extracellular matrix hydrogel derived from decellularized tissues enables endodermal organoid culture. Nat. Commun. 2019, 10, 5658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, A.; Eder, A.; Bonstrup, M.; Flato, M.; Mewe, M.; Schaaf, S.; Aksehirlioglu, B.; Schwoerer, A.P.; Uebeler, J.; Eschenhagen, T. Development of a drug screening platform based on engineered heart tissue. Circ. Res. 2010, 107, 35–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masumoto, H.; Yamashita, J.K. Human iPS cell-engineered three-dimensional cardiac tissues perfused by capillary networks between host and graft. Inflamm. Regen. 2018, 38, 26. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, I.C.; Karakikes, I.; Serrao, G.W.; Backeris, P.; Lee, J.J.; Xie, C.; Senyei, G.; Gordon, R.E.; Li, R.A.; Akar, F.G.; et al. Advancing functional engineered cardiac tissues toward a preclinical model of human myocardium. FASEB J. 2014, 28, 644–654. [Google Scholar] [CrossRef] [Green Version]
- Jackman, C.P.; Carlson, A.L.; Bursac, N. Dynamic culture yields engineered myocardium with near-adult functional output. Biomaterials 2016, 111, 66–79. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Yang, H.; Bai, A.; Jiang, W.; Li, X.; Wang, X.; Mao, Y.; Lu, C.; Qian, R.; Guo, F.; et al. Functional engineered human cardiac patches prepared from nature’s platform improve heart function after acute myocardial infarction. Biomaterials 2016, 105, 52–65. [Google Scholar] [CrossRef]
- VeDepo, M.C.; Detamore, M.S.; Hopkins, R.A.; Converse, G.L. Recellularization of decellularized heart valves: Progress toward the tissue-engineered heart valve. J. Tissue Eng. 2017, 8, 2041731417726327. [Google Scholar] [CrossRef]
- Navaei, A.; Truong, D.; Heffernan, J.; Cutts, J.; Brafman, D.; Sirianni, R.W.; Vernon, B.; Nikkhah, M. PNIPAAm-based biohybrid injectable hydrogel for cardiac tissue engineering. Acta Biomater. 2016, 32, 10–23. [Google Scholar] [CrossRef]
- Blazeski, A.; Lowenthal, J.; Zhu, R.; Ewoldt, J.; Boheler, K.R.; Tung, L. Functional Properties of Engineered Heart Slices Incorporating Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Stem Cell Rep. 2019, 12, 982–995. [Google Scholar] [CrossRef] [Green Version]
- Tulloch, N.L.; Muskheli, V.; Razumova, M.V.; Korte, F.S.; Regnier, M.; Hauch, K.D.; Pabon, L.; Reinecke, H.; Murry, C.E. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ. Res. 2011, 109, 47–59. [Google Scholar] [CrossRef]
- Zhang, W.; Kong, C.W.; Tong, M.H.; Chooi, W.H.; Huang, N.; Li, R.A.; Chan, B.P. Maturation of human embryonic stem cell-derived cardiomyocytes (hESC-CMs) in 3D collagen matrix: Effects of niche cell supplementation and mechanical stimulation. Acta Biomater. 2017, 49, 204–217. [Google Scholar] [CrossRef] [PubMed]
- Burridge, P.W.; Metzler, S.A.; Nakayama, K.H.; Abilez, O.J.; Simmons, C.S.; Bruce, M.A.; Matsuura, Y.; Kim, P.; Wu, J.C.; Butte, M.; et al. Multi-cellular interactions sustain long-term contractility of human pluripotent stem cell-derived cardiomyocytes. Am. J. Transl. Res. 2014, 6, 724–735. [Google Scholar] [PubMed]
- Caspi, O.; Lesman, A.; Basevitch, Y.; Gepstein, A.; Arbel, G.; Habib, I.H.; Gepstein, L.; Levenberg, S. Tissue engineering of vascularized cardiac muscle from human embryonic stem cells. Circ. Res. 2007, 100, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Majdi, M.; Xia, P.; Wei, K.A.; Talantova, M.; Spiering, S.; Nelson, B.; Mercola, M.; Chen, H.S. Non-cardiomyocytes influence the electrophysiological maturation of human embryonic stem cell-derived cardiomyocytes during differentiation. Stem Cells Dev. 2010, 19, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Giacomelli, E.; Bellin, M.; Sala, L.; van Meer, B.J.; Tertoolen, L.G.; Orlova, V.V.; Mummery, C.L. Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development 2017, 144, 1008–1017. [Google Scholar] [CrossRef] [Green Version]
- Giacomelli, E.; Meraviglia, V.; Campostrini, G.; Cochrane, A.; Cao, X.; van Helden, R.W.J.; Krotenberg Garcia, A.; Mircea, M.; Kostidis, S.; Davis, R.P.; et al. Human-iPSC-Derived Cardiac Stromal Cells Enhance Maturation in 3D Cardiac Microtissues and Reveal Non-cardiomyocyte Contributions to Heart Disease. Cell Stem Cell 2020, 26, 862–879.e811. [Google Scholar] [CrossRef]
- Abilez, O.J.; Tzatzalos, E.; Yang, H.; Zhao, M.T.; Jung, G.; Zollner, A.M.; Tiburcy, M.; Riegler, J.; Matsa, E.; Shukla, P.; et al. Passive Stretch Induces Structural and Functional Maturation of Engineered Heart Muscle as Predicted by Computational Modeling. Stem Cells 2018, 36, 265–277. [Google Scholar] [CrossRef] [Green Version]
