Abstract
Gallic acid is a natural phenolic acid that displays potent anti-cancer activity in a large variety of cell types and rodent cancer xenograft models. Although research has focused on determining the efficacy of gallic acid against various types of human cancer cells, the molecular mechanisms governing the anti-cancer properties of gallic acid remain largely unclear, and a transcriptomic study of gallic acid-induced cancer cell death has rarely been reported. Therefore, we applied time-course bulk RNA-sequencing to elucidate the molecular signature of gallic acid-induced cell death in human cervical cancer HeLa cells, as this is a widely used in vitro model in the field. Our RNA-sequencing dataset covers the early (2nd hour), middle (4th, 6th hour), and late (9th hour) stages of the cell death process after exposure of HeLa cells to gallic acid, and the untreated (0th hour) cells served as controls. Differential expression of messenger RNAs (mRNAs) and long non-coding RNAs (lncRNAs) was identified at each time point in the dataset. In summary, this dataset is a unique and valuable resource with which the scientific community can explore the molecular mechanisms and identify druggable regulators of the gallic acid-induced cell death process in cancer.
Dataset: The RNA-sequencing dataset has been deposited to the Gene Expression Omnibus (GEO) database with the accession number GSE158788, and is associated with published data with Digital Object Identifier (DOI) number 10.1038/s41598-021-96174-1 and PubMed Identifier (PMID) number 34408198. The GEO accession numbers of the raw FASTQ files for all individual samples are listed in Table 1.
Dataset License: License under which the dataset is made available (CC-BY)
Keywords:
gallic acid; HeLa cells; RNA-seq; cell death; apoptosis; ferroptosis; necroptosis; mRNA; lncRNA; gene expression 1. Summary
Gallic acid (GA, also known as 3,4,5-trihydroxybenzoic acid) is a natural bioactive substance that displays anti-cancer properties [1,2,3,4]. This low molecular weight phenolic compound (C7H6O5) is commonly found in edible fungi, fruits, nuts, vegetables, tea leaves, and herbs [5,6]. Accumulating evidence from cell culture and clinically relevant rodent models has demonstrated the anti-cancer bioactivity of gallic acid and its derivatives in various human cancer cell types [1,2,3,4], such as adenocarcinoma [7], bone cancer [8], breast cancer [9], cervical cancer [10], colon cancer [11], gastric cancer [12], leukemia [13], lung cancer [14], lymphoma [15], ovarian cancer [16], prostate cancer [17], and skin cancer [18]. Importantly, gallic acid is well-tolerated by rodent models at the dosages that display an anti-cancer effect [19,20,21,22,23]. Thus, both in vitro and in vivo studies reveal the pharmacological potential of developing gallic acid as a broad-spectrum anti-cancer agent for humans, one that is expected to be safely applied to treat cancers, with minimal side effects.
Resisting cell death is the hallmark of cancer [24,25,26]. To overcome the obstacle in drug resistance, the ideal anti-cancer agents should therefore be able to simultaneously activate multiple pathways that trigger cancer cell death [27,28]. The three major targets of cancer therapy are a cell suicide pathway called apoptosis [29], an iron-dependent form of non-apoptotic cell death called ferroptosis [30], and a regulated form of necrotic cell death called necroptosis [31]. Interestingly, we have recently shown that gallic acid can trigger concurrent apoptosis, ferroptosis, and necroptosis in human cancer cell lines [32,33]. Our finding provides new mechanistic insights into the high potency of gallic acid in killing cancer cells reported by other investigators in a diversity of in vitro and in vivo models [1,2,3,4]. As apoptosis, ferroptosis, and necroptosis regulate cell death by distinctive mechanisms [34,35,36], the cancer cells that resist one of those cell death mechanisms can still be killed by gallic acid because it activates all three of these cell death pathways.
To unleash the potential of gallic acid in clinical application, it is important to elucidate the molecular mechanisms that govern gallic acid-induced cancer cell death. Therefore, we applied a time-course RNA-sequencing (RNA-Seq) analysis to elucidate the transcriptomic profile of the early (2nd hour), middle (4th, 6th hour), and late (9th hour) stages of the gallic acid-induced cell death process [33]. We used human cervical cancer HeLa cells as the study model here, as this cell line is commonly used in studies of gallic acid and cancer biology [1,37]. We applied 50 μg/mL of gallic acid to trigger HeLa cell death in this study, as this is the physiologically relevant dosage that is lower than the no-observed-adverse-effect level (NOAEL) in the rodent models used for studying gallic acid [19,20,21], and this dosage displays anti-cancer activity to a wide range of cancer cell types in vivo and in vitro [1,2,3,4].
Of particular interest, our RNA-Seq dataset [33] covers both the well-characterized protein-coding RNAs/messenger RNAs (mRNAs) [38] and the long non-coding RNAs (lncRNAs) [39], in which their functions are being emerged. Therefore, our dataset will serve as a unique and valuable resource for further studying the molecular mechanisms that regulate the anti-cancer activity of gallic acid, thereby facilitating the translation of this natural anti-cancer agent into clinical applications.
2. Data Description
2.1. Overview of Dataset
We performed quality controls on all the 15 samples in our dataset (Figure 1 and Table 1, with five time points, and three replicates for each time point, covering GA0hr, GA2hr, GA4hr, GA6hr, and GA9hr) to evaluate the accuracy of raw RNA-sequencing reads and read depth (Figure 2), the distribution and coverage breakdown of raw counts (Figure 3), and the sample clustering and differential gene expression analyses (Figure 4) of the dataset [40].
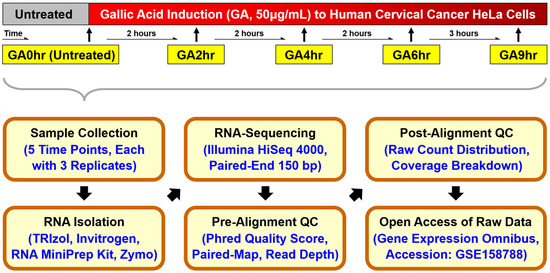
Figure 1.
Schematic of the workflow for the time-course RNA-sequencing study. Human cervical cancer HeLa cells were exposed to 50 μg/mL gallic acid for 2 hours (GA2hr), 4 hours (GA4hr), 6 hours (GA6hr), and 9 hours (GA9hr) to induce the cell death process. Untreated cells served as a control (GA0hr). Cell samples of three biological replicates from each condition were collected for total RNA extraction, analyzed using Illumina HiSeq 4000 for RNA-sequencing, and subjected to pre-alignment and post-alignment quality checks. The raw RNA-sequencing dataset of all 15 samples is publicly available at the Gene Expression Omnibus (GEO accession: GSE158788) [40].
Figure 1.
