Modulation of Swine Gut Microbiota by Phytogenic Blends and High Concentrations of Casein in a Validated Swine Large Intestinal In Vitro Model
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Product
2.2. Fecal Samples Collection and Preparation
2.3. Experimental Set up and the TIM-2
2.4. Gut Microbiota Composition
2.5. SCFA and BCFA Production
2.6. Statistical Analysis
3. Results
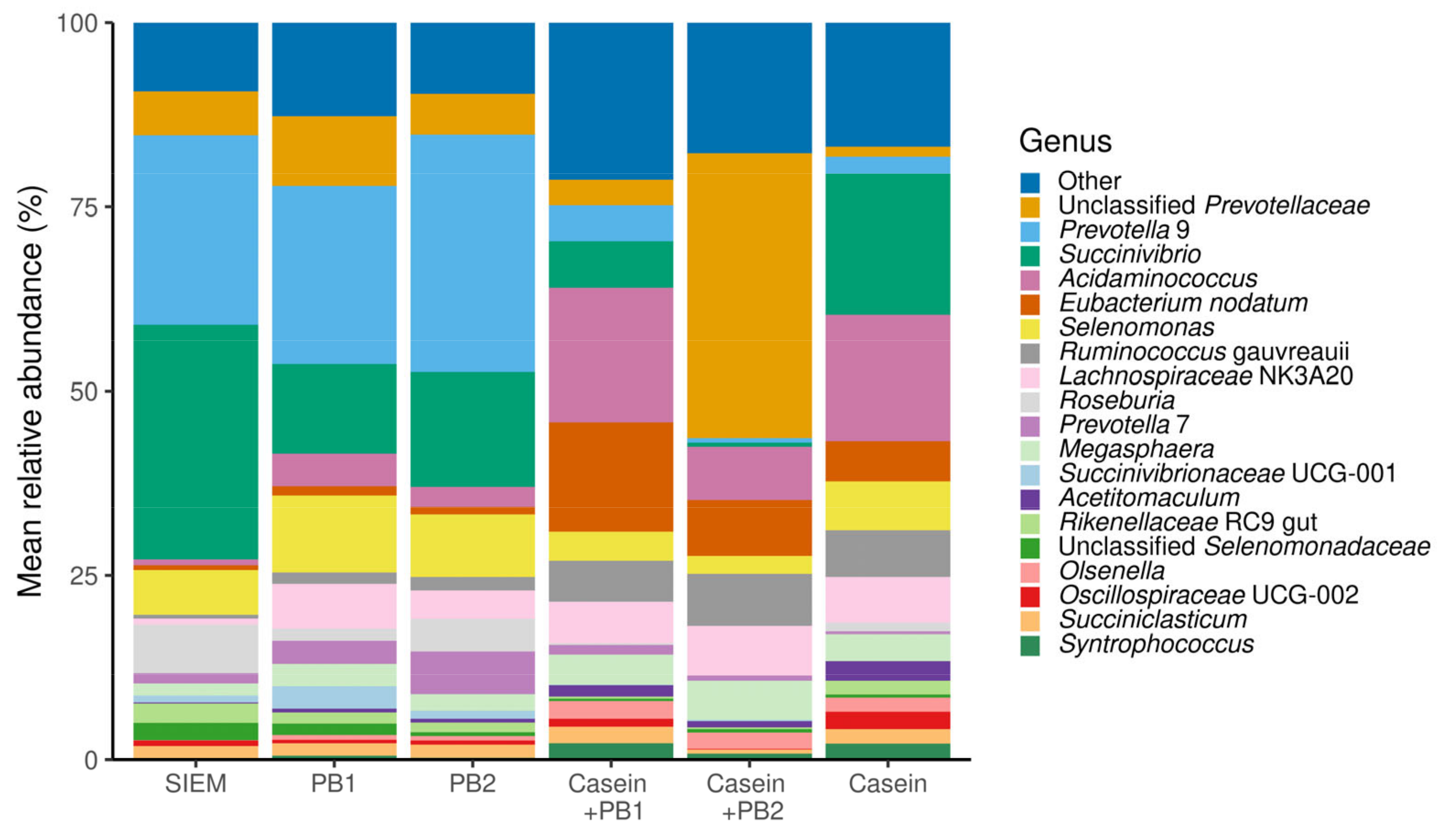
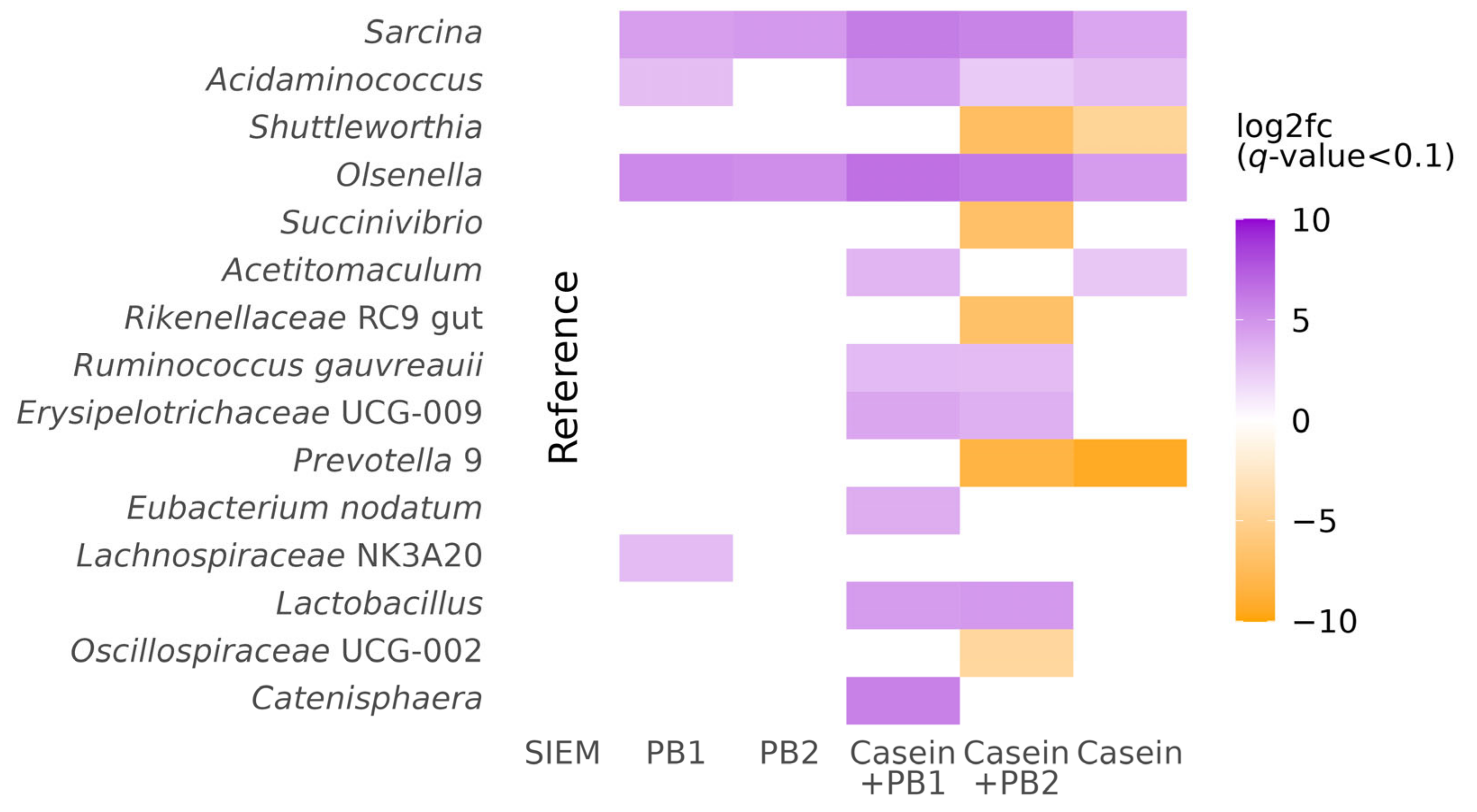
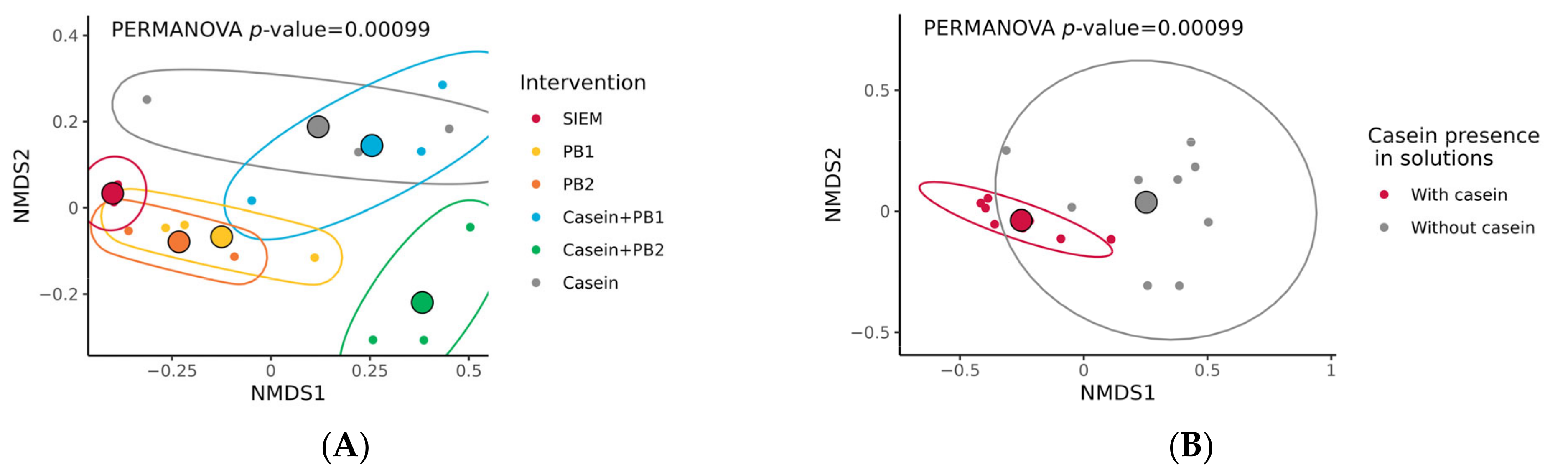
3.1. Changes in Microbiota Composition
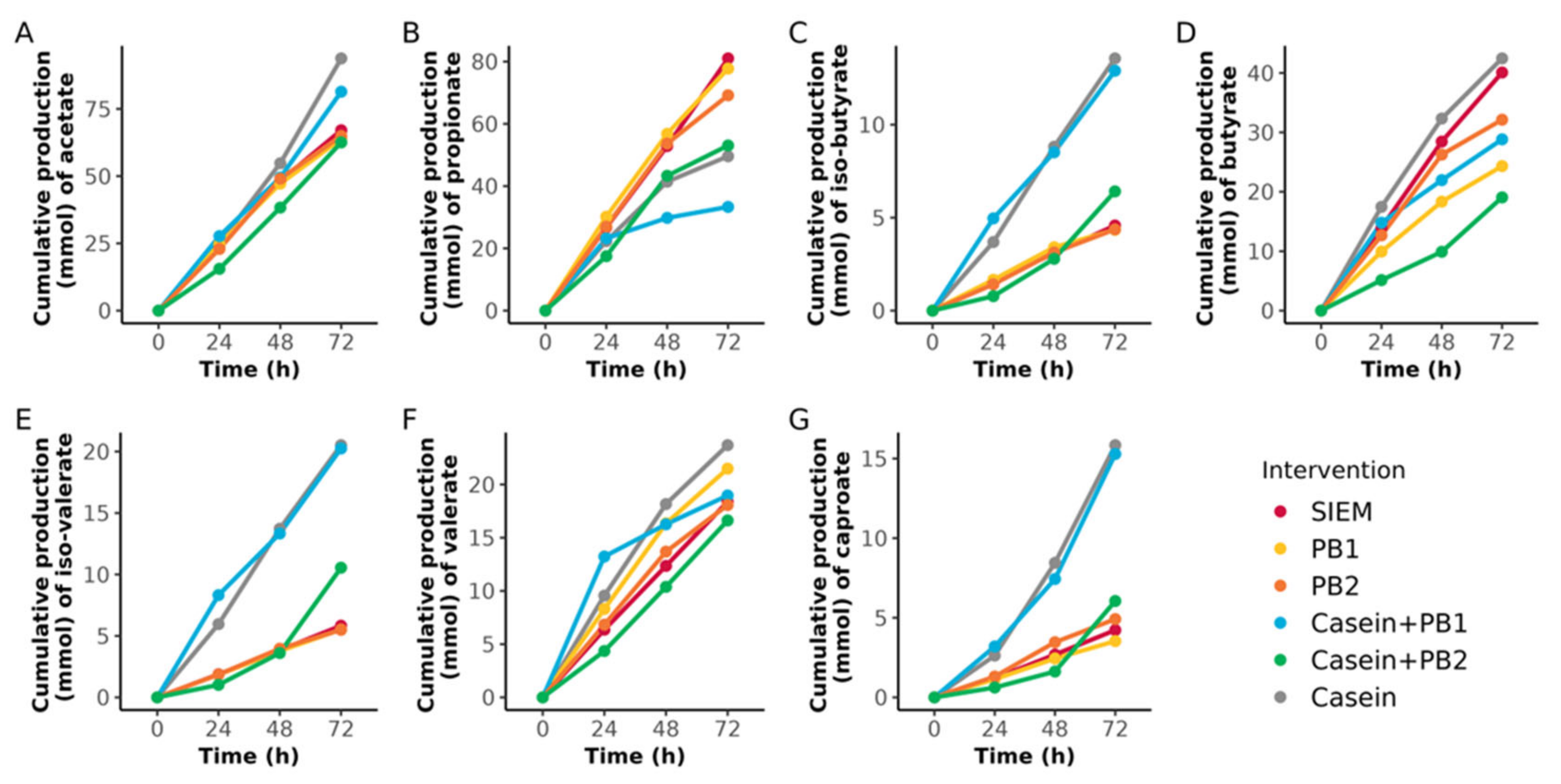
3.2. Production of SCFA and BCFA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oh, J.; Wall, E.H.; Bravo, D.M.; Hristov, A.N. Host-mediated effects of phytonutrients in ruminants: A review. J. Dairy Sci. 2017, 100, 5974–5983. [Google Scholar] [CrossRef]
- Rafiq, K.; Tofazzal Hossain, M.; Ahmed, R.; Hasan, M.M.; Islam, R.; Hossen, M.I.; Shaha, S.N.; Islam, M.R. Role of Different Growth Enhancers as Alternative to In-feed Antibiotics in Poultry Industry. Front. Vet. Sci. 2022, 8, 794588. [Google Scholar] [CrossRef] [PubMed]
- Taha-Abdelaziz, K.; Hodgins, D.C.; Lammers, A.; Alkie, T.N.; Sharif, S. Effects of early feeding and dietary interventions on development of lymphoid organs and immune competence in neonatal chickens: A review. Vet. Immunol. Immunopathol. 2018, 201, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kikusato, M. Phytobiotics to improve health and production of broiler chickens: Functions beyond the antioxidant activity. Anim. Biosci. 2021, 34, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Chowdhury, M.A.; Huo, Y.; Gong, J. Phytogenic compounds as alternatives to in-feed antibiotics: Potentials and challenges in application. Pathogens 2015, 4, 137–156. [Google Scholar] [CrossRef] [PubMed]
- Nowak, P.; Kasprowicz-Potocka, M.; Zaworska, A.; Nowak, W.; Stefańska, B.; Sip, A.; Grajek, W.; Juzwa, W.; Taciak, M.; Barszcz, M.; et al. The effect of eubiotic feed additives on the performance of growing pigs and the activity of intestinal microflora. Arch. Anim. Nutr. 2017, 71, 455–469. [Google Scholar] [CrossRef] [PubMed]
- Duarte, M.E.; Kim, S.W. Phytobiotics from Oregano Extracts Enhance the Intestinal Health and Growth Performance of Pigs. Antioxidants 2022, 11, 2066. [Google Scholar] [CrossRef] [PubMed]