- Leonard, A.; Bertero, A.; Powers, J.D.; Beussman, K.M.; Bhandari, S.; Regnier, M.; Murry, C.E.; Sniadecki, N.J. Afterload promotes maturation of human induced pluripotent stem cell derived cardiomyocytes in engineered heart tissues. J. Mol. Cell Cardiol. 2018, 118, 147–158. [Google Scholar] [CrossRef]
- Ronaldson-Bouchard, K.; Ma, S.P.; Yeager, K.; Chen, T.; Song, L.; Sirabella, D.; Morikawa, K.; Teles, D.; Yazawa, M.; Vunjak-Novakovic, G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 2018, 556, 239–243. [Google Scholar] [CrossRef]
- Nunes, S.S.; Miklas, J.W.; Liu, J.; Aschar-Sobbi, R.; Xiao, Y.; Zhang, B.; Jiang, J.; Masse, S.; Gagliardi, M.; Hsieh, A.; et al. Biowire: A platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat. Methods 2013, 10, 781–787. [Google Scholar] [CrossRef] [Green Version]
- Eng, G.; Lee, B.W.; Protas, L.; Gagliardi, M.; Brown, K.; Kass, R.S.; Keller, G.; Robinson, R.B.; Vunjak-Novakovic, G. Autonomous beating rate adaptation in human stem cell-derived cardiomyocytes. Nat. Commun. 2016, 7, 10312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Rodriguez, M.; Pabon, L.; Fischer, K.A.; Reinecke, H.; Regnier, M.; Sniadecki, N.J.; Ruohola-Baker, H.; Murry, C.E. Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. J. Mol. Cell. Cardiol. 2014, 72, 296–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, M.C.; Tertoolen, L.G.; Guadix, J.A.; Bellin, M.; Kosmidis, G.; D’Aniello, C.; Monshouwer-Kloots, J.; Goumans, M.J.; Wang, Y.L.; Feinberg, A.W.; et al. Functional maturation of human pluripotent stem cell derived cardiomyocytes in vitro—Correlation between contraction force and electrophysiology. Biomaterials 2015, 51, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Lieu, D.K.; Fu, J.D.; Chiamvimonvat, N.; Tung, K.C.; McNerney, G.P.; Huser, T.; Keller, G.; Kong, C.W.; Li, R.A. Mechanism-based facilitated maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Arrhythm. Electrophysiol. 2013, 6, 191–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Lieu, D.K.; Siu, C.W.; Fu, J.D.; Tse, H.F.; Li, R.A. Facilitated maturation of Ca2+ handling properties of human embryonic stem cell-derived cardiomyocytes by calsequestrin expression. Am. J. Physiol. Cell Physiol. 2009, 297, C152–C159. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.S.; Chen, J.H.; Lundy, D.J.; Liu, C.H.; Hwang, S.M.; Pabon, L.; Shieh, R.C.; Chen, C.C.; Wu, S.N.; Yan, Y.T.; et al. Defined MicroRNAs Induce Aspects of Maturation in Mouse and Human Embryonic-Stem-Cell-Derived Cardiomyocytes. Cell Rep. 2015, 12, 1960–1967. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.Y.; Peres Moreno Maia-Joca, R.; Ong, C.S.; Wilson, I.; DiSilvestre, D.; Tomaselli, G.F.; Reich, D.H. Enhancement of human iPSC-derived cardiomyocyte maturation by chemical conditioning in a 3D environment. J. Mol. Cell. Cardiol. 2020, 138, 1–11. [Google Scholar] [CrossRef]
- Skorska, A.; Johann, L.; Chabanovska, O.; Vasudevan, P.; Kussauer, S.; Hillemanns, M.; Wolfien, M.; Jonitz-Heincke, A.; Wolkenhauer, O.; Bader, R.; et al. Monitoring the maturation of the sarcomere network: A super-resolution microscopy-based approach. Cell. Mol. Life Sci. 2022, 79, 149. [Google Scholar] [CrossRef]
- Feyen, D.A.M.; McKeithan, W.L.; Bruyneel, A.A.N.; Spiering, S.; Hormann, L.; Ulmer, B.; Zhang, H.; Briganti, F.; Schweizer, M.; Hegyi, B.; et al. Metabolic Maturation Media Improve Physiological Function of Human iPSC-Derived Cardiomyocytes. Cell Rep. 2020, 32, 107925. [Google Scholar] [CrossRef]
- Seguret, M.; Vermersch, E.; Jouve, C.; Hulot, J.S. Cardiac Organoids to Model and Heal Heart Failure and Cardiomyopathies. Biomedicines 2021, 9, 563. [Google Scholar] [CrossRef]
- Visone, R.; Talo, G.; Lopa, S.; Rasponi, M.; Moretti, M. Enhancing all-in-one bioreactors by combining interstitial perfusion, electrical stimulation, on-line monitoring and testing within a single chamber for cardiac constructs. Sci. Rep. 2018, 8, 16944. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.L.; Tulloch, N.L.; Razumova, M.V.; Saiget, M.; Muskheli, V.; Pabon, L.; Reinecke, H.; Regnier, M.; Murry, C.E. Mechanical Stress Conditioning and Electrical Stimulation Promote Contractility and Force Maturation of Induced Pluripotent Stem Cell-Derived Human Cardiac Tissue. Circulation 2016, 134, 1557–1567. [Google Scholar] [CrossRef] [PubMed]
- LaBarge, W.; Mattappally, S.; Kannappan, R.; Fast, V.G.; Pretorius, D.; Berry, J.L.; Zhang, J. Maturation of three-dimensional, hiPSC-derived cardiomyocyte spheroids utilizing cyclic, uniaxial stretch and electrical stimulation. PLoS ONE 2019, 14, e0219442. [Google Scholar] [CrossRef]