Schematic of the workflow for the time-course RNA-sequencing study. Human cervical cancer HeLa cells were exposed to 50 μg/mL gallic acid for 2 hours (GA2hr), 4 hours (GA4hr), 6 hours (GA6hr), and 9 hours (GA9hr) to induce the cell death process. Untreated cells served as a control (GA0hr). Cell samples of three biological replicates from each condition were collected for total RNA extraction, analyzed using Illumina HiSeq 4000 for RNA-sequencing, and subjected to pre-alignment and post-alignment quality checks. The raw RNA-sequencing dataset of all 15 samples is publicly available at the Gene Expression Omnibus (GEO accession: GSE158788) [40].
2.2. Accuracy of Raw RNA Sequences
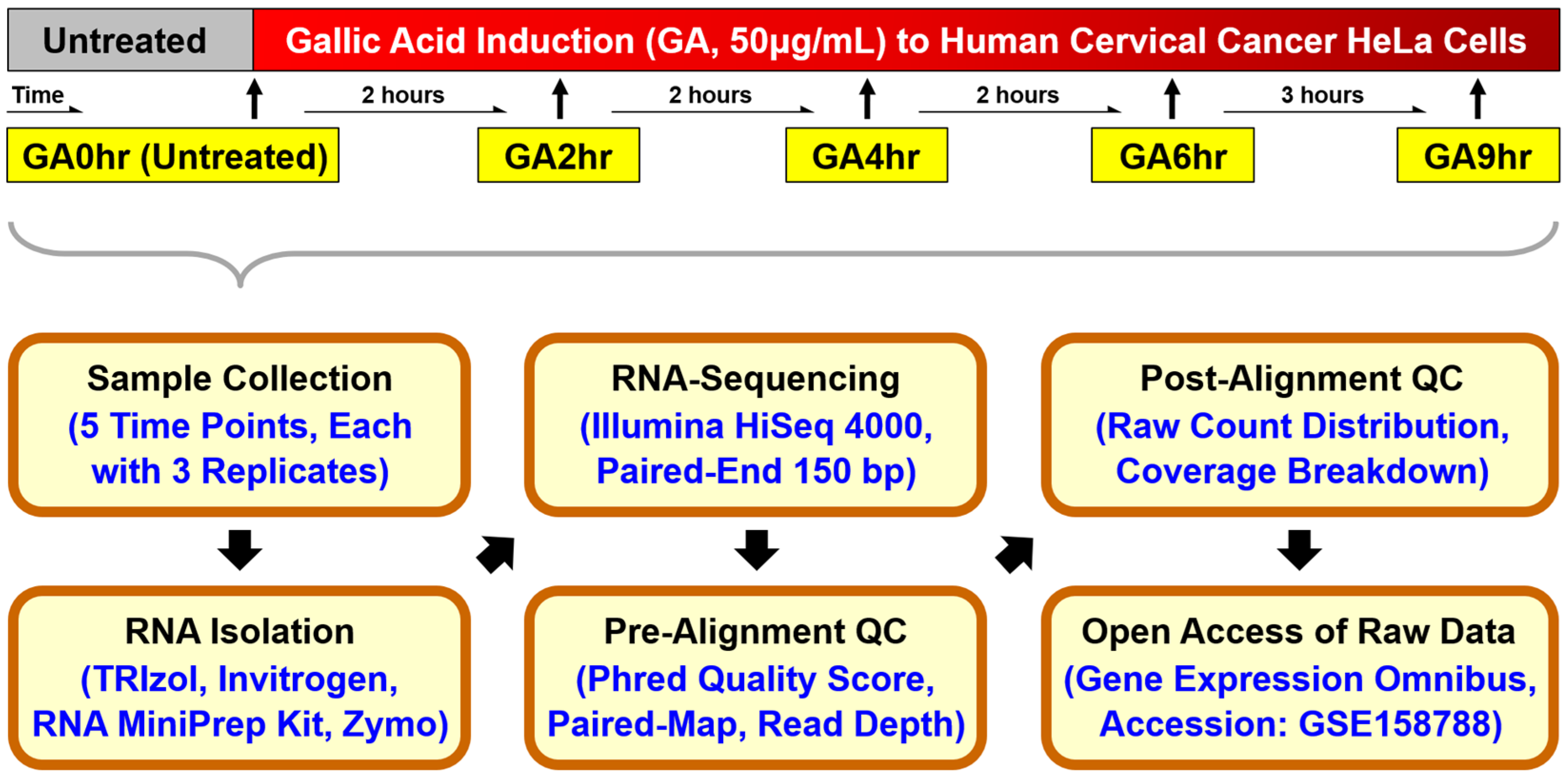
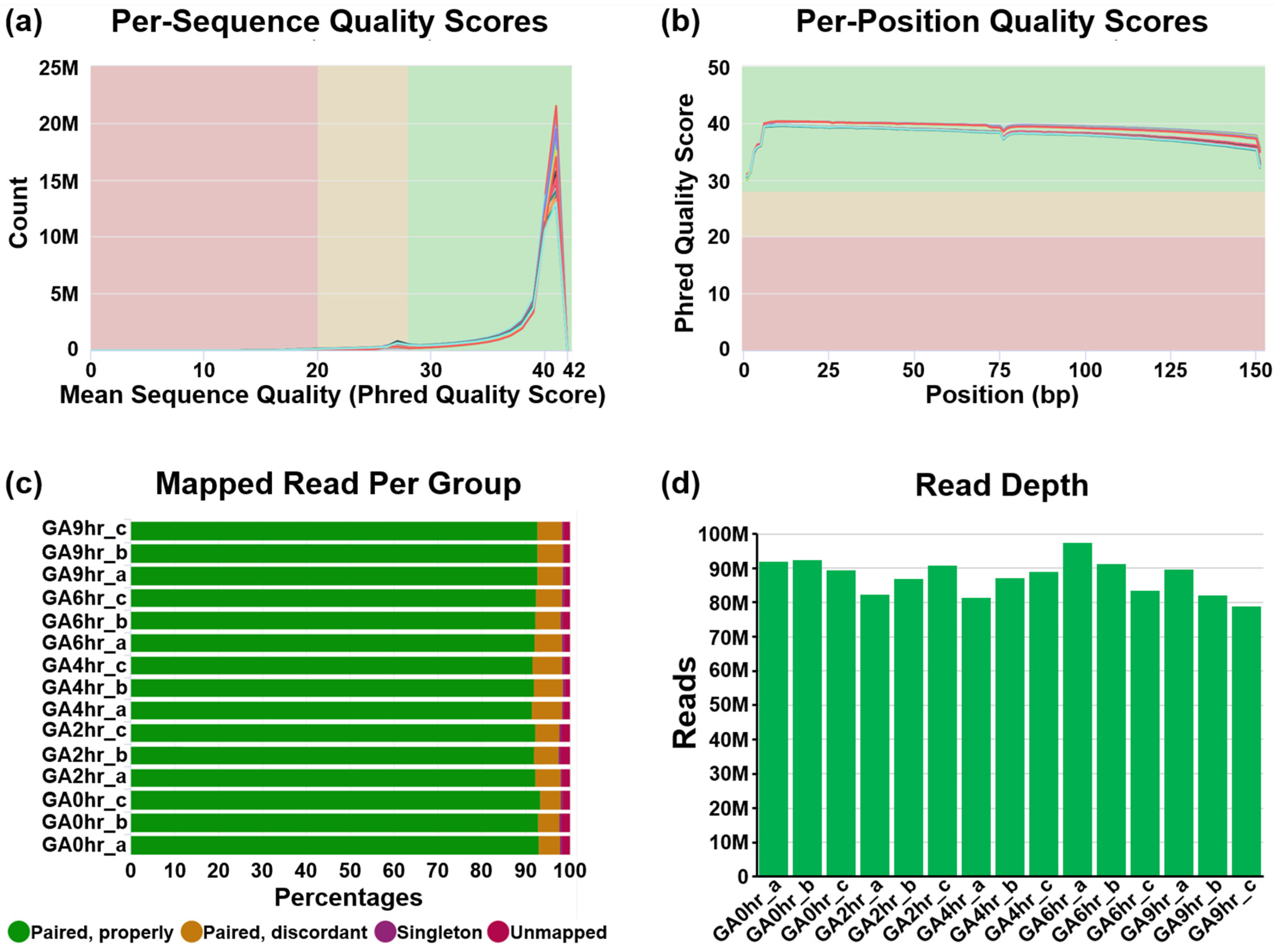
We conducted a pre-alignment quality check of our dataset using MultiQC analysis (Figure 2). The Phred quality score (Q Score) is a standard method widely used to measure the base quality in RNA-sequencing [41,42]. In our dataset, the per-sequence Q Score (Figure 2a) and per-position Q Score (Figure 2b) of most raw reads were significantly higher than Q30 (correct base call by the sequencer is 99.9%, standard from Illumina) and are approaching Q40 (99.99%). The result of the high correct base call is further validated by the high rate of mapped reads per read group analysis of the paired-end sequencing (Figure 2c), which was higher than 90% for each of the 15 samples.

