Transcriptomic Changes and satP Gene Function Analysis in Pasteurella multocida with Different Levels of Resistance to Enrofloxacin
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Antibiotics, Culture Media, Bacterial Strains and Growth Conditions
2.2. Determination of the MIC of Fluoroquinolones
2.3. Amplification and Sequencing of the QRDR Genes
2.4. RNA Extraction, Library Construction, and Sequencing
2.5. Quantitative Reverse Transcription PCR (RT-qPCR) Analysis
2.6. Construction satP of the Deletion Strains and the Complemented Strains
2.7. Tolerance of Wild-Type and satP Mutant
2.8. Animal Studies
2.8.1. Pathogenicity Test in Mice
2.8.2. Acute Pathogenicity Protection Test in Mice
3. Results
3.1. Selection of Fluoroquinolone-Resistant Mutants
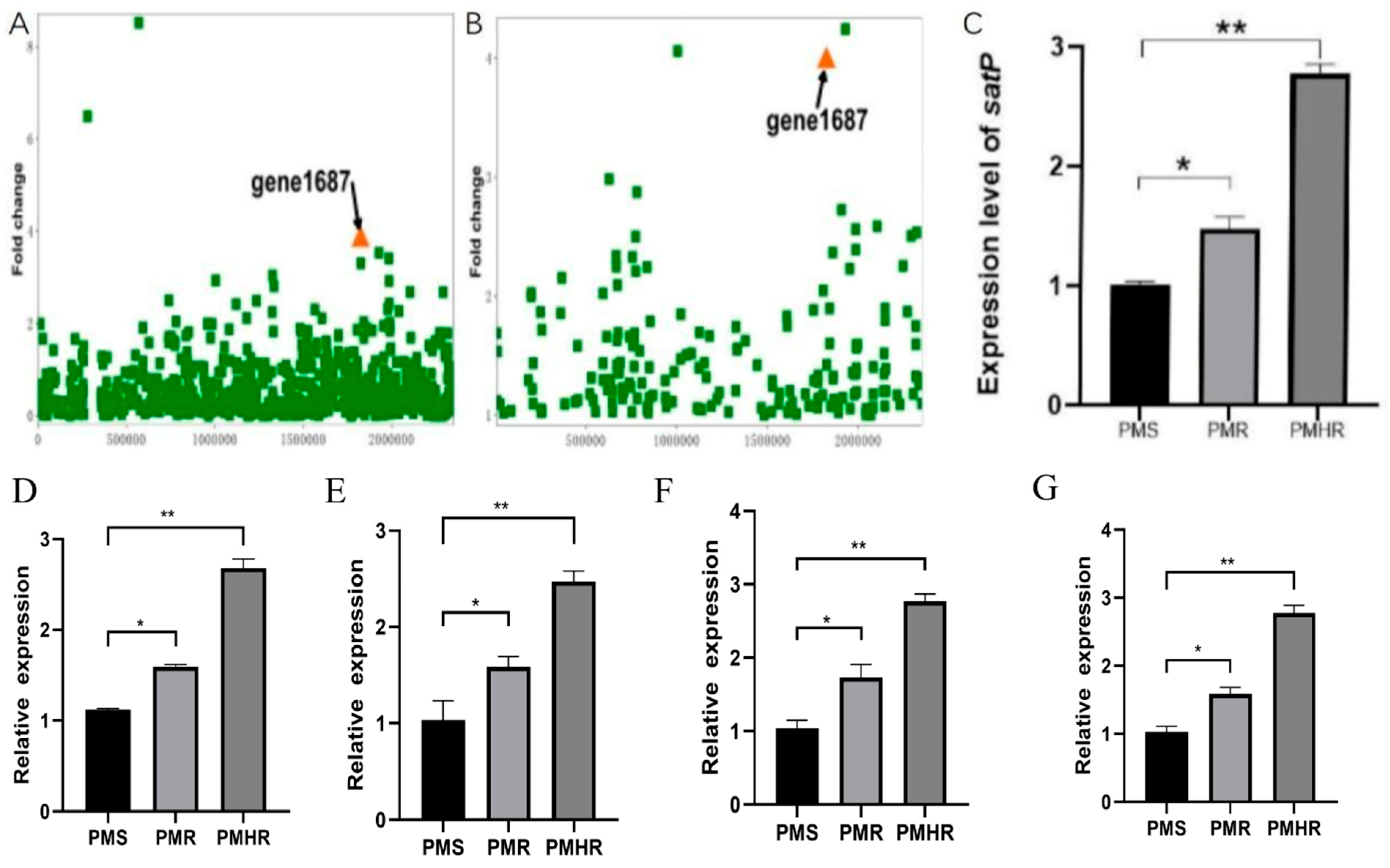
3.2. Transcriptional Profiling of PmS, PmR and PmHR by RNA Sequencing
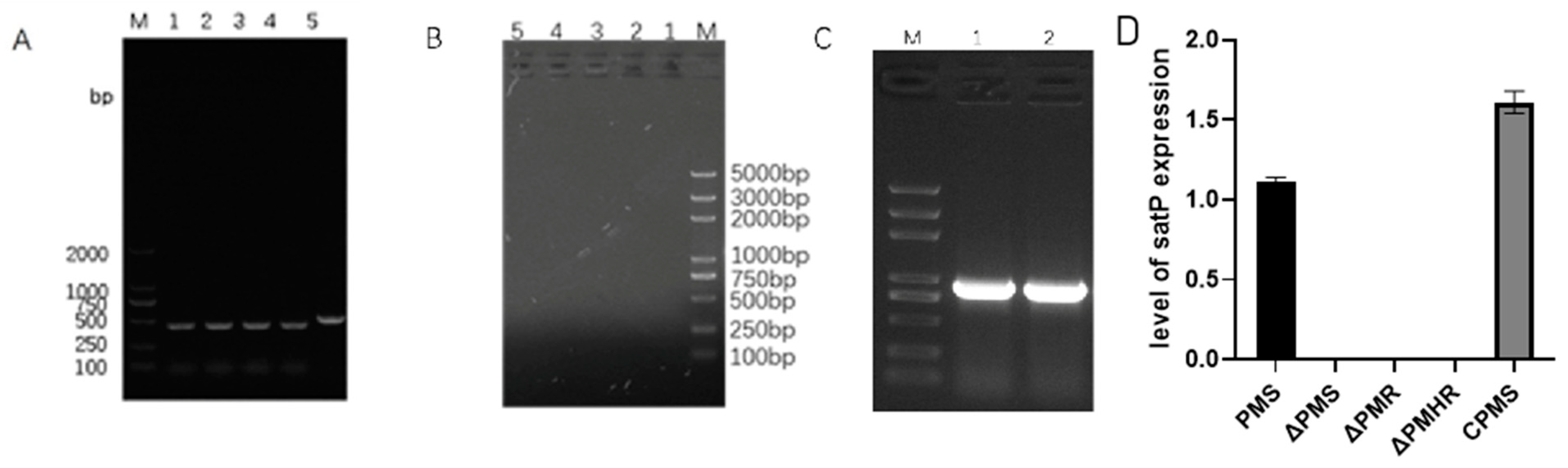
3.3. Construction and Detection of satP Strains
3.4. Effect of satP Deletion on Drug Resistance Formation
3.5. Effect of ΔPmS on Drug Tolerance Formation
3.6. Determination of Median Lethal Dose
3.7. Acute Pathogenicity Protection Test in Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Townsend, K.M.; Boyce, J.D.; Chung, J.Y.; Frost, A.J.; Adler, B. Genetic organization of Pasteurella multocida cap Loci and development of a multiplex capsular PCR typing system. J. Clin. Microbiol. 2001, 39, 924–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christenson, E.S.; Ahmed, H.M.; Durand, C.M. Pasteurella multocida infection in solid organ transplantation. Lancet Infect. Dis. 2015, 15, 235–240. [Google Scholar] [CrossRef]
- Zhao, G.; He, H.; Wang, H. Use of a recombinase polymerase amplification commercial kit for rapid visual detection of Pasteurella multocida. BMC Vet. Res. 2019, 15, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanford, K.; Zaheer, R.; Klima, C.; McAllister, T.; Peters, D.; Niu, Y.D.; Ralston, B. Antimicrobial resistance in members of the bacterial bovine respiratory disease complex isolated from lung tissue of cattle mortalities managed with or without the use of antimicrobials. Microorganisms 2020, 8, 288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elsayed, M.; Eldsouky, S.M.; Roshdy, T.; Said, L.; Thabet, N.; Allam, T.; Mohammed, A.B.A.; Nasr, G.M.; Basiouny, M.S.M.; Akl, B.A.; et al. Virulence determinants and antimicrobial profiles of Pasteurella multocida isolated from cattle and humans in Egypt. Antibiotics 2021, 10, 480. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Wang, H.; Liang, W.; Chen, Y.; Tang, X.; Chen, H.; Wu, B. A capsule/lipopolysaccharide/MLST genotype D/L6/ST11 of Pasteurella multocida is likely to be strongly associated with swine respiratory disease in China. Arch. Microbiol. 2018, 200, 107–118. [Google Scholar] [CrossRef]