- de Lange, W.J.; Farrell, E.T.; Kreitzer, C.R.; Jacobs, D.R.; Lang, D.; Glukhov, A.V.; Ralphe, J.C. Human iPSC-engineered cardiac tissue platform faithfully models important cardiac physiology. Am. J. Physiol. Heart Circ. Physiol. 2021, 320, H1670–H1686. [Google Scholar] [CrossRef] [PubMed]
- Funakoshi, S.; Fernandes, I.; Mastikhina, O.; Wilkinson, D.; Tran, T.; Dhahri, W.; Mazine, A.; Yang, D.; Burnett, B.; Lee, J.; et al. Generation of mature compact ventricular cardiomyocytes from human pluripotent stem cells. Nat. Commun. 2021, 12, 3155. [Google Scholar] [CrossRef]
- Yuan, Q.; Maas, R.G.C.; Brouwer, E.C.J.; Pei, J.; Blok, C.S.; Popovic, M.A.; Paauw, N.J.; Bovenschen, N.; Hjortnaes, J.; Harakalova, M.; et al. Sarcomere Disassembly and Transfection Efficiency in Proliferating Human iPSC-Derived Cardiomyocytes. J. Cardiovasc. Dev. Dis. 2022, 9, 43. [Google Scholar] [CrossRef]
- Murrow, L.M.; Weber, R.J.; Gartner, Z.J. Dissecting the stem cell niche with organoid models: An engineering-based approach. Development 2017, 144, 998–1007. [Google Scholar] [CrossRef] [Green Version]
- Smith, E.; Cochrane, W. Cystic organoid teratoma:(report of a case). Can. Med. Assoc. J. 1946, 55, 151. [Google Scholar]
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef] [Green Version]
- Sasai, Y. Cytosystems dynamics in self-organization of tissue architecture. Nature 2013, 493, 318–326. [Google Scholar] [CrossRef]
- Sasai, Y. Next-generation regenerative medicine: Organogenesis from stem cells in 3D culture. Cell Stem Cell 2013, 12, 520–530. [Google Scholar] [CrossRef] [Green Version]
- Woodford, C.; Zandstra, P.W. Tissue engineering 2.0: Guiding self-organization during pluripotent stem cell differentiation. Curr. Opin. Biotechnol. 2012, 23, 810–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Jiao, Y.; Qin, S.; Zhao, W.; Chu, Q.; Wu, K. Organoid technology in disease modelling, drug development, personalized treatment and regeneration medicine. Exp. Hematol. Oncol. 2018, 7, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef] [PubMed]
- Forsythe, S.D.; Devarasetty, M.; Shupe, T.; Bishop, C.; Atala, A.; Soker, S.; Skardal, A. Environmental Toxin Screening Using Human-Derived 3D Bioengineered Liver and Cardiac Organoids. Front. Public Health 2018, 6, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nugraha, B.; Buono, M.F.; von Boehmer, L.; Hoerstrup, S.P.; Emmert, M.Y. Human Cardiac Organoids for Disease Modeling. Clin. Pharmacol. Ther. 2019, 105, 79–85. [Google Scholar] [CrossRef] [Green Version]
- Eiraku, M.; Watanabe, K.; Matsuo-Takasaki, M.; Kawada, M.; Yonemura, S.; Matsumura, M.; Wataya, T.; Nishiyama, A.; Muguruma, K.; Sasai, Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 2008, 3, 519–532. [Google Scholar] [CrossRef] [Green Version]
- Eiraku, M.; Takata, N.; Ishibashi, H.; Kawada, M.; Sakakura, E.; Okuda, S.; Sekiguchi, K.; Adachi, T.; Sasai, Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011, 472, 51–56. [Google Scholar] [CrossRef]
- Nakano, T.; Ando, S.; Takata, N.; Kawada, M.; Muguruma, K.; Sekiguchi, K.; Saito, K.; Yonemura, S.; Eiraku, M.; Sasai, Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 2012, 10, 771–785. [Google Scholar] [CrossRef] [Green Version]
- Miller, A.J.; Dye, B.R.; Ferrer-Torres, D.; Hill, D.R.; Overeem, A.W.; Shea, L.D.; Spence, J.R. Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc. 2019, 14, 518–540. [Google Scholar] [CrossRef]
- Zeng, Z.; Huang, B.; Parvez, R.K.; Li, Y.; Chen, J.; Vonk, A.C.; Thornton, M.E.; Patel, T.; Rutledge, E.A.; Kim, A.D.; et al. Generation of patterned kidney organoids that recapitulate the adult kidney collecting duct system from expandable ureteric bud progenitors. Nat. Commun. 2021, 12, 3641. [Google Scholar] [CrossRef] [PubMed]
- Takebe, T.; Sekine, K.; Enomura, M.; Koike, H.; Kimura, M.; Ogaeri, T.; Zhang, R.R.; Ueno, Y.; Zheng, Y.W.; Koike, N.; et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 2013, 499, 481–484. [Google Scholar] [CrossRef] [PubMed]
- Takebe, T.; Zhang, R.R.; Koike, H.; Kimura, M.; Yoshizawa, E.; Enomura, M.; Koike, N.; Sekine, K.; Taniguchi, H. Generation of a vascularized and functional human liver from an iPSC-derived organ bud transplant. Nat. Protoc. 2014, 9, 396–409. [Google Scholar] [CrossRef] [PubMed]
- Lukonin, I.; Serra, D.; Challet Meylan, L.; Volkmann, K.; Baaten, J.; Zhao, R.; Meeusen, S.; Colman, K.; Maurer, F.; Stadler, M.B.; et al. Phenotypic landscape of intestinal organoid regeneration. Nature 2020, 586, 275–280. [Google Scholar] [CrossRef]
- Watanabe, S.; Kobayashi, S.; Ogasawara, N.; Okamoto, R.; Nakamura, T.; Watanabe, M.; Jensen, K.B.; Yui, S. Transplantation of intestinal organoids into a mouse model of colitis. Nat. Protoc. 2022, 17, 649–671. [Google Scholar] [CrossRef]