Table 1.
Gene Expression Omnibus (GEO) accession number of all 15 samples in the time-course transcriptome dataset for gallic acid-induced HeLa cell death. GEO accession number of the dataset is GSE1587881 [40].
Table 1.
Gene Expression Omnibus (GEO) accession number of all 15 samples in the time-course transcriptome dataset for gallic acid-induced HeLa cell death. GEO accession number of the dataset is GSE1587881 [40].
| Sample | Sample Description (Treatment to HeLa Cells) | Read Length (bp) | GEO Accession |
|---|---|---|---|
| GA0hr_a | Untreated control—replicate 1 | Paired-end 150 | GSM4810497 |
| GA0hr_b | Untreated control—replicate 2 | Paired-end 150 | GSM4810498 |
| GA0hr_c | Untreated control—replicate 3 | Paired-end 150 | GSM4810499 |
| GA2hr_a | 50 μg/mL gallic acid, 2 hours—replicate 1 | Paired-end 150 | GSM4810500 |
| GA2hr_b | 50 μg/mL gallic acid, 2 hours—replicate 2 | Paired-end 150 | GSM4810501 |
| GA2hr_c | 50 μg/mL gallic acid, 2 hours—replicate 3 | Paired-end 150 | GSM4810502 |
| GA4hr_a | 50 μg/mL gallic acid, 4 hours—replicate 1 | Paired-end 150 | GSM4810503 |
| GA4hr_b | 50 μg/mL gallic acid, 4 hours—replicate 2 | Paired-end 150 | GSM4810504 |
| GA4hr_c | 50 μg/mL gallic acid, 4 hours—replicate 3 | Paired-end 150 | GSM4810505 |
| GA6hr_a | 50 μg/mL gallic acid, 6 hours—replicate 1 | Paired-end 150 | GSM4810506 |
| GA6hr_b | 50 μg/mL gallic acid, 6 hours—replicate 2 | Paired-end 150 | GSM4810507 |
| GA6hr_c | 50 μg/mL gallic acid, 6 hours—replicate 3 | Paired-end 150 | GSM4810508 |
| GA9hr_a | 50 μg/mL gallic acid, 9 hours—replicate 1 | Paired-end 150 | GSM4810509 |
| GA9hr_b | 50 μg/mL gallic acid, 9 hours—replicate 2 | Paired-end 150 | GSM4810510 |
| GA9hr_c | 50 μg/mL gallic acid, 9 hours—replicate 3 | Paired-end 150 | GSM4810511 |
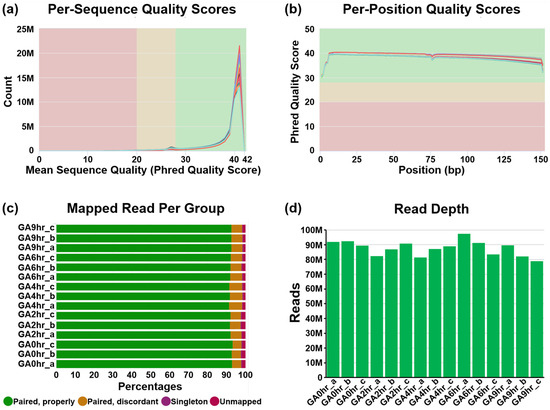
Figure 2.
Pre-alignment quality check of dataset. The quality of the raw RNA reads in all FASTQ files of 15 samples in the dataset GSE158788 was evaluated to determine (a) mean per-sequence Phred quality scores, (b) mean per-position Phred quality scores, (c) percentage of paired reads, and (d) total number of reads of each sample.
Figure 2.
Pre-alignment quality check of dataset. The quality of the raw RNA reads in all FASTQ files of 15 samples in the dataset GSE158788 was evaluated to determine (a) mean per-sequence Phred quality scores, (b) mean per-position Phred quality scores, (c) percentage of paired reads, and (d) total number of reads of each sample.