- Diao, H.; Zheng, P.; Yu, B.; He, J.; Mao, X.; Yu, J.; Chen, D. Effects of benzoic Acid and thymol on growth performance and gut characteristics of weaned piglets. Asian-Australas. J. Anim. Sci. 2015, 28, 827–839. [Google Scholar] [CrossRef]
- Gardiner, G.E.; Metzler-Zebeli, B.U.; Lawlor, P.G. Impact of Intestinal Microbiota on Growth and Feed Efficiency in Pigs: A Review. Microorganisms 2020, 8, 1886. [Google Scholar] [CrossRef]
- Vasquez, R.; Oh, J.K.; Song, J.H.; Kang, D.K. Gut microbiome-produced metabolites in pigs: A review on their biological functions and the influence of probiotics. J. Anim. Sci. Technol. 2022, 64, 671–695. [Google Scholar] [CrossRef]
- Wang, C.; Wei, S.; Chen, N.; Xiang, Y.; Wang, Y.; Jin, M. Characteristics of gut microbiota in pigs with different breeds, growth periods and genders. Microb. Biotechnol. 2022, 15, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Venema, K.; van den Abbeele, P. Experimental models of the gut microbiome. Best Pract. Res. Clin. Gastroenterol. 2013, 27, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Long, C.; de Vries, S.; Venema, K. Polysaccharide source altered ecological network, functional profile, and short-chain fatty acid production in a porcine gut microbiota. Benef. Microbes 2020, 11, 591–610. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Tena, M.; Alegria, A.; Lagarda, M.J.; Venema, K. Impact of plant sterols enrichment dose on gut microbiota from lean and obese subjects using TIM-2 in vitro fermentation model. J. Funct. Foods 2019, 54, 164–174. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Shannon, C.E. The mathematical theory of communication. 1963. MD Comput. 1997, 14, 306–317. [Google Scholar]
- Chao, A. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Pielou, E.C. The Measurement of Diversity in Different Types of Biological Collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An ordination of the upland Forest communities of southern Wisconsin. Ecol. Mon. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 2021, 17, e1009442. [Google Scholar] [CrossRef]
- Nearing, J.T.; Douglas, G.M.; Hayes, M.G.; MacDonald, J.; Desai, D.K.; Allward, N.; Jones, C.M.A.; Wright, B.J.; Dhanani, A.S.; Comeau, A.M.; et al. Microbiome differential abundance methods produce different results across 38 datasets. Nat. Commun. 2022, 13, 777. