Temporal Variability of the Dominant Fecal Microbiota in Healthy Adult Cats
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal and Sample Collection
2.2. Quantitative PCR Analysis and Feline Dysbiosis Index
2.3. Fecal Concentrations of Unconjugated Bile Acids
2.4. Statistical Analysis
3. Results
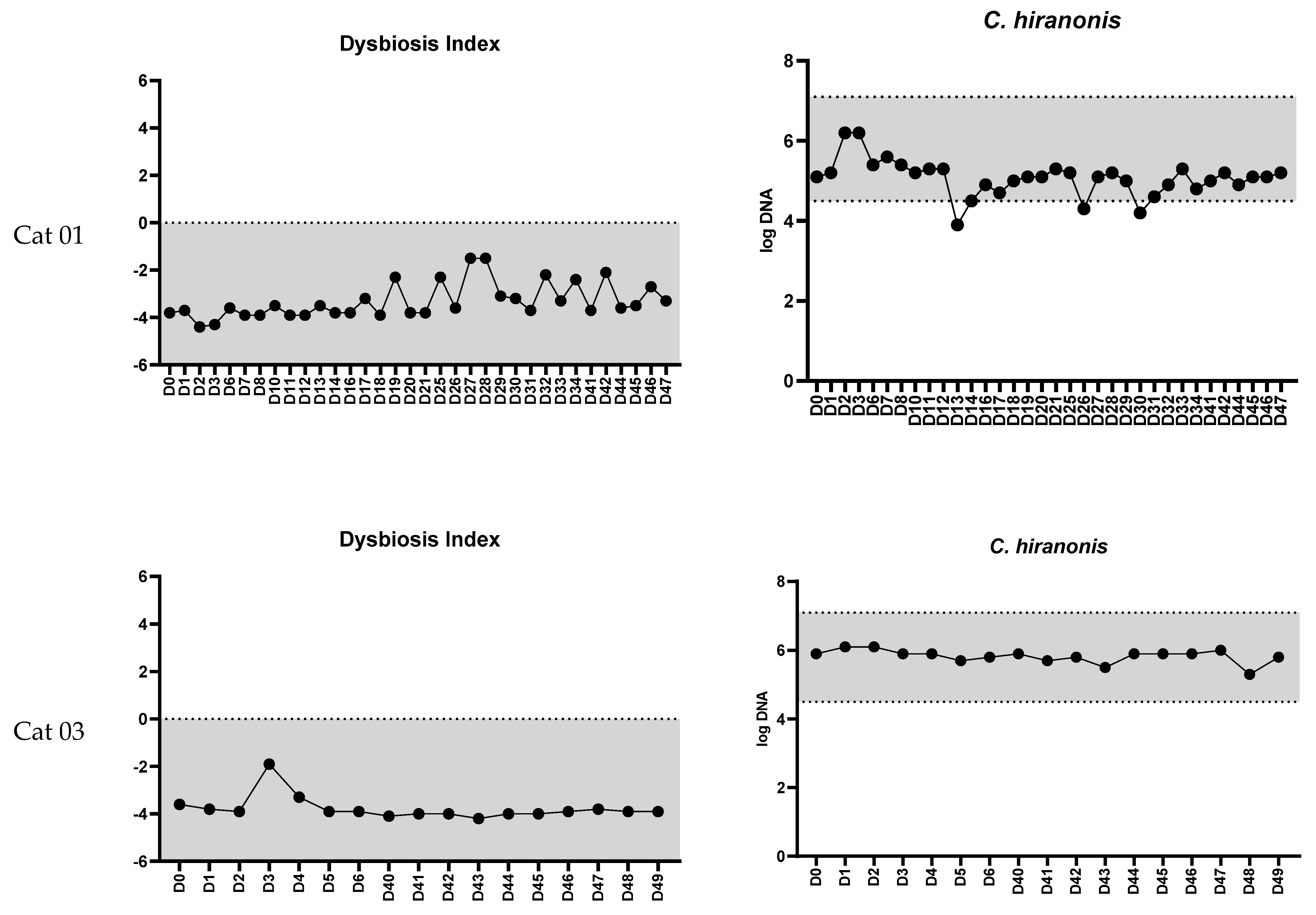
3.1. Feline Dysbiosis Index and the Abundances of Targeted Bacterial Groups
3.2. Relationship between the Fecal Bile Acid Profile and the Abundance of Clostridium hiranonis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tizard, I.R.; Jones, S.W. The microbiota regulates immunity and immunologic diseases in dogs and cats. Vet. Clin. N. Am. Small Anim. Pract. 2018, 48, 307–322. [Google Scholar] [CrossRef]
- Lyu, Y.; Su, C.; Verbrugghe, A.; Van de Wiele, T.; Martos Martinez-Caja, A.; Hesta, M. Past, present, and future of gastrointestinal microbiota research in cats. Front. Microbiol. 2020, 11, 1661. [Google Scholar] [CrossRef]
- Marsilio, S.; Pilla, R.; Sarawichitr, B.; Chow, B.; Hill, S.L.; Ackermann, M.R.; Estep, J.S.; Lidbury, J.A.; Steiner, J.M.; Suchodolski, J.S. Characterization of the fecal microbiome in cats with inflammatory bowel disease or alimentary small cell lymphoma. Sci. Rep. 2019, 9, 19208. [Google Scholar] [CrossRef]
- Kathrani, A.; Yen, S.; Swann, J.R.; Hall, E.J. The effect of a hydrolyzed protein diet on the fecal microbiota in cats with chronic enteropathy. Sci. Rep. 2022, 12, 2746. [Google Scholar] [CrossRef]
- Bai, H.; Liu, T.; Wang, S.; Gong, W.; Shen, L.; Zhang, S.; Wang, Z. Identification of gut microbiome and metabolites associated with acute diarrhea in cats. Microbiol. Spectr. 2023, 11, e0059023. [Google Scholar] [CrossRef]
- Kim, Y.; Carrai, M.; Leung, M.H.Y.; Chin, J.; Li, J.; Lee, P.K.H.; Beatty, J.A.; Pfeiffer, D.U.; Barrs, V.R. Dysbiosis of the urinary bladder microbiome in cats with chronic kidney disease. mSystems 2021, 6, e0051021. [Google Scholar] [CrossRef]
- Stavroulaki, E.M.; Suchodolski, J.S.; Pilla, R.; Fosgate, G.T.; Sung, C.-H.; Lidbury, J.A.; Steiner, J.M.; Xenoulis, P.G. Short-and long-term effects of amoxicillin/clavulanic acid or doxycycline on the gastrointestinal microbiome of growing cats. PLoS ONE 2021, 16, e0253031. [Google Scholar] [CrossRef]
- Claesson, M.J.; Cusack, S.; O’Sullivan, O.; Greene-Diniz, R.; de Weerd, H.; Flannery, E.; Marchesi, J.R.; Falush, D.; Dinan, T.; Fitzgerald, G.; et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4586–4591. [Google Scholar] [CrossRef]