- Jourquin, S.; Lowie, T.; Debruyne, F.; Chantillon, L.; Vereecke, N.; Boyen, F.; Boone, R.; Bokma, J.; Pardon, B. Dynamics of subclinical pneumonia in male dairy calves in relation to antimicrobial therapy and production outcomes. J. Dairy Sci. 2023, 106, 676–689. [Google Scholar] [CrossRef]
- Becker, J.; Perreten, V.; Schüpbach-Regula, G.; Stucki, D.; Steiner, A.; Meylan, M. Associations of antimicrobial use with antimicrobial susceptibility at the calf level in bacteria isolated from the respiratory and digestive tracts of veal calves before slaughter. J. Antimicrob. Chemother. 2022, 77, 2859–2866. [Google Scholar] [CrossRef]
- Wang, Z.; Kong, L.C.; Jia, B.Y.; Liu, S.M.; Jiang, X.Y.; Ma, H.X. Aminoglycoside susceptibility of Pasteurella multocida isolates from bovine respiratory infections in China and mutations in ribosomal protein S5 associated with high-level induced spectinomycin resistance. J. Vet. Med. Sci. 2017, 79, 1678–1681. [Google Scholar] [CrossRef] [Green Version]
- Kong, L.C.; Wang, Z.; Wang, Y.M.; Dong, W.L.; Jia, B.Y.; Gao, D.; Jiang, X.Y.; Ma, H.X. Antimicrobial susceptibility and molecular typing of Pasteurella multocida isolated from six provinces in China. Trop. Anim. Health Prod. 2019, 51, 987–992. [Google Scholar] [CrossRef]
- Khamesipour, F.; Momtaz, H.; Azhdary Mamoreh, M. Occurrence of virulence factors and antimicrobial resistance in Pasteurella multocida strains isolated from slaughter cattle in Iran. Front. Microbiol. 2014, 5, 536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coetzee, J.F.; Magstadt, D.R.; Sidhu, P.K.; Follett, L.; Schuler, A.M.; Krull, A.C.; Cooper, V.L.; Engelken, T.J.; Kleinhenz, M.D.; O’Connor, A.M. Association between antimicrobial drug class for treatment and retreatment of bovine respiratory disease (BRD) and frequency of resistant BRD pathogen isolation from veterinary diagnostic laboratory samples. PLoS ONE 2019, 14, e0219104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, D.; Yuan, D.; Wang, M.; Jia, R.; Chen, S.; Liu, M.; Zhao, X.; Yang, Q.; Wu, Y.; Zhang, S.; et al. Emergence of a multidrug-resistant hypervirulent Pasteurella multocida ST342 strain with a floR-carrying plasmid. J. Glob. Antimicrob. Resist. 2020, 20, 348–350. [Google Scholar] [CrossRef] [PubMed]
- Timsit, E.; Hallewell, J.; Booker, C.; Tison, N.; Amat, S.; Alexander, T.W. Prevalence and antimicrobial susceptibility of Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni isolated from the lower respiratory tract of healthy feedlot cattle and those diagnosed with bovine respiratory disease. Vet. Microbiol. 2017, 208, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Blondeau, J.M.; Fitch, S.D. Mutant prevention and minimum inhibitory concentration drug values for enrofloxacin, ceftiofur, florfenicol, tilmicosin and tulathromycin tested against swine pathogens Actinobacillus pleuropneumoniae, Pasteurella multocida and Streptococcus suis. PLoS ONE 2019, 14, e0210154. [Google Scholar] [CrossRef] [Green Version]