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, M.; Ramiro-Garcia, J.; Koenen, M.E.; Venema, K. To pool or not to pool? Impact of the use of individual and pooled fecal samples for in vitro fermentation studies. J. Microbiol. Methods 2014, 107, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Duque-Soto, C.; Quintriqueo-Cid, A.; Rueda-Robles, A.; Robert, P.; Borrás-Linares, I.; Lozano-Sánchez, J. Evaluation of Different Advanced Approaches to Simulation of Dynamic In Vitro Digestion of Polyphenols from Different Food Matrices—A Systematic Review. Antioxidants 2022, 12, 101. [Google Scholar] [CrossRef]
- Gościniak, A.; Eder, P.; Walkowiak, J.; Cielecka-Piontek, J. Artificial Gastrointestinal Models for Nutraceuticals Research-Achievements and Challenges: A Practical Review. Nutrients 2022, 14, 2560. [Google Scholar] [CrossRef] [PubMed]
- Falduto, M.; Smedile, F.; Zhang, M.; Zheng, T.; Zhu, J.; Huang, Q.; Weeks, R.; Ermakov, A.M.; Chikindas, M.L. Anti-obesity effects of Chenpi: An artificial gastrointestinal system study. Microb. Biotechnol. 2022, 15, 874–885. [Google Scholar] [CrossRef]
- Maas, E.; Penders, J.; Venema, K. Modelling the Gut Fungal-Community in TIM-2 with a Microbiota from Healthy Individuals. J. Fungi 2023, 9, 104. [Google Scholar] [CrossRef]
- Aguirre, M.; Jonkers, D.M.; Troost, F.J.; Roeselers, G.; Venema, K. In vitro characterization of the impact of different substrates on metabolite production, energy extraction and composition of gut microbiota from lean and obese subjects. PLoS ONE 2014, 9, e113864. [Google Scholar] [CrossRef]
- Long, C.; de Vries, S.; Venema, K. Differently Pre-treated Rapeseed Meals Affect in vitro Swine Gut Microbiota Composition. Front. Microbiol. 2020, 11, 570985. [Google Scholar] [CrossRef]
- Long, C.; Venema, K. Pretreatment of Rapeseed Meal Increases Its Recalcitrant Fiber Fermentation and Alters the Microbial Community in an In Vitro Model of Swine Large Intestine. Front. Microbiol. 2020, 11, 588264. [Google Scholar] [CrossRef] [PubMed]
- Long, C.; Rösch, C.; de Vries, S.; Schols, H.; Venema, K. Cellulase and Alkaline Treatment Improve Intestinal Microbial Degradation of Recalcitrant Fibers of Rapeseed Meal in Pigs. J. Agric. Food Chem. 2020, 68, 11011–11025. [Google Scholar] [CrossRef] [PubMed]
- Amat, S.; Lantz, H.; Munyaka, P.M.; Willing, B.P. Prevotella in Pigs: The Positive and Negative Associations with Production and Health. Microorganisms 2020, 8, 1584. [Google Scholar] [CrossRef] [PubMed]