- Faith, J.J.; Guruge, J.L.; Charbonneau, M.; Subramanian, S.; Seedorf, H.; Goodman, A.L.; Clemente, J.C.; Knight, R.; Heath, A.C.; Leibel, R.L. The long-term stability of the human gut microbiota. Science 2013, 341, 1237439. [Google Scholar] [CrossRef]
- Jeffery, I.B.; Lynch, D.B.; O’Toole, P.W. Composition and temporal stability of the gut microbiota in older persons. ISME J. 2016, 10, 170–182. [Google Scholar] [CrossRef]
- Fragiadakis, G.K.; Wastyk, H.C.; Robinson, J.L.; Sonnenburg, E.D.; Sonnenburg, J.L.; Gardner, C.D. Long-term dietary intervention reveals resilience of the gut microbiota despite changes in diet and weight. Am. J. Clin. Nutr. 2020, 111, 1127–1136. [Google Scholar] [CrossRef] [PubMed]
- De La Cochetière, M.-F.; Durand, T.; Lepage, P.; Bourreille, A.; Galmiche, J.; Dore, J. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J. Clin. Microbiol. 2005, 43, 5588–5592. [Google Scholar] [CrossRef] [PubMed]
- Jalanka-Tuovinen, J.; Salonen, A.; Nikkilä, J.; Immonen, O.; Kekkonen, R.; Lahti, L.; Palva, A.; De Vos, W.M. Intestinal microbiota in healthy adults: Temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS ONE 2011, 6, e23035. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; Relman, D.A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. USA 2011, 108, 4554–4561. [Google Scholar] [CrossRef] [PubMed]
- Schmid, S.M.; Suchodolski, J.S.; Price, J.M.; Tolbert, M.K. Omeprazole minimally alters the fecal microbial community in six cats: A pilot study. Front. Vet. Sci. 2018, 5, 79. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mazcorro, J.F.; Lanerie, D.J.; Dowd, S.E.; Paddock, C.G.; Grutzner, N.; Steiner, J.M.; Ivanek, R.; Suchodolski, J.S. Effect of a multi-species synbiotic formulation on fecal bacterial microbiota of healthy cats and dogs as evaluated by pyrosequencing. FEMS Microbiol. Ecol. 2011, 78, 542–554. [Google Scholar] [CrossRef] [PubMed]
- Whittemore, J.C.; Stokes, J.E.; Laia, N.L.; Price, J.M.; Suchodolski, J.S. Short and long-term effects of a synbiotic on clinical signs, the fecal microbiome, and metabolomic profiles in healthy research cats receiving clindamycin: A randomized, controlled trial. PeerJ 2018, 6, e5130. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, I.B.; Das, A.; O’Herlihy, E.; Coughlan, S.; Cisek, K.; Moore, M.; Bradley, F.; Carty, T.; Pradhan, M.; Dwibedi, C.; et al. Differences in fecal microbiomes and metabolomes of people with vs. without irritable bowel syndrome and bile acid malabsorption. Gastroenterology 2020, 158, 1016–1028.e8. [Google Scholar] [CrossRef]
- Filimoniuk, A.; Daniluk, U.; Samczuk, P.; Wasilewska, N.; Jakimiec, P.; Kucharska, M.; Lebensztejn, D.M.; Ciborowski, M. Metabolomic profiling in children with inflammatory bowel disease. Adv. Med. Sci. 2020, 65, 65–70. [Google Scholar] [CrossRef]