2.3. Read Depth and Coverage
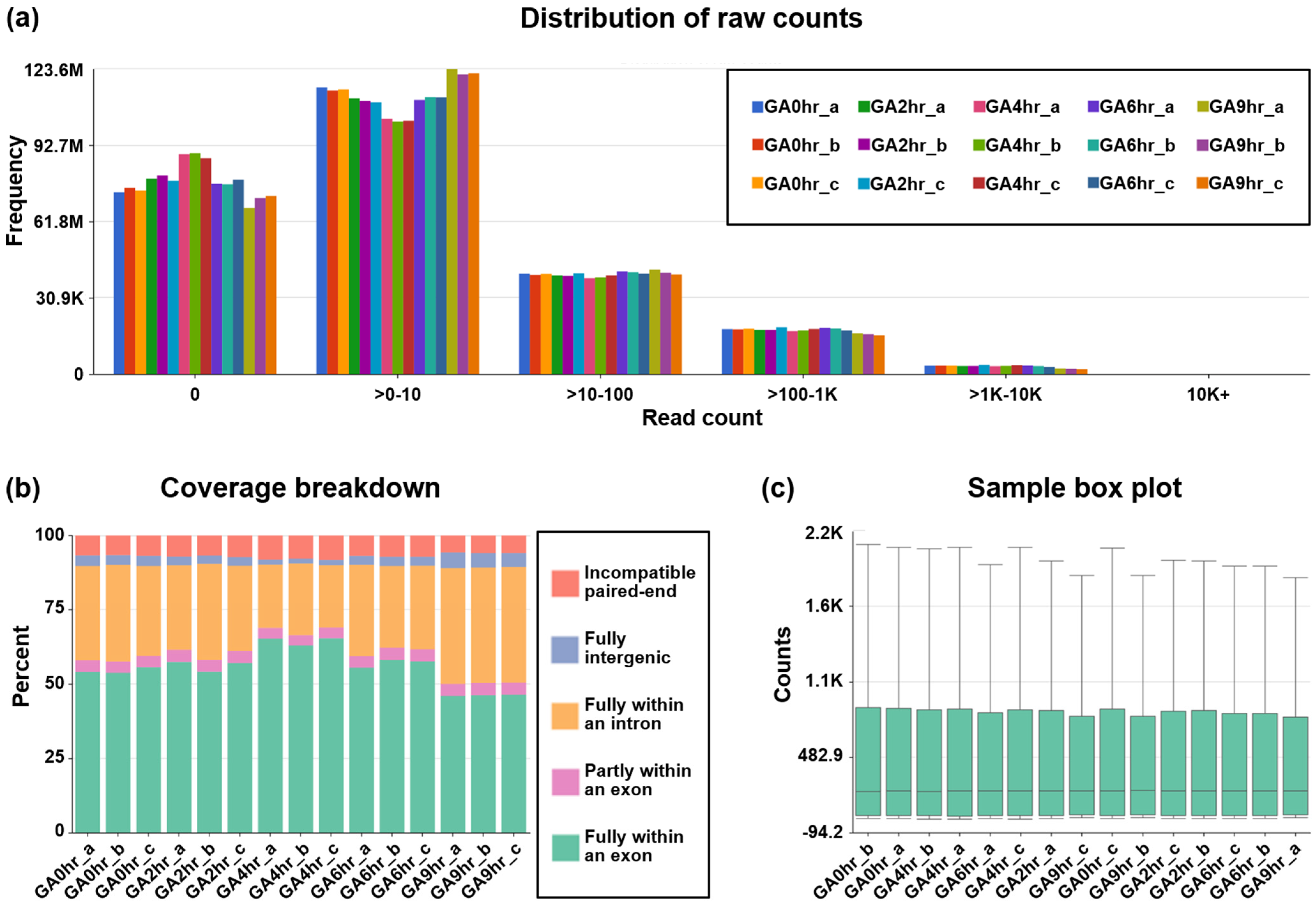
Our dataset contains at least 78.8 million reads per sample (Figure 2d), which indicates a high read depth that is suitable for studying the molecular mechanisms of cellular processes such as differential gene expression [43]. The post-alignment quality check was performed using Partek Flow analysis, which revealed the wide distribution of raw counts (Figure 3a) and the even distribution of the reads that were aligned within exon (e.g., mature RNA) and intron (premature RNA) regions (Figure 3b). Furthermore, the sample box plot illustrates the similar distribution of the counts of all samples in the dataset (Figure 3c). In summary, these validation analyses indicate the high quality of the raw RNA-sequencing dataset in our study.
2.4. Sample Clustering
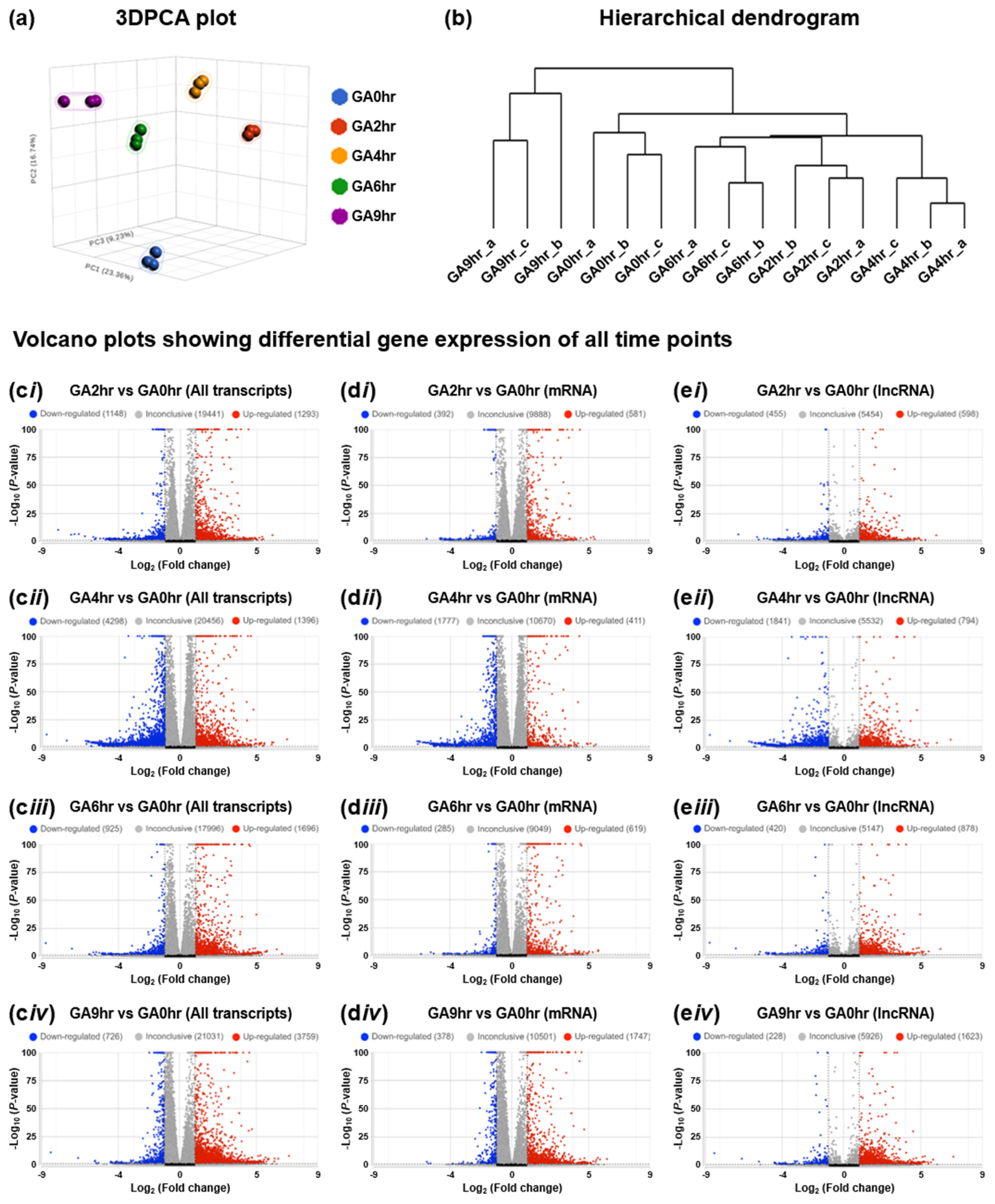
To illustrate the overall characteristic of the dataset, we evaluated all 15 samples using the three-dimensional scatterplot of principal component analysis (3DPCA) to determine the similarity or difference in each sample in the overall dataset. Our data reveals that all samples from the same time points (biological replicates) closely cluster, while samples that belong to different time points are distinct from others (Figure 4a) [33]. This validation analysis indicates that the gene expression profile of samples at the same time point is similar to each other, and is distinct from those at the different time points. This result is also supported by our hierarchical dendrogram (Figure 4b) [33], which demonstrates that all the biological replicates are connected with the shortest branch at their corresponding time points, which indicates the highest similarity in their gene expression patterns.