- Mach, N.; Berri, M.; Estellé, J.; Levenez, F.; Lemonnier, G.; Denis, C.; Leplat, J.; Chevaleyre, C.; Billon, Y.; Doré, J.; et al. Early-life establishment of the swine gut microbiome and impact on host phenotypes. Environ. Microbiol. Rep. 2015, 7, 554–569. [Google Scholar] [CrossRef] [PubMed]
- Dou, S.; Gadonna-Widehem, P.; Rome, V.; Hamoudi, D.; Rhazi, L.; Lakhal, L.; Larcher, T.; Bahi-Jaber, N.; Pinon-Quintana, A.; Guyonvarch, A.; et al. Characterisation of Early-Life Fecal Microbiota in Susceptible and Healthy Pigs to Post-Weaning Diarrhoea. PLoS ONE 2017, 12, e0169851. [Google Scholar] [CrossRef] [PubMed]
- Rist, V.T.; Weiss, E.; Sauer, N.; Mosenthin, R.; Eklund, M. Effect of dietary protein supply originating from soybean meal or casein on the intestinal microbiota of piglets. Anaerobe 2014, 25, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Li, R.W.; Wu, S.; Li, W.; Navarro, K.; Couch, R.D.; Hill, D.; Urban, J.F. Alterations in the porcine colon microbiota induced by the gastrointestinal nematode Trichuris suis. Infect. Immun. 2012, 80, 2150–2157. [Google Scholar] [CrossRef]
- Buzoianu, S.G.; Walsh, M.C.; Rea, M.C.; O’Sullivan, O.; Cotter, P.D.; Ross, R.P.; Gardiner, G.E.; Lawlor, P.G. High-throughput sequence-based analysis of the intestinal microbiota of weanling pigs fed genetically modified MON810 maize expressing Bacillus thuringiensis Cry1Ab (Bt maize) for 31 days. Appl. Environ. Microbiol. 2012, 78, 4217–4224. [Google Scholar] [CrossRef]
- Ordiz, M.I.; Stephenson, K.; Agapova, S.; Wylie, K.M.; Maleta, K.; Martin, J.; Trehan, I.; Tarr, P.I.; Manary, M.J. Environmental Enteric Dysfunction and the Fecal Microbiota in Malawian Children. Am. J. Trop. Med. Hyg. 2017, 96, 473–476. [Google Scholar] [CrossRef]
- Hernandez-Sanabria, E.; Goonewardene, L.A.; Wang, Z.; Durunna, O.N.; Moore, S.S.; Guan, L.L. Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl. Environ. Microbiol. 2012, 78, 1203–1214. [Google Scholar] [CrossRef]
- Shili, C.N.; Broomhead, J.N.; Spring, S.C.; Lanahan, M.B.; Pezeshki, A. A Novel Corn-Expressed Phytase Improves Daily Weight Gain, Protein Efficiency Ratio and Nutrients Digestibility and Alters Fecal Microbiota in Pigs Fed with Very Low Protein Diets. Animals 2020, 10, 1926. [Google Scholar] [CrossRef] [PubMed]