- Balaje, R.M.; Sidhu, P.K.; Kaur, G.; Rampal, S. Mutant prevention concentration and PK-PD relationships of enrofloxacin for Pasteurella multocida in buffalo calves. Res. Vet. Sci. 2013, 95, 1114–1124. [Google Scholar] [CrossRef]
- Michael, G.B.; Kadlec, K.; Sweeney, M.T.; Brzuszkiewicz, E.; Liesegang, H.; Daniel, R.; Murray, R.W.; Watts, J.L.; Schwarz, S. ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: Analysis of the regions that comprise 12 antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 84–90. [Google Scholar] [CrossRef]
- Hooper, D.C.; Jacoby, G.A. Mechanisms of drug resistance: Quinolone resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 12–31. [Google Scholar] [CrossRef] [Green Version]
- Cárdenas, M.; Barbé, J.; Llagostera, M.; Miró, E.; Navarro, F.; Mirelis, B.; Prats, G.; Badiola, I. Quinolone resistance-determining regions of gyrA and parC in Pasteurella multocida strains with different levels of nalidixic acid resistance. Antimicrob. Agents Chemother. 2001, 45, 990–991. [Google Scholar] [CrossRef] [Green Version]
- Fuzi, M.; Szabo, D.; Csercsik, R. Double-serine fluoroquinolone resistance mutations advance major international clones and lineages of various multi-drug resistant bacteria. Front. Microbiol. 2017, 8, 2261. [Google Scholar] [CrossRef]
- Kong, L.C.; Gao, D.; Gao, Y.H.; Liu, S.M.; Ma, H.X. Fluoroquinolone resistance mechanism of clinical isolates and selected mutants of Pasteurella multocida from bovine respiratory disease in China. J. Vet. Med. Sci. 2014, 76, 1655–1657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosas, N.C.; Lithgow, T. Targeting bacterial outer-membrane remodelling to impact antimicrobial drug resistance. Trends Microbiol. 2022, 30, 544–552. [Google Scholar] [CrossRef]
- Rodrigues, I.C.; Rodrigues, S.C.; Duarte, F.V.; Costa, P.M.D.; Costa, P.M.D. The role of outer membrane proteins in UPEC antimicrobial resistance: A systematic review. Membranes 2022, 12, 981. [Google Scholar] [CrossRef] [PubMed]
- Rattanapanadda, P.; Kuo, H.C.; Chang, S.K.; Tell, L.A.; Shia, W.Y.; Chou, C.C. Effect of carbonyl cyanide chlorophenylhydrazone on intrabacterial concentration and antimicrobial activity of amphenicols against swine resistant Actinobacillus pleuropneumoniae and Pasteurella multocida. Vet. Res. Commun. 2022, 46, 903–916. [Google Scholar] [CrossRef]