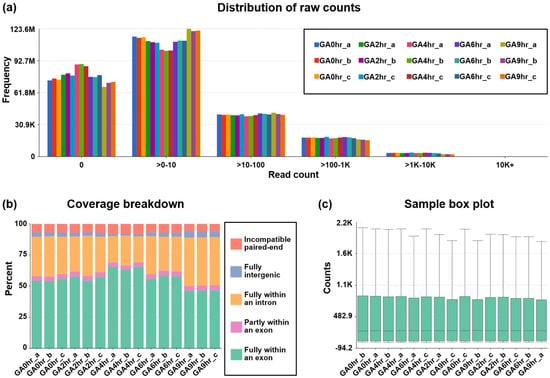
Figure 3.
Post-alignment quality check of dataset. The post-alignment quality check of RNA-sequencing data from all 15 samples in the dataset GSE158788 was evaluated to determine (a) frequency distribution of raw counts, (b) coverage breakdown of the counts, and (c) sample box plot that reveals 10th, 25th, 50th, 75th, and 90th percentiles of the counts after normalization of each sample.
Figure 3.
Post-alignment quality check of dataset. The post-alignment quality check of RNA-sequencing data from all 15 samples in the dataset GSE158788 was evaluated to determine (a) frequency distribution of raw counts, (b) coverage breakdown of the counts, and (c) sample box plot that reveals 10th, 25th, 50th, 75th, and 90th percentiles of the counts after normalization of each sample.
2.5. Differential Gene Expression
To validate that our dataset [33] is suitable for differential gene expression analysis, we applied volcano plots to illustrate the differential expression of all transcripts (Figure 4c) for all four individual time points (GA2hr, GA4hr, GA6hr, and GA9hr) for comparison with the control (GA0hr). Our further validation reveals that the differential expression occurs in both mRNAs (Figure 4d) and lncRNAs (Figure 4e) at each time point. These analyses demonstrate that the gene expression profile at each stage of the gallic acid-induced cell death process is distinct from the control cells that have not been exposed to the gallic acid, thereby indicating that our dataset is suitable for differential gene expression analysis to study the molecular signature of this cell death process.
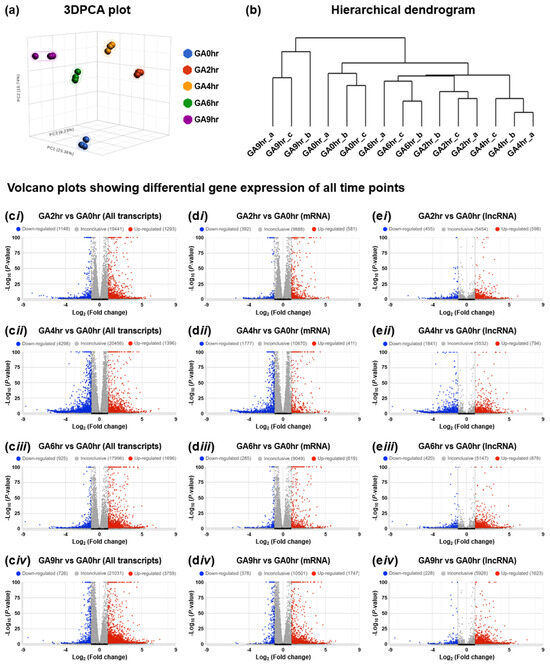
Figure 4.
Time-course transcriptomic profiling of gallic acid-induced HeLa cell death. (a) Three-dimensional scatterplot of principal component analysis (3DPCA), (b) hierarchical dendrogram of all 15 samples in the dataset GSE158788, and (c–e) volcano plots to show the differential gene expression of (c) all transcripts, (d) mRNA, and (e) lncRNA of the 2nd hour (i GA2hr vs. GA0hr), 4th hour (ii GA4hr vs. GA0hr), 6th hour (iii GA6hr vs. GA0hr), and 9th hour (iv GA9hr vs. GA0hr) of the gallic acid-induced HeLa cell death process.
Figure 4.
Time-course transcriptomic profiling of gallic acid-induced HeLa cell death. (a) Three-dimensional scatterplot of principal component analysis (3DPCA), (b) hierarchical dendrogram of all 15 samples in the dataset GSE158788, and (c–e) volcano plots to show the differential gene expression of (c) all transcripts, (d) mRNA, and (e) lncRNA of the 2nd hour (i GA2hr vs. GA0hr), 4th hour (ii GA4hr vs. GA0hr), 6th hour (iii GA6hr vs. GA0hr), and 9th hour (iv GA9hr vs. GA0hr) of the gallic acid-induced HeLa cell death process.
3. Methods
3.1. Cell Culture
The human cervical cancer HeLa cell line was obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA; catalog # CCL-2). This cell line was cultured in Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F-12; catalog # 10565042) supplemented with 10% heat inactivated fetal bovine serum (FBS; catalog # 16140071), 2 mM GlutaMAX (catalog # 35050061), 100 U/mL penicillin, and 100 μg/mL streptomycin (catalog # 15070063) purchased from Thermo Fisher Scientific (Waltham, MA, USA), at 37 degrees Celsius (°C) under an atmosphere of 5% carbon dioxide (CO2). Before the start of the experiment, HeLa cells were plated onto a 100 mm Corning tissue culture dish (Corning, NY, USA; catalog # 430167) for 24 hours to reach 80% cell confluency.
3.2. Cell Death Induction by Gallic Acid
Gallic acid (catalog # G7384) was first dissolved in dimethyl sulfoxide (DMSO; catalog # D2438) purchased from Sigma-Aldrich (Burlington, MA, USA) to make a 20 mg/mL stock solution. This was then applied to the cell culture medium (described above) to achieve the 50 μg/mL working concentration before being further applied to the cells to induce cell death. The stock solution was freshly made before each cell death induction experiment.