- Betancur-Murillo, C.L.; Aguilar-Marín, S.B.; Jovel, J. Prevotella: A Key Player in Ruminal Metabolism. Microorganisms 2022, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Long, W.; Zhang, C.; Liu, S.; Zhao, L.; Hamaker, B.R. Fiber-Utilizing Capacity Varies in Prevotella- versus Bacteroides-Dominated Gut Microbiota. Sci. Rep. 2017, 7, 2594. [Google Scholar] [CrossRef] [PubMed]
- Sopha, S.C.; Manejwala, A.; Boutros, C.N. Sarcina, a new threat in the bariatric era. Hum. Pathol. 2015, 46, 1405–1407. [Google Scholar] [CrossRef]
- Lam-Himlin, D.; Tsiatis, A.C.; Montgomery, E.; Pai, R.K.; Brown, J.A.; Razavi, M.; Lamps, L.; Eshleman, J.R.; Bhagavan, B.; Anders, R.A. Sarcina organisms in the gastrointestinal tract: A clinicopathologic and molecular study. Am. J. Surg. Pathol. 2011, 35, 1700–1705. [Google Scholar] [CrossRef] [PubMed]
- Helm, E.T.; Gabler, N.K.; Burrough, E.R. Highly Fermentable Fiber Alters Fecal Microbiota and Mitigates Swine Dysentery Induced by Brachyspira hyodysenteriae. Animals 2021, 11, 396. [Google Scholar] [CrossRef]
- Quan, J.; Cai, G.; Ye, J.; Yang, M.; Ding, R.; Wang, X.; Zheng, E.; Fu, D.; Li, S.; Zhou, S.; et al. A global comparison of the microbiome compositions of three gut locations in commercial pigs with extreme feed conversion ratios. Sci. Rep. 2018, 8, 4536. [Google Scholar] [CrossRef]
- Horvathova, K.; Modrackova, N.; Splichal, I.; Splichalova, A.; Amin, A.; Ingribelli, E.; Killer, J.; Doskocil, I.; Pechar, R.; Kodesova, T.; et al. Defined Pig Microbiota with a Potential Protective Effect against Infection with Salmonella Typhimurium. Microorganisms 2023, 11, 1007. [Google Scholar] [CrossRef]
- Aalbaek, B. Gram-negative anaerobes in the intestinal flora of pigs. Acta Vet. Scand. 1972, 13, 228–237. [Google Scholar] [CrossRef]
- Bearson, S.M.D.; Trachsel, J.M.; Bearson, B.L.; Loving, C.L.; Kerr, B.J.; Shippy, D.C.; Kiros, T.G. Effects of β-glucan on Salmonella enterica serovar Typhimurium swine colonization and microbiota alterations. Porcine Health Manag. 2023, 9, 7. [Google Scholar] [CrossRef]
- Lima, J.; Manning, T.; Rutherford, K.M.; Baima, E.; Dewhurst, R.; Walsh, P.; Roehe, R. Taxonomic annotation of 16S rRNA sequences of pig intestinal samples using MG-RAST and QIIME2 generated different microbiota compositions. J. Microbiol. Methods 2021, 186, 106235. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Van Dixhoorn, I.; Schokker, D.; Woelders, H.; Stockhofe-Zurwieden, N.; Rebel, J.M.; Smidt, H. Environmentally enriched housing conditions affect pig welfare, immune system and gut microbiota in early life. Anim. Microbiome 2021, 3, 52. [Google Scholar] [CrossRef] [PubMed]