- Nishino, K.; Yamasaki, S.; Nakashima, R.; Zwama, M.; Hayashi-Nishino, M. Function and inhibitory mechanisms of multidrug efflux pumps. Front. Microbiol. 2021, 12, 737288. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Plésiat, P.; Nikaido, H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef] [Green Version]
- Shin, B.; Park, C.; Park, W. Stress responses linked to antimicrobial resistance in Acinetobacter species. Appl. Microbiol. Biotechnol. 2020, 104, 1423–1435. [Google Scholar] [CrossRef]
- Cao, L.; Wang, J.; Sun, L.; Kong, Z.; Wu, Q.; Wang, Z. Transcriptional analysis reveals the relativity of acid tolerance and antimicrobial peptide resistance of Salmonella. Microb. Pathog. 2019, 136, 103701. [Google Scholar] [CrossRef]
- Lewis, K.; Shan, Y. Why tolerance invites resistance. Science 2017, 355, 796. [Google Scholar] [CrossRef]
- Beaber, J.W.; Hochhut, B.; Waldor, M.K. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 2004, 427, 72–74. [Google Scholar] [CrossRef]
- Alam, M.K.; Alhhazmi, A.; DeCoteau, J.F.; Luo, Y.; Geyer, C.R. RecA inhibitors potentiate antibiotic activity and block evolution of antibiotic resistance. Cell Chem. Biol. 2016, 23, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Thomas, E.; Grandemange, E.; Pommier, P.; Wessel-Robert, S.; Davot, J.L. Field evaluation of efficacy and tolerance of a 2% marbofloxacin injectable solution for the treatment of respiratory disease in fattening pigs. Vet. Q. 2000, 22, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Westfall, C.; Flores-Mireles, A.L.; Robinson, J.I.; Lynch, A.J.L.; Hultgren, S.; Henderson, J.P.; Levin, P.A. The widely used antimicrobial triclosan induces high levels of antibiotic tolerance in vitro and reduces antibiotic efficacy up to 100-fold in vivo. Antimicrob. Agents Chemother. 2019, 63, e02312-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cirz, R.T.; Chin, J.K.; Andes, D.R.; de Crécy-Lagard, V.; Craig, W.A.; Romesberg, F.E. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005, 3, e176. [Google Scholar] [CrossRef] [Green Version]
- Zuluaga, A.F.; Salazar, B.E.; Rodriguez, C.A.; Zapata, A.X.; Agudelo, M.; Vesga, O. Neutropenia induced in outbred mice by a simplified low-dose cyclophosphamide regimen: Characterization and applicability to diverse experimental models of infectious diseases. BMC Infect. Dis. 2006, 6, 55. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Zhao, Z.; Hu, J.; Wu, B.; Cai, X.; He, Q.; Chen, H. Isolation, antimicrobial resistance, and virulence genes of Pasteurella multocida strains from swine in China. J. Clin. Microbiol. 2009, 47, 951–958. [Google Scholar] [CrossRef] [Green Version]