3.3. Total RNA Isolation
Total RNA was isolated from the HeLa cells that were exposed to 50 μg/mL gallic acid for 0, 2, 4, 6, and 9 hours (Figure 1). At each of the time points, the cell culture medium was removed from the cell culture dish, and the cells were lysed on the dish using 2 mL of ice-cooled TRIzol by thorough pipetting. Total RNA was extracted using the Direct-zol™ RNA MiniPrep kit (Zymo Research, Irvine, CA, USA; catalog # R2070T) according to the manufacturer’s instructions, eluted in nuclease-free water, quantified by a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), and stored at −80 °C.
3.4. RNASequencing
The RNA-sequencing analysis was conducted at the Novogene Core Facility (Novogene Corporation Inc., Sacramento, CA, USA) using the standard protocols from Novogene and Illumina (San Diego, CA, USA) [33]. The concentration and the purity of each RNA sample was measured using a NanoDrop spectrophotometer and Qubit RNA Assay Kit in a Qubit 2.0 fluorometer (Thermo Fisher Scientific; catalog # Q32852). The integrity of RNA was determined using the RNA Nano 600 Assay Kit on the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Ribosomal RNA depletion of the samples was performed using the Ribo-zero Magnetic Gold Kit (Illumina; catalog # 20037135). NEBNext Ultra™ II Directional RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA; catalog # E7760L) was used to generate sequencing libraries, which were sequenced using the Illumina HiSeq 4000 sequencer for paired-end mode with 150 base-pair read length (paired-end 150, 2x150). Corresponding raw RNA-sequencing data was generated by the sequencer as FASTQ files for 15 RNA samples (Table 1) [33].
3.5. Pre-Alignment Quality Check of Dataset
The quality of raw RNA reads in all FASTQ files was evaluated using MultiQC analysis [44] through the Illumina’s DRAGEN (Dynamic Read Analysis for GENomics) platform (Code Availability 1-2). This evaluation includes per-sequence (Figure 2a) and per-position (Figure 2b) Phred quality score (Q Score) which indicates the accuracy of the raw reads generated by a sequencer [41,42], the percentage of mapped reads per read group of each sample (Figure 2c) which indicates the quality of the paired-end sequencing [38], and the total number of sequencing reads generated per sample (Figure 2d) which reveals an overview of the read depth of the dataset [38].
3.6. Post-Alignment Quality Check of Dataset
The post-alignment quality check of the RNA reads in all the FASTQ files was assessed using Partek Flow bioinformatics software (Partek Inc., St. Louis, MO, USA) (Code Availability 3) with the STAR 2.7.8a aligner package (Code Availability 4) to perform transcript alignment of each sample with the human reference genome hg38 [45]. This quality check includes the distribution of raw counts of all samples (Figure 3a), the coverage breakdown of the counts to reveal the percentage of the reads that fully or partly align with exons or introns (Figure 3b), and a sample box plot that illustrates the 10th, 25th, 50th, 75th, and 90th percentiles of the counts to show the distribution of the normalized gene expression level of each sample (Figure 3c).
3.7. Differential Gene Expression Analysis
The characteristics of the dataset and the differential gene expression of coding RNA (messenger RNAs, mRNAs) and long non-coding RNAs (lncRNAs) were assessed using Partek Flow (Code Availability 3) with the DESeq2 statistical package [46] (Code Availability 5). These analyses include the three-dimensional scatterplot of principal component analysis (3DPCA) [47,48] that illustrates the similarity or difference in each sample in the overall dataset with percentages listed on the planes of “X”, “Y”, and “Z” that refer to their corresponding contribution rate (Figure 4a), the hierarchical dendrogram [41,49,50,51] that reveals the similarity of gene expression patterns between samples (Figure 4b), and differential gene expression of all transcripts (Figure 4c), mRNAs (Figure 4d), and lncRNAs (Figure 4e) at the 2nd hour (b GA2hr vs. GA0hr), 4th hour (ii GA4hr vs. GA0hr), 6th hour (iii GA6hr vs. GA0hr) and 9th hour (iv GA9hr vs. GA0hr) of the HeLa cell death process induced by gallic acid in the dataset.
3.8. Code Availability
The pre-alignment quality check, post-alignment quality check, cell clustering, and differential gene expression data analyses were performed using the following software and their default parameters. No custom code was used in the present study.
(1) MultiQC, version 1.18: https://multiqc.info/.
(2) Illumina’s DRAGEN, version 4.2: https://www.illumina.com/products/by-type/informatics-products/dragen-secondary-analysis.html.
(3) Partek Flow, version 10.0: https://www.partek.com/partek-flow/.
(4) STAR aligner, version 2.7.8a: https://github.com/alexdobin/STAR.
(5) DESeq2, version 3.18: https://www.bioconductor.org/packages/release/bioc/html/DESeq2.html.
4. User Notes
The present study demonstrates that we have generated a high-quality time-course RNA-sequencing dataset revealing the gene expression profile of gallic acid-induced HeLa cell death [33]. Gallic acid is a broad-spectrum anti-cancer compound [1,2,3,4]. By comparing the molecular signature of the present dataset generated with human cervical cancer HeLa cells and with those generated in the past and in the future for the cell death response of other human cancer cell types [19,20,21,22,23] to gallic acid, it will be able to reveal the molecular signature shared by different cancer models, and therefore identify the druggable regulators and therapeutic targets that mediate gallic acid-induced cancer cell death mechanisms.
Multiple factors could impact the transcriptomic changes that can be taken into consideration for studying the molecular signature of gallic acid. For example, different cancer cell types may respond to gallic acid differently [1,2,3,4]. In our present studies, our RNA-Seq detected striking changes in transcription of genes at the early, middle, and late stages of cell death in HeLa cells that were exposed to 50 μg/mL of gallic acid, in which apoptosis, ferroptosis, and necroptosis were identified [33]. However, differential gene expression was detected in the colon cancer HCT116 cells treated with 50 μM of gallic acid for an extended time (12th and 24th hour), in which activation of apoptotic cell death pathways was identified [52]. Heterogeneity is the hallmark of cancer cells [25], and therefore different cancer cell types could display different responses to the same dosage of gallic acid.
The molecular profile of gallic acid-induced cell death can be illustrated by determining the differential gene expression and RNA splicing of the datasets using TopHat [53], Cufflinks [54,55], HISAT [56], StringTie [57], Ballgown [58,59], and HTSeq [60]. Functional and pathway analyses can be performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) [61,62], Gene Ontology (GO) [63,64], Ingenuity Pathway Analysis (IPA) [65], and WebGestalt [66]. Future analyses of the transcriptomic changes in response to gallic acid in different cancer cell types will reveal the molecular mechanisms governing the anti-cancer property of gallic acid. Therefore, our dataset is a valuable resource with high reuse potential for future studies, and thus facilitates the translation of this natural phenolic anti-cancer compound into clinical cancer treatment.