- González-Solé, F.; Solà-Oriol, D.; Ramayo-Caldas, Y.; Rodriguez-Prado, M.; Ortiz, G.G.; Bedford, M.R.; Pérez, J.F. Supplementation of xylo-oligosaccharides to suckling piglets promotes the growth of fiber-degrading gut bacterial populations during the lactation and nursery periods. Sci. Rep. 2022, 12, 11594. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Huang, S.; Jiang, L.; Dai, Z.; Li, T.; Han, D.; Wang, J. Characterization of the Early Life Microbiota Development and Predominant Lactobacillus Species at Distinct Gut Segments of Low- and Normal-Birth-Weight Piglets. Front. Microbiol. 2019, 10, 797. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Pu, G.; Fan, L.; Gao, C.; Lan, T.; Liu, C.; Du, T.; Kim, S.W.; Niu, P.; Zhang, Z.; et al. Identification of Gut Microbiota Affecting Fiber Digestibility in Pigs. Curr. Issues Mol. Biol. 2022, 44, 4557–4569. [Google Scholar] [CrossRef]
- Hall, H.N.; Wilkinson, D.J.; Le Bon, M. Oregano essential oil improves piglet health and performance through maternal feeding and is associated with changes in the gut microbiota. Anim. Microbiome 2021, 3, 2. [Google Scholar] [CrossRef]
- Hou, L.; Wang, J.; Zhang, W.; Quan, R.; Wang, D.; Zhu, S.; Jiang, H.; Wei, L.; Liu, J. Dynamic Alterations of Gut Microbiota in Porcine Circovirus Type 3-Infected Piglets. Front. Microbiol. 2020, 11, 1360. [Google Scholar] [CrossRef]
- Shin, D.; Chang, S.Y.; Bogere, P.; Won, K.; Choi, J.-Y.; Choi, Y.-J.; Lee, H.K.; Hur, J.; Park, B.-Y.; Kim, Y.; et al. Beneficial roles of probiotics on the modulation of gut microbiota and immune response in pigs. PLoS ONE 2019, 14, e0220843. [Google Scholar] [CrossRef]
- Hu, J.; Ma, L.; Nie, Y.; Chen, J.; Zheng, W.; Wang, X.; Xie, C.; Zheng, Z.; Wang, Z.; Yang, T.; et al. A Microbiota-Derived Bacteriocin Targets the Host to Confer Diarrhea Resistance in Early-Weaned Piglets. Cell Host Microbe 2018, 24, 817–832.e8. [Google Scholar] [CrossRef]
- Suda, Y.; Kagawa, K.; Fukuyama, K.; Elean, M.; Zhou, B.; Tomokiyo, M.; Islam, M.A.; Rajoka, M.S.R.; Kober, A.H.; Shimazu, T.; et al. Soymilk-fermented with Lactobacillus delbrueckii subsp. delbrueckii TUA4408L improves immune-health in pigs. Benef. Microbes 2022, 13, 61–72. [Google Scholar] [CrossRef]