- Kathrani, A.; Yen, S.; Hall, E.J.; Swann, J.R. The effects of a hydrolyzed protein diet on the plasma, fecal and urine metabolome in cats with chronic enteropathy. Sci. Rep. 2023, 13, 19979. [Google Scholar] [CrossRef]
- Huijghebaert, S.M.; Hofmann, A.F. Influence of the amino acid moiety on deconjugation of bile acid amidates by cholylglycine hydrolase or human fecal cultures. J. Lipid Res. 1988, 27, 742–752. [Google Scholar] [CrossRef]
- Stellwag, E.; Hylemon, P. 7alpha-dehydroxylation of cholic acid and chenodeoxycholic acid by Clostridium leptum. J. Lipid Res. 1979, 20, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.B.; Lajczak, N.K.; Kelly, O.B.; O’Dwyer, A.M.; Giddam, A.K.; Ní Gabhann, J.; Franco, P.; Tambuwala, M.M.; Jefferies, C.A.; Keely, S. Ursodeoxycholic acid and lithocholic acid exert anti-inflammatory actions in the colon. Am. J. Physiol.—Gastrointest. Liver Physiol. 2017, 312, G550–G558. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Sun, X.; Oh, S.F.; Wu, M.; Zhang, Y.; Zheng, W.; Geva-Zatorsky, N.; Jupp, R.; Mathis, D.; Benoist, C.; et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 2020, 577, 410–415. [Google Scholar] [CrossRef]
- Sinha, S.R.; Haileselassie, Y.; Nguyen, L.P.; Tropini, C.; Wang, M.; Becker, L.S.; Sim, D.; Jarr, K.; Spear, E.T.; Singh, G.; et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe 2020, 27, 659–670.e5. [Google Scholar] [CrossRef] [PubMed]
- Manchester, A.C.; Chow, L.; Wheat, W.; Dow, S. Modulation of in vitro macrophage responses via primary and secondary bile acids in dogs. Animals 2023, 13, 3714. [Google Scholar] [CrossRef] [PubMed]
- Marion, S.; Studer, N.; Desharnais, L.; Menin, L.; Escrig, S.; Meibom, A.; Hapfelmeier, S.; Bernier-Latmani, R. In Vitro and in vivo characterization of Clostridium scindens bile acid transformations. Gut Microbes 2019, 10, 481–503. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.-J.; Hylemon, P.B. Isolation and characterization of a bile acid inducible 7α-dehydroxylating operon in Clostridium hylemonae TN271. Anaerobe 2010, 16, 137–146. [Google Scholar] [CrossRef]
- Kitahara, M.; Takamine, F.; Imamura, T.; Benno, Y. Clostridium hiranonis sp. nov., a human intestinal bacterium with bile acid 7alpha-dehydroxylating activity. Int. J. Syst. Evol. Microbiol. 2001, 51, 39–44. [Google Scholar] [CrossRef]
- Comito, R.; Porru, E.; Interino, N.; Conti, M.; Terragni, R.; Gotti, R.; Candela, M.; Simoni, P.; Roda, A.; Fiori, J. Metabolic bile acid profile impairments in dogs affected by chronic inflammatory enteropathy. Metabolites 2023, 13, 980. [Google Scholar] [CrossRef]