Author Contributions
Conceptualization, H.M.T. and P.C.K.C.; methodology, H.M.T. and P.C.K.C.; validation, H.M.T. and P.C.K.C.; formal analysis, H.M.T. and P.C.K.C.; resources, H.M.T. and P.C.K.C.; data curation, H.M.T. and P.C.K.C.; writing—original draft preparation, H.M.T. and P.C.K.C.; writing—review and editing, H.M.T. and P.C.K.C.; visualization, H.M.T. and P.C.K.C.; supervision, H.M.T. and P.C.K.C.; project administration, H.M.T. and P.C.K.C.; funding acquisition, P.C.K.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Direct Grant (4053367) of the Chinese University of Hong Kong (CUHK) Research Committee to Dr. Peter Chi Keung Cheung.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the raw FASTQ files of the dataset are publicly available at the Gene Expression Omnibus (GEO) database with the accession number GSE158788 [40]. The dataset is available for public access through the following link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE158788.
Acknowledgments
We thank all members of Peter Chi Keung Cheung’s laboratory for their great support in this investigation.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Subramanian, A.P.; John, A.A.; Vellayappan, M.V.; Balaji, A.; Jaganathan, S.K.; Supriyanto, E.; Yusof, M. Gallic acid: Prospects and molecular mechanisms of its anticancer activity. Rsc Adv. 2015, 5, 35608–35621. [Google Scholar] [CrossRef]
- Jiang, Y.; Pei, J.; Zheng, Y.; Miao, Y.J.; Duan, B.Z.; Huang, L.F. Gallic Acid: A Potential Anti-Cancer Agent. Chin. J. Integr. Med. 2022, 28, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Ashrafizadeh, M.; Zarrabi, A.; Mirzaei, S.; Hashemi, F.; Samarghandian, S.; Zabolian, A.; Hushmandi, K.; Ang, H.L.; Sethi, G.; Kumar, A.P.; et al. Gallic acid for cancer therapy: Molecular mechanisms and boosting efficacy by nanoscopical delivery. Food Chem. Toxicol. 2021, 157, 112576. [Google Scholar] [CrossRef]
- Tuli, H.S.; Mistry, H.; Kaur, G.; Aggarwal, D.; Garg, V.K.; Mittal, S.; Yerer, M.B.; Sak, K.; Khan, M.A. Gallic Acid: A Dietary Polyphenol that Exhibits Anti-neoplastic Activities by Modulating Multiple Oncogenic Targets. Anticancer Agents Med. Chem. 2022, 22, 499–514. [Google Scholar] [CrossRef]
- Serrano, J.; Puupponen-Pimia, R.; Dauer, A.; Aura, A.M.; Saura-Calixto, F. Tannins: Current knowledge of food sources, intake, bioavailability and biological effects. Mol. Nutr. Food Res. 2009, 53 (Suppl. 2), S310–S329. [Google Scholar] [CrossRef] [PubMed]
- Daglia, M.; Di Lorenzo, A.; Nabavi, S.F.; Talas, Z.S.; Nabavi, S.M. Polyphenols: Well Beyond The Antioxidant Capacity: Gallic Acid and Related Compounds as Neuroprotective Agents: You are What You Eat! Curr. Pharm. Biotechnol. 2014, 15, 362–372. [Google Scholar] [CrossRef]
- Jara, J.A.; Castro-Castillo, V.; Saavedra-Olavarria, J.; Peredo, L.; Pavanni, M.; Jana, F.; Letelier, M.E.; Parra, E.; Becker, M.I.; Morello, A.; et al. Antiproliferative and uncoupling effects of delocalized, lipophilic, cationic gallic acid derivatives on cancer cell lines. Validation in vivo in singenic mice. J. Med. Chem. 2014, 57, 2440–2454. [Google Scholar] [CrossRef]
- Liang, C.Z.; Zhang, X.; Li, H.; Tao, Y.Q.; Tao, L.J.; Yang, Z.R.; Zhou, X.P.; Shi, Z.L.; Tao, H.M. Gallic acid induces the apoptosis of human osteosarcoma cells in vitro and in vivo via the regulation of mitogen-activated protein kinase pathways. Cancer Biother. Radiopharm. 2012, 27, 701–710. [Google Scholar] [CrossRef]
- Wang, K.; Zhu, X.; Zhang, K.; Zhu, L.; Zhou, F. Investigation of gallic acid induced anticancer effect in human breast carcinoma MCF-7 cells. J. Biochem. Mol. Toxicol. 2014, 28, 387–393. [Google Scholar] [CrossRef]
- Zhao, B.; Hu, M. Gallic acid reduces cell viability, proliferation, invasion and angiogenesis in human cervical cancer cells. Oncol. Lett. 2013, 6, 1749–1755. [Google Scholar] [CrossRef]
- Khaledi, H.; Alhadi, A.A.; Yehye, W.A.; Ali, H.M.; Abdulla, M.A.; Hassandarvish, P. Antioxidant, cytotoxic activities, and structure-activity relationship of gallic acid-based indole derivatives. Arch. Pharm. 2011, 344, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.H.; Chang, C.S.; Ho, W.C.; Liao, S.Y.; Lin, W.L.; Wang, C.J. Gallic acid inhibits gastric cancer cells metastasis and invasive growth via increased expression of RhoB, downregulation of AKT/small GTPase signals and inhibition of NF-kappaB activity. Toxicol. Appl. Pharmacol. 2013, 266, 76–85. [Google Scholar] [CrossRef]
- Inoue, M.; Suzuki, R.; Koide, T.; Sakaguchi, N.; Ogihara, Y.; Yabu, Y. Antioxidant, gallic acid, induces apoptosis in HL-60RG cells. Biochem. Biophys. Res. Commun. 1994, 204, 898–904. [Google Scholar] [CrossRef] [PubMed]
- Ohno, Y.; Fukuda, K.; Takemura, G.; Toyota, M.; Watanabe, M.; Yasuda, N.; Xinbin, Q.; Maruyama, R.; Akao, S.; Gotou, K.; et al. Induction of apoptosis by gallic acid in lung cancer cells. Anticancer Drugs 1999, 10, 845–851. [Google Scholar] [CrossRef]
- Kim, N.S.; Jeong, S.I.; Hwang, B.S.; Lee, Y.E.; Kang, S.H.; Lee, H.C.; Oh, C.H. Gallic acid inhibits cell viability and induces apoptosis in human monocytic cell line U937. J. Med. Food 2011, 14, 240–246. [Google Scholar] [CrossRef]
- Varela-Rodriguez, L.; Sanchez-Ramirez, B.; Hernandez-Ramirez, V.I.; Varela-Rodriguez, H.; Castellanos-Mijangos, R.D.; Gonzalez-Horta, C.; Chavez-Munguia, B.; Talamas-Rohana, P. Effect of Gallic acid and Myricetin on ovarian cancer models: A possible alternative antitumoral treatment. BMC Complement. Med. Ther. 2020, 20, 110. [Google Scholar] [CrossRef]
- Kaur, M.; Velmurugan, B.; Rajamanickam, S.; Agarwal, R.; Agarwal, C. Gallic acid, an active constituent of grape seed extract, exhibits anti-proliferative, pro-apoptotic and anti-tumorigenic effects against prostate carcinoma xenograft growth in nude mice. Pharm. Res. 2009, 26, 2133–2140. [Google Scholar] [CrossRef] [PubMed]