- McCauley, H.A.; Wells, J.M. Pluripotent stem cell-derived organoids: Using principles of developmental biology to grow human tissues in a dish. Development 2017, 144, 958–962. [Google Scholar] [CrossRef] [Green Version]
- McCracken, K.W.; Cata, E.M.; Crawford, C.M.; Sinagoga, K.L.; Schumacher, M.; Rockich, B.E.; Tsai, Y.H.; Mayhew, C.N.; Spence, J.R.; Zavros, Y.; et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014, 516, 400–404. [Google Scholar] [CrossRef] [Green Version]
- Richards, D.J.; Coyle, R.C.; Tan, Y.; Jia, J.; Wong, K.; Toomer, K.; Menick, D.R.; Mei, Y. Inspiration from heart development: Biomimetic development of functional human cardiac organoids. Biomaterials 2017, 142, 112–123. [Google Scholar] [CrossRef]
- Zhao, D.; Lei, W.; Hu, S. Cardiac organoid—A promising perspective of preclinical model. Stem Cell Res. Ther. 2021, 12, 272. [Google Scholar] [CrossRef]
- Hulot, J.S. Modeling Cardiac Arrhythmias With Organoids. J. Am. Coll. Cardiol. 2019, 73, 2325–2327. [Google Scholar] [CrossRef]
- Polonchuk, L.; Chabria, M.; Badi, L.; Hoflack, J.C.; Figtree, G.; Davies, M.J.; Gentile, C. Cardiac spheroids as promising in vitro models to study the human heart microenvironment. Sci. Rep. 2017, 7, 7005. [Google Scholar] [CrossRef] [PubMed]
- Brassard, J.A.; Lutolf, M.P. Engineering Stem Cell Self-organization to Build Better Organoids. Cell Stem Cell 2019, 24, 860–876. [Google Scholar] [CrossRef] [PubMed]
- Dang, S.M.; Kyba, M.; Perlingeiro, R.; Daley, G.Q.; Zandstra, P.W. Efficiency of embryoid body formation and hematopoietic development from embryonic stem cells in different culture systems. Biotechnol. Bioeng. 2002, 78, 442–453. [Google Scholar] [CrossRef]
- Shkumatov, A.; Baek, K.; Kong, H. Matrix rigidity-modulated cardiovascular organoid formation from embryoid bodies. PLoS ONE 2014, 9, e94764. [Google Scholar] [CrossRef] [PubMed]
- Lewis-Israeli, Y.R.; Wasserman, A.H.; Gabalski, M.A.; Volmert, B.D.; Ming, Y.; Ball, K.A.; Yang, W.; Zou, J.; Ni, G.; Pajares, N.; et al. Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nat. Commun. 2021, 12, 5142. [Google Scholar] [CrossRef]
- Andersen, P.; Tampakakis, E.; Jimenez, D.V.; Kannan, S.; Miyamoto, M.; Shin, H.K.; Saberi, A.; Murphy, S.; Sulistio, E.; Chelko, S.P.; et al. Precardiac organoids form two heart fields via Bmp/Wnt signaling. Nat. Commun. 2018, 9, 3140. [Google Scholar] [CrossRef]
- Hofbauer, P.; Jahnel, S.M.; Papai, N.; Giesshammer, M.; Deyett, A.; Schmidt, C.; Penc, M.; Tavernini, K.; Grdseloff, N.; Meledeth, C.; et al. Cardioids reveal self-organizing principles of human cardiogenesis. Cell 2021, 184, 3299–3317.e22. [Google Scholar] [CrossRef]
- Varzideh, F.; Pahlavan, S.; Ansari, H.; Halvaei, M.; Kostin, S.; Feiz, M.S.; Latifi, H.; Aghdami, N.; Braun, T.; Baharvand, H. Human cardiomyocytes undergo enhanced maturation in embryonic stem cell-derived organoid transplants. Biomaterials 2019, 192, 537–550. [Google Scholar] [CrossRef]
- Kerr, C.M.; Richards, D.; Menick, D.R.; Deleon-Pennell, K.Y.; Mei, Y. Multicellular Human Cardiac Organoids Transcriptomically Model Distinct Tissue-Level Features of Adult Myocardium. Int. J. Mol. Sci. 2021, 22, 8482. [Google Scholar] [CrossRef]
- Hubert, C.G.; Rivera, M.; Spangler, L.C.; Wu, Q.; Mack, S.C.; Prager, B.C.; Couce, M.; McLendon, R.E.; Sloan, A.E.; Rich, J.N. A Three-Dimensional Organoid Culture System Derived from Human Glioblastomas Recapitulates the Hypoxic Gradients and Cancer Stem Cell Heterogeneity of Tumors Found In Vivo. Cancer Res. 2016, 76, 2465–2477. [Google Scholar] [CrossRef] [Green Version]
- Daviaud, N.; Chevalier, C.; Friedel, R.H.; Zou, H. Distinct Vulnerability and Resilience of Human Neuroprogenitor Subtypes in Cerebral Organoid Model of Prenatal Hypoxic Injury. Front. Cell. Neurosci. 2019, 13, 336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, J.; Wen, S.; Cao, W.; Yue, P.; Xu, X.; Zhang, Y.; Luo, L.; Chen, T.; Li, L.; Wang, F.; et al. Lung organoids, useful tools for investigating epithelial repair after lung injury. Stem Cell Res. Ther. 2021, 12, 95. [Google Scholar] [CrossRef]
- Zhao, X.; Xu, Z.; Xiao, L.; Shi, T.; Xiao, H.; Wang, Y.; Li, Y.; Xue, F.; Zeng, W. Review on the Vascularization of Organoids and Organoids-on-a-Chip. Front. Bioeng. Biotechnol. 2021, 9, 637048. [Google Scholar] [CrossRef] [PubMed]
- Yu, J. Vascularized Organoids: A More Complete Model. Int. J. Stem Cells 2021, 14, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.P.; Toksoy, Z.; Davis, B.A.; Geibel, J.P. 3D Bioprinting of Vascularized Tissues for in vitro and in vivo Applications. Front. Bioeng. Biotechnol. 2021, 9, 664188. [Google Scholar] [CrossRef]