- Sung, C.H.; Pilla, R.; Marsilio, S.; Chow, B.; Zornow, K.A.; Slovak, J.E.; Lidbury, J.A.; Steiner, J.M.; Hill, S.L.; Suchodolski, J.S. Fecal Concentrations of Long-Chain Fatty Acids, Sterols, and Unconjugated Bile Acids in Cats with Chronic Enteropathy. Animals 2023, 13, 2753. [Google Scholar] [CrossRef]
- Stavroulaki, E.M.; Suchodolski, J.S.; Pilla, R.; Fosgate, G.T.; Sung, C.-H.; Lidbury, J.; Steiner, J.M.; Xenoulis, P.G. The serum and fecal metabolomic profiles of growing kittens treated with amoxicillin/clavulanic acid or doxycycline. Animals 2022, 12, 330. [Google Scholar] [CrossRef] [PubMed]
- Weingarden, A.R.; Dosa, P.I.; DeWinter, E.; Steer, C.J.; Shaughnessy, M.K.; Johnson, J.R.; Khoruts, A.; Sadowsky, M.J. Changes in colonic bile acid composition following fecal microbiota transplantation are sufficient to control Clostridium difficile germination and growth. PLoS ONE 2016, 11, e0147210. [Google Scholar] [CrossRef] [PubMed]
- Staley, C.; Weingarden, A.R.; Khoruts, A.; Sadowsky, M.J. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl. Microbiol. Biotechnol. 2017, 101, 47–64. [Google Scholar] [CrossRef] [PubMed]
- Islam, K.S.; Fukiya, S.; Hagio, M.; Fujii, N.; Ishizuka, S.; Ooka, T.; Ogura, Y.; Hayashi, T.; Yokota, A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 2011, 141, 1773–1781. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Larouche-Lebel, É.; Loughran, K.A.; Huh, T.P.; Suchodolski, J.S.; Oyama, M.A. Gut dysbiosis and its associations with gut microbiota-derived metabolites in dogs with myxomatous mitral valve disease. mSystems 2021, 6, e00111–e00121. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Martins, R.; Sullivan, M.C.; Friedman, E.S.; Misic, A.M.; El-Fahmawi, A.; De Martinis, E.C.P.; O’Brien, K.; Chen, Y.; Bradley, C.; et al. Diet-induced remission in chronic enteropathy is associated with altered microbial community structure and synthesis of secondary bile acids. Microbiome 2019, 7, 126. [Google Scholar] [CrossRef]
- Tremblay, J.; Singh, K.; Fern, A.; Kirton, E.S.; He, S.; Woyke, T.; Lee, J.; Chen, F.; Dangl, J.L.; Tringe, S.G. Primer and platform effects on 16S rRNA tag sequencing. Front. Microbiol. 2015, 6, 771. [Google Scholar] [CrossRef]
- D’Amore, R.; Ijaz, U.Z.; Schirmer, M.; Kenny, J.G.; Gregory, R.; Darby, A.C.; Shakya, M.; Podar, M.; Quince, C.; Hall, N. A comprehensive benchmarking study of protocols and sequencing platforms for 16S rRNA community profiling. BMC Genom. 2016, 17, 55. [Google Scholar] [CrossRef]