- Ortega, E.; Sadaba, M.C.; Ortiz, A.I.; Cespon, C.; Rocamora, A.; Escolano, J.M.; Roy, G.; Villar, L.M.; Gonzalez-Porque, P. Tumoricidal activity of lauryl gallate towards chemically induced skin tumours in mice. Br. J. Cancer 2003, 88, 940–943. [Google Scholar] [CrossRef]
- van der Heijden, C.A.; Janssen, P.J.; Strik, J.J. Toxicology of gallates: A review and evaluation. Food Chem. Toxicol. 1986, 24, 1067–1070. [Google Scholar] [CrossRef]
- Rajalakshmi, K.; Devaraj, H.; Niranjali Devaraj, S. Assessment of the no-observed-adverse-effect level (NOAEL) of gallic acid in mice. Food Chem. Toxicol. 2001, 39, 919–922. [Google Scholar] [CrossRef]
- Niho, N.; Shibutani, M.; Tamura, T.; Toyoda, K.; Uneyama, C.; Takahashi, N.; Hirose, M. Subchronic toxicity study of gallic acid by oral administration in F344 rats. Food Chem. Toxicol. 2001, 39, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Vijaya Padma, V.; Sowmya, P.; Arun Felix, T.; Baskaran, R.; Poornima, P. Protective effect of gallic acid against lindane induced toxicity in experimental rats. Food Chem. Toxicol. 2011, 49, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.Y.; Hu, J.X.; Hu, D.Y.; Yang, X. A Role of Gallic Acid in Oxidative Damage Diseases: A Comprehensive Review. Nat. Prod. Commun. 2019, 14, 1934578X19874174. [Google Scholar] [CrossRef]
- Vasan, N.; Baselga, J.; Hyman, D.M. A view on drug resistance in cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Swanton, C.; Bernard, E.; Abbosh, C.; Andre, F.; Auwerx, J.; Balmain, A.; Bar-Sagi, D.; Bernards, R.; Bullman, S.; DeGregori, J.; et al. Embracing cancer complexity: Hallmarks of systemic disease. Cell 2024, 187, 1589–1616. [Google Scholar] [CrossRef]
- Szakacs, G.; Paterson, J.K.; Ludwig, J.A.; Booth-Genthe, C.; Gottesman, M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006, 5, 219–234. [Google Scholar] [CrossRef]
- Scott, E.C.; Baines, A.C.; Gong, Y.; Moore, R., Jr.; Pamuk, G.E.; Saber, H.; Subedee, A.; Thompson, M.D.; Xiao, W.; Pazdur, R.; et al. Trends in the approval of cancer therapies by the FDA in the twenty-first century. Nat. Rev. Drug Discov. 2023, 22, 625–640. [Google Scholar] [CrossRef]
- Carneiro, B.A.; El-Deiry, W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020, 17, 395–417. [Google Scholar] [CrossRef]
- Lei, G.; Zhuang, L.; Gan, B. Targeting ferroptosis as a vulnerability in cancer. Nat. Rev. Cancer 2022, 22, 381–396. [Google Scholar] [CrossRef]
- Gong, Y.; Fan, Z.; Luo, G.; Yang, C.; Huang, Q.; Fan, K.; Cheng, H.; Jin, K.; Ni, Q.; Yu, X.; et al. The role of necroptosis in cancer biology and therapy. Mol. Cancer 2019, 18, 100. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.M.; Cheung, P.C.K. Gallic Acid Triggers Iron-Dependent Cell Death with Apoptotic, Ferroptotic, and Necroptotic Features. Toxins 2019, 11, 492. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.M.; Cheung, P.C.K. Gene expression profile analysis of gallic acid-induced cell death process. Sci. Rep. 2021, 11, 16743. [Google Scholar] [CrossRef]
- Vitale, I.; Pietrocola, F.; Guilbaud, E.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostini, M.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; et al. Apoptotic cell death in disease-Current understanding of the NCCD 2023. Cell Death Differ. 2023, 30, 1097–1154. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, B.R. Ferroptosis turns 10: Emerging mechanisms, physiological functions, and therapeutic applications. Cell 2022, 185, 2401–2421. [Google Scholar] [CrossRef]
- Shan, B.; Pan, H.; Najafov, A.; Yuan, J. Necroptosis in development and diseases. Genes Dev. 2018, 32, 327–340. [Google Scholar] [CrossRef]
- Masters, J.R. HeLa cells 50 years on: The good, the bad and the ugly. Nat. Rev. Cancer 2002, 2, 315–319. [Google Scholar] [CrossRef]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Tang, H.M.; Cheung, P.C.K. Gene Expression Profile Analysis of Gallic Acid-induced Cell Death Process. Gene Expression Omnibus, 2021. [Google Scholar]
- Li, W.V.; Li, J.J. Modeling and analysis of RNA-seq data: A review from a statistical perspective. Quant. Biol. 2018, 6, 195–209. [Google Scholar] [CrossRef]
- Richterich, P. Estimation of errors in “raw” DNA sequences: A validation study. Genome Res. 1998, 8, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Sims, D.; Sudbery, I.; Ilott, N.E.; Heger, A.; Ponting, C.P. Sequencing depth and coverage: Key considerations in genomic analyses. Nat. Rev. Genet. 2014, 15, 121–132. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Genome Assembly GRCh38.p14. National Library of Medicine, 2022.
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Ringner, M. What is principal component analysis? Nat. Biotechnol. 2008, 26, 303–304. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szczesniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef]
- Van den Berge, K.; Hembach, K.M.; Soneson, C.; Tiberi, S.; Clement, L.; Love, M.I.; Patro, R.; Robinson, M.D. RNA Sequencing Data: Hitchhiker’s Guide to Expression Analysis. Annu. Rev. Biomed. Data Sci. 2019, 2, 139–173. [Google Scholar] [CrossRef]
- Boutros, P.C.; Okey, A.B. Unsupervised pattern recognition: An introduction to the whys and wherefores of clustering microarray data. Brief. Bioinform. 2005, 6, 331–343. [Google Scholar] [CrossRef]
- Yang, C.; Xie, X.; Tang, H.; Dong, X.; Zhang, X.; Huang, F. Transcriptome analysis reveals GA induced apoptosis in HCT116 human colon cancer cells through calcium and p53 signal pathways. Rsc Adv. 2018, 8, 12449–12458. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Frazee, A.C.; Pertea, G.; Jaffe, A.E.; Langmead, B.; Salzberg, S.L.; Leek, J.T. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 2015, 33, 243–246. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology, C.; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