- Narmoneva, D.A.; Vukmirovic, R.; Davis, M.E.; Kamm, R.D.; Lee, R.T. Endothelial cells promote cardiac myocyte survival and spatial reorganization: Implications for cardiac regeneration. Circulation 2004, 110, 962–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brutsaert, D.L. Cardiac endothelial-myocardial signaling: Its role in cardiac growth, contractile performance, and rhythmicity. Physiol. Rev. 2003, 83, 59–115. [Google Scholar] [CrossRef]
- Nolan, D.J.; Ginsberg, M.; Israely, E.; Palikuqi, B.; Poulos, M.G.; James, D.; Ding, B.S.; Schachterle, W.; Liu, Y.; Rosenwaks, Z.; et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev. Cell 2013, 26, 204–219. [Google Scholar] [CrossRef] [Green Version]
- Saini, H.; Navaei, A.; Van Putten, A.; Nikkhah, M. 3D cardiac microtissues encapsulated with the co-culture of cardiomyocytes and cardiac fibroblasts. Adv. Healthc. Mater. 2015, 4, 1961–1971. [Google Scholar] [CrossRef]
- Roberts, M.A.; Tran, D.; Coulombe, K.L.; Razumova, M.; Regnier, M.; Murry, C.E.; Zheng, Y. Stromal Cells in Dense Collagen Promote Cardiomyocyte and Microvascular Patterning in Engineered Human Heart Tissue. Tissue Eng. Part A 2016, 22, 633–644. [Google Scholar] [CrossRef]
- Peters, E.B. Endothelial Progenitor Cells for the Vascularization of Engineered Tissues. Tissue Eng. Part B Rev. 2018, 24, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.M.; Liu, X. Advancing biomaterials of human origin for tissue engineering. Prog. Polym. Sci. 2016, 53, 86–168. [Google Scholar] [CrossRef] [Green Version]
- Song, M.H.; Choi, S.C.; Noh, J.M.; Joo, H.J.; Park, C.Y.; Cha, J.J.; Ahn, T.H.; Ko, T.H.; Choi, J.I.; Na, J.E.; et al. LEFTY-PITX2 signaling pathway is critical for generation of mature and ventricular cardiac organoids in human pluripotent stem cell-derived cardiac mesoderm cells. Biomaterials 2021, 278, 121133. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Velasco, E.; Garcia-Padilla, C.; Del Mar Munoz-Gallardo, M.; Martinez-Amaro, F.J.; Cano-Carrillo, S.; Castillo-Casas, J.M.; Sanchez-Fernandez, C.; Aranega, A.E.; Franco, D. Post-Transcriptional Regulation of Molecular Determinants during Cardiogenesis. Int. J. Mol. Sci. 2022, 23, 2839. [Google Scholar] [CrossRef] [PubMed]
- Kitsuka, T.; Itoh, M.; Amamoto, S.; Arai, K.I.; Oyama, J.; Node, K.; Toda, S.; Morita, S.; Nishida, T.; Nakayama, K. 2-Cl-C.OXT-A stimulates contraction through the suppression of phosphodiesterase activity in human induced pluripotent stem cell-derived cardiac organoids. PLoS ONE 2019, 14, e0213114. [Google Scholar] [CrossRef] [Green Version]
- Mills, R.J.; Titmarsh, D.M.; Koenig, X.; Parker, B.L.; Ryall, J.G.; Quaife-Ryan, G.A.; Voges, H.K.; Hodson, M.P.; Ferguson, C.; Drowley, L.; et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. USA 2017, 114, E8372–E8381. [Google Scholar] [CrossRef] [Green Version]
- Mills, R.J.; Parker, B.L.; Quaife-Ryan, G.A.; Voges, H.K.; Needham, E.J.; Bornot, A.; Ding, M.; Andersson, H.; Polla, M.; Elliott, D.A.; et al. Drug Screening in Human PSC-Cardiac Organoids Identifies Pro-proliferative Compounds Acting via the Mevalonate Pathway. Cell Stem Cell 2019, 24, 895–907.e6. [Google Scholar] [CrossRef]
- Silva, A.C.; Matthys, O.B.; Joy, D.A.; Kauss, M.A.; Natarajan, V.; Lai, M.H.; Turaga, D.; Blair, A.P.; Alexanian, M.; Bruneau, B.G.; et al. Co-emergence of cardiac and gut tissues promotes cardiomyocyte maturation within human iPSC-derived organoids. Cell Stem Cell 2021, 28, 2137–2152.e6. [Google Scholar] [CrossRef]
- Kozaniti, F.K.; Metsiou, D.N.; Manara, A.E.; Athanassiou, G.; Deligianni, D.D. Recent Advancements in 3D Printing and Bioprinting Methods for Cardiovascular Tissue Engineering. Bioengineering 2021, 8, 133. [Google Scholar] [CrossRef]
- Hu, J.B.; Tomov, M.L.; Buikema, J.W.; Chen, C.; Mahmoudi, M.; Wu, S.M.; Serpooshan, V. Cardiovascular tissue bioprinting: Physical and chemical processes. Appl. Phys. Rev. 2018, 5, 041106. [Google Scholar] [CrossRef]
- Cui, H.; Miao, S.; Esworthy, T.; Zhou, X.; Lee, S.J.; Liu, C.; Yu, Z.X.; Fisher, J.P.; Mohiuddin, M.; Zhang, L.G. 3D bioprinting for cardiovascular regeneration and pharmacology. Adv. Drug Deliv. Rev. 2018, 132, 252–269. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, Y.; Akagi, T.; Akashi, M. Vascularized cardiac tissue construction with orientation by layer-by-layer method and 3D printer. Sci. Rep. 2020, 10, 5484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puluca, N.; Lee, S.; Doppler, S.; Munsterer, A.; Dressen, M.; Krane, M.; Wu, S.M. Bioprinting Approaches to Engineering Vascularized 3D Cardiac Tissues. Curr. Cardiol. Rep. 2019, 21, 90. [Google Scholar] [CrossRef] [PubMed]