- Kong, L.C.; Gao, D.; Jia, B.Y.; Wang, Z.; Gao, Y.H.; Pei, Z.H.; Liu, S.M.; Xin, J.Q.; Ma, H.X. Antimicrobial susceptibility and molecular characterization of macrolide resistance of Mycoplasma bovis isolates from multiple provinces in China. J. Vet. Med. Sci. 2016, 78, 293–296. [Google Scholar] [CrossRef] [Green Version]
- Sun, P.; Li, J.; Zhang, X.; Guan, Z.; Xiao, Q.; Zhao, C.; Song, M.; Zhou, Y.; Mou, L.; Ke, M.; et al. Crystal structure of the bacterial acetate transporter SatP reveals that it forms a hexameric channel. J. Biol. Chem. 2018, 293, 19492–19500. [Google Scholar] [CrossRef] [Green Version]
- Indrajith, S.; Mukhopadhyay, A.K.; Chowdhury, G.; Farraj, D.A.A.; Alkufeidy, R.M.; Natesan, S.; Meghanathan, V.; Gopal, S.; Muthupandian, S. Molecular insights of carbapenem resistance Klebsiella pneumoniae isolates with focus on multidrug resistance from clinical samples. J. Infect. Public Health 2021, 14, 131–138. [Google Scholar] [CrossRef]
- Yamamoto, K.; Nakata, N.; Mukai, T.; Kawagishi, I.; Ato, M. Coexpression of MmpS5 and MmpL5 contributes to both efflux transporter MmpL5 trimerization and drug resistance in Mycobacterium tuberculosis. mSphere 2021, 6. [Google Scholar] [CrossRef]
- Li, W.; Wang, G.; Zhang, S.; Fu, Y.; Jiang, Y.; Yang, X.; Lin, X. An integrated quantitative proteomic and metabolomics approach to reveal the negative regulation mechanism of LamB in antibiotics resistance. J. Proteom. 2019, 194, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Safi, H.; Gopal, P.; Lingaraju, S.; Ma, S.; Levine, C.; Dartois, V.; Yee, M.; Li, L.; Blanc, L.; Ho Liang, H.P.; et al. Phase variation in Mycobacterium tuberculosis glpK produces transiently heritable drug tolerance. Proc. Natl. Acad. Sci. USA 2019, 116, 19665–19674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petes, C.; Mintsopoulos, V.; Finnen, R.L.; Banfield, B.W.; Gee, K. The effects of CD14 and IL-27 on induction of endotoxin tolerance in human monocytes and macrophages. J. Biol. Chem. 2018, 293, 17631–17645. [Google Scholar] [CrossRef] [Green Version]
- Poudyal, B.; Sauer, K. The ABC of Biofilm Drug Tolerance: The MerR-like regulator BrlR is an activator of ABC transport systems, with PA1874-77 contributing to the tolerance of Pseudomonas aeruginosa biofilms to tobramycin. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef]
- Ronneau, S.; Hallez, R. Make and break the alarmone: Regulation of (p)ppGpp synthetase/hydrolase enzymes in bacteria. FEMS Microbiol. Rev. 2019, 43, 389–400. [Google Scholar] [CrossRef] [Green Version]
- Niu, Y.; Wang, K.; Zheng, S.; Wang, Y.; Ren, Q.; Li, H.; Ding, L.; Li, W.; Zhang, L. Antibacterial effect of caffeic acid phenethyl ester on Cariogenic Bacteria and Streptococcus mutans biofilms. Antimicrob. Agents Chemother. 2020, 64, e00251-2. [Google Scholar] [CrossRef] [PubMed]