- Wei, H.K.; Xue, H.X.; Zhou, Z.X.; Peng, J. A carvacrol-thymol blend decreased intestinal oxidative stress and influenced selected microbes without changing the messenger RNA levels of tight junction proteins in jejunal mucosa of weaning piglets. Animal 2017, 11, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Piao, X.; Ru, Y.; Han, X.; Xue, L.; Zhang, H. Effects of adding essential oil to the diet of weaned pigs on performance, nutrient utilization, immune response and intestinal health. Asian-Australas. J. Anim. Sci. 2012, 25, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, T.G.; Arfken, A.M.; Summers, K.L. Enteroendocrine peptides, growth, and the microbiome during the porcine weaning transition. Anim. Microbiome 2022, 4, 56. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ma, L.; Li, Z.; Yin, J.; Tan, B.; Chen, J.; Jiang, Q.; Ma, X. Evolution of the Gut Microbiota and Its Fermentation Characteristics of Ningxiang Pigs at the Young Stage. Animals 2021, 11, 638. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wang, Y.; Liu, S.; Huang, J.; Zhai, Z.; He, C.; Ding, J.; Wang, J.; Wang, H.; Fan, W.; et al. The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PLoS ONE 2015, 10, e0117441. [Google Scholar] [CrossRef]
- Rossi, B.; Toschi, A.; Piva, A.; Grilli, E. Single components of botanicals and nature-identical compounds as a non-antibiotic strategy to ameliorate health status and improve performance in poultry and pigs. Nutr. Res. Rev. 2020, 33, 218–234. [Google Scholar] [CrossRef]
- Rebucci, R.; Staurenghi, V.; Marchetti, L.; Giromini, C.; Bontempo, V. Effects of nature identical essential oils (carvacrol, thymol and cinnamaldehyde) on growth performance of piglets and non-invasive markers of antioxidant status and calprotectin release. Livest. Sci. 2022, 263, 104959. [Google Scholar] [CrossRef]
- Grilli, E.; Tugnoli, B.; Passey, J.L.; Stahl, C.H.; Piva, A.; Moeser, A.J. Impact of dietary organic acids and botanicals on intestinal integrity and inflammation in weaned pigs. BMC Vet. Res. 2015, 11, 96. [Google Scholar] [CrossRef]
- Dowley, A.; O’doherty, J.V.; Mukhopadhya, A.; Conway, E.; Vigors, S.; Maher, S.; Ryan, M.T.; Sweeney, T. Maternal supplementation with a casein hydrolysate and yeast beta-glucan from late gestation through lactation improves gastrointestinal health of piglets at weaning. Sci. Rep. 2022, 12, 17407. [Google Scholar] [CrossRef]