- Hatami, J.; Silva, S.G.; Oliveira, M.B.; Costa, R.R.; Reis, R.L.; Mano, J.F. Multilayered Films Produced by Layer-by-Layer Assembly of Chitosan and Alginate as a Potential Platform for the Formation of Human Adipose-Derived Stem Cell aggregates. Polymers 2017, 9, 440. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhang, J. Layer-By-Layer Fabrication of Thicker and Larger Human Cardiac Muscle Patches for Cardiac Repair in Mice. Front. Cardiovasc. Med. 2021, 8, 800667. [Google Scholar] [CrossRef]
- Al-Jubori, A.A.; Sulaiman, G.M.; Tawfeeq, A.T.; Mohammed, H.A.; Khan, R.A.; Mohammed, S.A.A. Layer-by-Layer Nanoparticles of Tamoxifen and Resveratrol for Dual Drug Delivery System and Potential Triple-Negative Breast Cancer Treatment. Pharmaceutics 2021, 13, 1098. [Google Scholar] [CrossRef]
- Takeda, M.; Miyagawa, S.; Akashi, M.; Sawa, Y. Construction of Three-Dimensional Cardiac Tissues Using Layer-by-Layer Method. Methods Mol. Biol. 2021, 2320, 75–79. [Google Scholar] [CrossRef]
- Amano, Y.; Nishiguchi, A.; Matsusaki, M.; Iseoka, H.; Miyagawa, S.; Sawa, Y.; Seo, M.; Yamaguchi, T.; Akashi, M. Development of vascularized iPSC derived 3D-cardiomyocyte tissues by filtration Layer-by-Layer technique and their application for pharmaceutical assays. Acta Biomater. 2016, 33, 110–121. [Google Scholar] [CrossRef]
- Tadano, K.; Miyagawa, S.; Takeda, M.; Tsukamoto, Y.; Kazusa, K.; Takamatsu, K.; Akashi, M.; Sawa, Y. Cardiotoxicity assessment using 3D vascularized cardiac tissue consisting of human iPSC-derived cardiomyocytes and fibroblasts. Mol. Ther. Methods Clin. Dev. 2021, 22, 338–349. [Google Scholar] [CrossRef]
- Schaefer, J.A.; Guzman, P.A.; Riemenschneider, S.B.; Kamp, T.J.; Tranquillo, R.T. A cardiac patch from aligned microvessel and cardiomyocyte patches. J. Tissue Eng. Regen. Med. 2018, 12, 546–556. [Google Scholar] [CrossRef]
- Ong, C.S.; Fukunishi, T.; Zhang, H.; Huang, C.Y.; Nashed, A.; Blazeski, A.; DiSilvestre, D.; Vricella, L.; Conte, J.; Tung, L.; et al. Biomaterial-Free Three-Dimensional Bioprinting of Cardiac Tissue using Human Induced Pluripotent Stem Cell Derived Cardiomyocytes. Sci. Rep. 2017, 7, 4566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.S.; Arneri, A.; Bersini, S.; Shin, S.R.; Zhu, K.; Goli-Malekabadi, Z.; Aleman, J.; Colosi, C.; Busignani, F.; Dell’Erba, V.; et al. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 2016, 110, 45–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaicharoenaudomrung, N.; Kunhorm, P.; Noisa, P. Three-dimensional cell culture systems as an in vitro platform for cancer and stem cell modeling. World J. Stem Cells 2019, 11, 1065–1083. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.; Cejas, C.M.; Pitingolo, G. Advances in microfluidic 3D cell culture for preclinical drug development. Prog. Mol. Biol. Transl. Sci. 2022, 187, 163–204. [Google Scholar] [CrossRef] [PubMed]
- Tibbitt, M.W.; Anseth, K.S. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 2009, 103, 655–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Lee, C.; Skylar-Scott, M.A.; Heilshorn, S.C.; Wu, J.C. Reconstructing the heart using iPSCs: Engineering strategies and applications. J. Mol. Cell. Cardiol. 2021, 157, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Doherty, E.L.; Aw, W.Y.; Hickey, A.J.; Polacheck, W.J. Microfluidic and Organ-on-a-Chip Approaches to Investigate Cellular and Microenvironmental Contributions to Cardiovascular Function and Pathology. Front. Bioeng. Biotechnol. 2021, 9, 624435. [Google Scholar] [CrossRef]
- Vollert, I.; Seiffert, M.; Bachmair, J.; Sander, M.; Eder, A.; Conradi, L.; Vogelsang, A.; Schulze, T.; Uebeler, J.; Holnthoner, W.; et al. In vitro perfusion of engineered heart tissue through endothelialized channels. Tissue Eng. Part A 2014, 20, 854–863. [Google Scholar] [CrossRef]
- Schneider, O.; Zeifang, L.; Fuchs, S.; Sailer, C.; Loskill, P. User-Friendly and Parallelized Generation of Human Induced Pluripotent Stem Cell-Derived Microtissues in a Centrifugal Heart-on-a-Chip. Tissue Eng. Part A 2019, 25, 786–798. [Google Scholar] [CrossRef] [Green Version]
- Mousavi Shaegh, S.A.; De Ferrari, F.; Zhang, Y.S.; Nabavinia, M.; Binth Mohammad, N.; Ryan, J.; Pourmand, A.; Laukaitis, E.; Banan Sadeghian, R.; Nadhman, A.; et al. A microfluidic optical platform for real-time monitoring of pH and oxygen in microfluidic bioreactors and organ-on-chip devices. Biomicrofluidics 2016, 10, 044111. [Google Scholar] [CrossRef] [PubMed]