- Zhan, L.; Zhang, J.; Zhao, B.; Li, X.; Zhang, X.; Hu, R.; Elken, E.M.; Kong, L.; Gao, Y. Genomic and transcriptomic analysis of bovine Pasteurella multocida serogroup a strain reveals insights into virulence attenuation. Front. Vet. Sci. 2021, 8, 765495. [Google Scholar] [CrossRef]
- Ma, D.; Mandell, J.B.; Donegan, N.P.; Cheung, A.L.; Ma, W.; Rothenberger, S.; Shanks, R.M.Q.; Richardson, A.R.; Urish, K.L. The Toxin-antitoxin MazEF drives Staphylococcus aureus biofilm formation, antibiotic tolerance, and chronic infection. mBio 2019, 10. [Google Scholar] [CrossRef] [Green Version]

| Strains | MIC Range (µg/mL) | Substitution in | ||||||
|---|---|---|---|---|---|---|---|---|
| CIP | ENR | ENR + CCCP | ENR + Verapamil | GyrA | GyrB | ParC | ParE | |
| PmS | 0.25 | 0.25 | 0.125 | 0.125 | - | - | - | - |
| PmR | 4 | 4 | 2 | 2 | Arg88Ile | Ser467Phe | Glu84Lys | - |
| PmHR | 64 | 64 | 64 | 64 | Arg88Ile | Ser467Phe | Glu84Lys | - |
| Strains | MIC Range (µg/mL) | Substitution in | ||||||
|---|---|---|---|---|---|---|---|---|
| CIP | ENR | ENR + CCCP | ENR + Verapamil | GyrA | GyrB | ParC | ParE | |
| ΔPmS | 0.25 | 0.25 | 0.125 | 0.125 | - | - | - | - |
| ΔPmR | 2 | 2 | 2 | 2 | Arg88Ile | Ser467Phe | Glu84Lys | - |
| ΔPmHR | 64 | 64 | 64 | 64 | Arg88Ile | Ser467Phe | Glu84Lys | - |
| Strain | Mutation Frequency (×10−8) (X ± SD) |
|---|---|
| PmS | 0.3 ± 0.1 |
| ΔPmS | - |
| PmR | 6.4 ± 2.8 |
| ΔPmR | 3.7 ± 1.5 |
| PmHR | 14.0 ± 5.8 |
| ΔPmHR | 8.2 ± 1.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.-S.; Qi, Y.; Xue, J.-Z.; Xu, G.-Y.; Xu, Y.-X.; Li, X.-Y.; Muhammad, I.; Kong, L.-C.; Ma, H.-X. Transcriptomic Changes and satP Gene Function Analysis in Pasteurella multocida with Different Levels of Resistance to Enrofloxacin. Vet. Sci. 2023, 10, 257. https://doi.org/10.3390/vetsci10040257
Li X-S, Qi Y, Xue J-Z, Xu G-Y, Xu Y-X, Li X-Y, Muhammad I, Kong L-C, Ma H-X. Transcriptomic Changes and satP Gene Function Analysis in Pasteurella multocida with Different Levels of Resistance to Enrofloxacin. Veterinary Sciences. 2023; 10(4):257. https://doi.org/10.3390/vetsci10040257
Chicago/Turabian StyleLi, Xue-Song, Yu Qi, Jun-Ze Xue, Guan-Yi Xu, Yu-Xuan Xu, Xuan-Yu Li, Inam Muhammad, Ling-Cong Kong, and Hong-Xia Ma. 2023. "Transcriptomic Changes and satP Gene Function Analysis in Pasteurella multocida with Different Levels of Resistance to Enrofloxacin" Veterinary Sciences 10, no. 4: 257. https://doi.org/10.3390/vetsci10040257
APA StyleLi, X.-S., Qi, Y., Xue, J.-Z., Xu, G.-Y., Xu, Y.-X., Li, X.-Y., Muhammad, I., Kong, L.-C., & Ma, H.-X. (2023). Transcriptomic Changes and satP Gene Function Analysis in Pasteurella multocida with Different Levels of Resistance to Enrofloxacin. Veterinary Sciences, 10(4), 257. https://doi.org/10.3390/vetsci10040257