- Gao, J.; Liu, Z.; Wang, C.; Ma, L.; Chen, Y.; Li, T. Effects of Dietary Protein Level on the Microbial Composition and Metabolomic Profile in Postweaning Piglets. Oxid. Med. Cell. Longev. 2022, 2022, 3355687. [Google Scholar] [CrossRef]
- Wang, H.; Xu, R.; Zhang, H.; Su, Y.; Zhu, W. Swine gut microbiota and its interaction with host nutrient metabolism. Anim. Nutr. 2020, 6, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Rist, V.T.; Weiss, E.; Eklund, M.; Mosenthin, R. Impact of dietary protein on microbiota composition and activity in the gastrointestinal tract of piglets in relation to gut health: A review. Animal 2013, 7, 1067–1078. [Google Scholar] [CrossRef] [PubMed]


| Run | Product | Concentration |
|---|---|---|
| 1 | SIEM | Standard [14] |
| 2 | SIEM + casein | 12 g casein * |
| 3 | SIEM + PB1 | 500 μL PB1 * |
| 4 | SIEM + PB1 + casein | 12 g casein + 500 μL PB1 * |
| 5 | SIEM + PB2 | 500 μL PB2 * |
| 6 | SIEM + PB2 + casein | 12 g casein + 500 μL PB2 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Popov, I.V.; Einhardt Manzke, N.; Sost, M.M.; Verhoeven, J.; Verbruggen, S.; Chebotareva, I.P.; Ermakov, A.M.; Venema, K. Modulation of Swine Gut Microbiota by Phytogenic Blends and High Concentrations of Casein in a Validated Swine Large Intestinal In Vitro Model. Vet. Sci. 2023, 10, 677. https://doi.org/10.3390/vetsci10120677
Popov IV, Einhardt Manzke N, Sost MM, Verhoeven J, Verbruggen S, Chebotareva IP, Ermakov AM, Venema K. Modulation of Swine Gut Microbiota by Phytogenic Blends and High Concentrations of Casein in a Validated Swine Large Intestinal In Vitro Model. Veterinary Sciences. 2023; 10(12):677. https://doi.org/10.3390/vetsci10120677
Chicago/Turabian StylePopov, Igor V., Naiana Einhardt Manzke, Mônica Maurer Sost, Jessica Verhoeven, Sanne Verbruggen, Iuliia P. Chebotareva, Alexey M. Ermakov, and Koen Venema. 2023. "Modulation of Swine Gut Microbiota by Phytogenic Blends and High Concentrations of Casein in a Validated Swine Large Intestinal In Vitro Model" Veterinary Sciences 10, no. 12: 677. https://doi.org/10.3390/vetsci10120677
APA StylePopov, I. V., Einhardt Manzke, N., Sost, M. M., Verhoeven, J., Verbruggen, S., Chebotareva, I. P., Ermakov, A. M., & Venema, K. (2023). Modulation of Swine Gut Microbiota by Phytogenic Blends and High Concentrations of Casein in a Validated Swine Large Intestinal In Vitro Model. Veterinary Sciences, 10(12), 677. https://doi.org/10.3390/vetsci10120677