- Mercader, A.; Ye, S.H.; Kim, S.; Orizondo, R.A.; Cho, S.K.; Wagner, W.R. PDMS-Zwitterionic Hybrid for Facile, Antifouling Microfluidic Device Fabrication. Langmuir 2022, 38, 3775–3784. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Lee, J.H.; Jeong, O.C. Pneumatically Driven Microfluidic Platform for Micro-Particle Concentration. J. Vis. Exp. 2022, 180. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Lim, Y.J.; Zheng, Y.; Jung, M.; Gaus, K.; Gardiner, E.E.; Lee, W.M. Modified inverted selective plane illumination microscopy for sub-micrometer imaging resolution in polydimethylsiloxane soft lithography devices. Lab Chip 2020, 20, 3960–3969. [Google Scholar] [CrossRef]
- Lai, B.F.L.; Lu, R.X.Z.; Davenport Huyer, L.; Kakinoki, S.; Yazbeck, J.; Wang, E.Y.; Wu, Q.; Zhang, B.; Radisic, M. A well plate-based multiplexed platform for incorporation of organoids into an organ-on-a-chip system with a perfusable vasculature. Nat. Protoc. 2021, 16, 2158–2189. [Google Scholar] [CrossRef]
- Marsano, A.; Conficconi, C.; Lemme, M.; Occhetta, P.; Gaudiello, E.; Votta, E.; Cerino, G.; Redaelli, A.; Rasponi, M. Beating heart on a chip: A novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip 2016, 16, 599–610. [Google Scholar] [CrossRef]
- Zhao, Y.; Rafatian, N.; Wang, E.Y.; Feric, N.T.; Lai, B.F.L.; Knee-Walden, E.J.; Backx, P.H.; Radisic, M. Engineering microenvironment for human cardiac tissue assembly in heart-on-a-chip platform. Matrix Biol. 2020, 85–86, 189–204. [Google Scholar] [CrossRef]
- Sakai, K.; Shimba, K.; Ishizuka, K.; Yang, Z.; Oiwa, K.; Takeuchi, A.; Kotani, K.; Jimbo, Y. Functional innervation of human induced pluripotent stem cell-derived cardiomyocytes by co-culture with sympathetic neurons developed using a microtunnel technique. Biochem. Biophys. Res. Commun. 2017, 494, 138–143. [Google Scholar] [CrossRef]
- Carnevale, D. Neuroimmune axis of cardiovascular control: Mechanisms and therapeutic implications. Nat. Rev. Cardiol. 2022. [Google Scholar] [CrossRef]
- Ellis, B.W.; Acun, A.; Can, U.I.; Zorlutuna, P. Human iPSC-derived myocardium-on-chip with capillary-like flow for personalized medicine. Biomicrofluidics 2017, 11, 024105. [Google Scholar] [CrossRef] [Green Version]
- Mathur, A.; Loskill, P.; Shao, K.; Huebsch, N.; Hong, S.; Marcus, S.G.; Marks, N.; Mandegar, M.; Conklin, B.R.; Lee, L.P.; et al. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci. Rep. 2015, 5, 8883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Rafatian, N.; Feric, N.T.; Cox, B.J.; Aschar-Sobbi, R.; Wang, E.Y.; Aggarwal, P.; Zhang, B.; Conant, G.; Ronaldson-Bouchard, K.; et al. A Platform for Generation of Chamber-Specific Cardiac Tissues and Disease Modeling. Cell 2019, 176, 913–927.e18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajan, S.A.P.; Aleman, J.; Wan, M.; Pourhabibi Zarandi, N.; Nzou, G.; Murphy, S.; Bishop, C.E.; Sadri-Ardekani, H.; Shupe, T.; Atala, A.; et al. Probing prodrug metabolism and reciprocal toxicity with an integrated and humanized multi-tissue organ-on-a-chip platform. Acta Biomater. 2020, 106, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Oleaga, C.; Bernabini, C.; Smith, A.S.; Srinivasan, B.; Jackson, M.; McLamb, W.; Platt, V.; Bridges, R.; Cai, Y.; Santhanam, N.; et al. Multi-Organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci. Rep. 2016, 6, 20030. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Peng, Y.; Li, H.; Chen, W. Organ-on-a-Chip: A New Paradigm for Drug Development. Trends Pharmacol. Sci. 2021, 42, 119–133. [Google Scholar] [CrossRef]
- Low, L.A.; Sutherland, M.; Lumelsky, N.; Selimovic, S.; Lundberg, M.S.; Tagle, D.A. Organs-on-a-Chip. Adv. Exp. Med. Biol. 2020, 1230, 27–42. [Google Scholar] [CrossRef]
- Young, R.E.; Huh, D.D. Organ-on-a-chip technology for the study of the female reproductive system. Adv. Drug Deliv. Rev. 2021, 173, 461–478. [Google Scholar] [CrossRef]
- Jalili-Firoozinezhad, S.; Miranda, C.C.; Cabral, J.M.S. Modeling the Human Body on Microfluidic Chips. Trends Biotechnol. 2021, 39, 838–852. [Google Scholar] [CrossRef]
- Jodat, Y.A.; Kang, M.G.; Kiaee, K.; Kim, G.J.; Martinez, A.F.H.; Rosenkranz, A.; Bae, H.; Shin, S.R. Human-Derived Organ-on-a-Chip for Personalized Drug Development. Curr. Pharm. Des. 2018, 24, 5471–5486. [Google Scholar] [CrossRef]
- Ajalik, R.E.; Alenchery, R.G.; Cognetti, J.S.; Zhang, V.Z.; McGrath, J.L.; Miller, B.L.; Awad, H.A. Human Organ-on-a-Chip Microphysiological Systems to Model Musculoskeletal Pathologies and Accelerate Therapeutic Discovery. Front. Bioeng. Biotechnol. 2022, 10, 846230. [Google Scholar] [CrossRef]
- Ingber, D.E. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat. Rev. Genet. 2022. [Google Scholar] [CrossRef] [PubMed]
- Parrotta, E.I.; Lucchino, V.; Scaramuzzino, L.; Scalise, S.; Cuda, G. Modeling Cardiac Disease Mechanisms Using Induced Pluripotent Stem Cell-Derived Cardiomyocytes: Progress, Promises and Challenges. Int. J. Mol. Sci. 2020, 21, 4354. [Google Scholar] [CrossRef] [PubMed]
- Hinson, J.T.; Chopra, A.; Nafissi, N.; Polacheck, W.J.; Benson, C.C.; Swist, S.; Gorham, J.; Yang, L.; Schafer, S.; Sheng, C.C.; et al. HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science 2015, 349, 982–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stillitano, F.; Turnbull, I.C.; Karakikes, I.; Nonnenmacher, M.; Backeris, P.; Hulot, J.S.; Kranias, E.G.; Hajjar, R.J.; Costa, K.D. Genomic correction of familial cardiomyopathy in human engineered cardiac tissues. Eur. Heart J. 2016, 37, 3282–3284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prondzynski, M.; Lemoine, M.D.; Zech, A.T.; Horvath, A.; Di Mauro, V.; Koivumaki, J.T.; Kresin, N.; Busch, J.; Krause, T.; Kramer, E.; et al. Disease modeling of a mutation in alpha-actinin 2 guides clinical therapy in hypertrophic cardiomyopathy. EMBO Mol. Med. 2019, 11, e11115. [Google Scholar] [CrossRef]
- Richards, D.J.; Li, Y.; Kerr, C.M.; Yao, J.; Beeson, G.C.; Coyle, R.C.; Chen, X.; Jia, J.; Damon, B.; Wilson, R.; et al. Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nat. Biomed. Eng. 2020, 4, 446–462. [Google Scholar] [CrossRef]
- Orita, K.; Sawada, K.; Matsumoto, N.; Ikegaya, Y. Machine-learning-based quality control of contractility of cultured human-induced pluripotent stem-cell-derived cardiomyocytes. Biochem. Biophys. Res. Commun. 2020, 526, 751–755. [Google Scholar] [CrossRef]
- Orita, K.; Sawada, K.; Koyama, R.; Ikegaya, Y. Deep learning-based quality control of cultured human-induced pluripotent stem cell-derived cardiomyocytes. J. Pharmacol. Sci. 2019, 140, 313–316. [Google Scholar] [CrossRef]
- Coronnello, C.; Francipane, M.G. Moving Towards Induced Pluripotent Stem Cell-based Therapies with Artificial Intelligence and Machine Learning. Stem Cell Rev. Rep. 2022, 18, 559–569. [Google Scholar] [CrossRef]
- Santulli, G.; Lewis, D.R.; Marks, A.R. Physiology and pathophysiology of excitation-contraction coupling: The functional role of ryanodine receptor. J. Muscle Res. Cell Motil. 2017, 38, 37–45. [Google Scholar] [CrossRef] [Green Version]
- Juhola, M.; Penttinen, K.; Joutsijoki, H.; Varpa, K.; Saarikoski, J.; Rasku, J.; Siirtola, H.; Iltanen, K.; Laurikkala, J.; Hyyro, H.; et al. Signal analysis and classification methods for the calcium transient data of stem cell-derived cardiomyocytes. Comput. Biol. Med. 2015, 61, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Juhola, M.; Joutsijoki, H.; Penttinen, K.; Aalto-Setala, K. Detection of genetic cardiac diseases by Ca(2+) transient profiles using machine learning methods. Sci. Rep. 2018, 8, 9355. [Google Scholar] [CrossRef] [PubMed]
- Joutsijoki, H.; Penttinen, K.; Juhola, M.; Aalto-Setala, K. Separation of HCM and LQT Cardiac Diseases with Machine Learning of Ca2+ Transient Profiles. Methods Inf. Med. 2019, 58, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Joutsijoki, H.; Haponen, M.; Rasku, J.; Aalto-Setala, K.; Juhola, M. Machine Learning Approach to Automated Quality Identification of Human Induced Pluripotent Stem Cell Colony Images. Comput. Math Methods Med. 2016, 2016, 3091039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juhola, M.; Penttinen, K.; Joutsijoki, H.; Aalto-Setala, K. Analysis of Drug Effects on iPSC Cardiomyocytes with Machine Learning. Ann. Biomed. Eng. 2021, 49, 129–138. [Google Scholar] [CrossRef]
- Aghasafari, P.; Yang, P.C.; Kernik, D.C.; Sakamoto, K.; Kanda, Y.; Kurokawa, J.; Vorobyov, I.; Clancy, C.E. A deep learning algorithm to translate and classify cardiac electrophysiology. eLife 2021, 10, e68335. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varzideh, F.; Mone, P.; Santulli, G. Bioengineering Strategies to Create 3D Cardiac Constructs from Human Induced Pluripotent Stem Cells. Bioengineering 2022, 9, 168. https://doi.org/10.3390/bioengineering9040168
Varzideh F, Mone P, Santulli G. Bioengineering Strategies to Create 3D Cardiac Constructs from Human Induced Pluripotent Stem Cells. Bioengineering. 2022; 9(4):168. https://doi.org/10.3390/bioengineering9040168
Chicago/Turabian StyleVarzideh, Fahimeh, Pasquale Mone, and Gaetano Santulli. 2022. "Bioengineering Strategies to Create 3D Cardiac Constructs from Human Induced Pluripotent Stem Cells" Bioengineering 9, no. 4: 168. https://doi.org/10.3390/bioengineering9040168
APA StyleVarzideh, F., Mone, P., & Santulli, G. (2022). Bioengineering Strategies to Create 3D Cardiac Constructs from Human Induced Pluripotent Stem Cells. Bioengineering, 9(4), 168. https://doi.org/10.3390/bioengineering9040168