- Sung, C.H.; Marsilio, S.; Chow, B.; Zornow, K.A.; Slovak, J.E.; Pilla, R.; Lidbury, J.A.; Steiner, J.M.; Park, S.Y.; Hong, M.P.; et al. Dysbiosis index to evaluate the fecal microbiota in healthy cats and cats with chronic enteropathies. J. Feline Med. Surg. 2022, 24, e1–e12. [Google Scholar] [CrossRef]
- Rajilić-Stojanović, M.; Heilig, H.G.; Tims, S.; Zoetendal, E.G.; de Vos, W.M. Long-term monitoring of the human intestinal microbiota composition. Environ. Microbiol. 2013, 15, 1146–1159. [Google Scholar] [CrossRef] [PubMed]
- Martinson, J.N.; Pinkham, N.V.; Peters, G.W.; Cho, H.; Heng, J.; Rauch, M.; Broadaway, S.C.; Walk, S.T. Rethinking gut microbiome residency and the Enterobacteriaceae in healthy human adults. ISME J. 2019, 13, 2306–2318. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.H.M.P. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar]
- Sung, C.-H.; Pilla, R.; Chen, C.-C.; Ishii, P.E.; Toresson, L.; Allenspach-Jorn, K.; Jergens, A.E.; Summers, S.; Swanson, K.S.; Volk, H. Correlation between Targeted qPCR Assays and Untargeted DNA Shotgun Metagenomic Sequencing for Assessing the Fecal Microbiota in Dogs. Animals 2023, 13, 2597. [Google Scholar] [CrossRef] [PubMed]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013, 62, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Gilliland, S.; Speck, M. Deconjugation of bile acids by intestinal lactobacilli. Appl. Environ. Microbiol. 1977, 33, 15–18. [Google Scholar] [CrossRef]
- Marteau, P.; Gerhardt, M.; Myara, A.; Bouvier, E.; Trivin, F.; Rambaud, J. Metabolism of bile salts by alimentary bacteria during transit in the human small intestine. Microb. Ecol. Health Dis. 1995, 8, 151–157. [Google Scholar] [CrossRef]
- Lewis, R.; Gorbach, S. Modification of bile acids by intestinal bacteria. AMA Arch. Intern. Med. 1972, 130, 545–549. [Google Scholar] [CrossRef]
- Guard, B.C.; Honneffer, J.B.; Jergens, A.E.; Jonika, M.M.; Toresson, L.; Lawrence, Y.A.; Webb, C.B.; Hill, S.; Lidbury, J.A.; Steiner, J.M. Longitudinal assessment of microbial dysbiosis, fecal unconjugated bile acid concentrations, and disease activity in dogs with steroid-responsive chronic inflammatory enteropathy. J. Vet. Intern. Med. 2019, 33, 1295–1305. [Google Scholar] [CrossRef]
- Pilla, R.; Gaschen, F.P.; Barr, J.W.; Olson, E.; Honneffer, J.; Guard, B.C.; Blake, A.B.; Villanueva, D.; Khattab, M.R.; AlShawaqfeh, M.K.; et al. Effects of metronidazole on the fecal microbiome and metabolome in healthy dogs. J. Vet. Intern. Med. 2020, 34, 1853–1866. [Google Scholar] [CrossRef]
- 2022 ACVIM Forum Research Abstract Program. J. Vet. Intern. Med. 2022, 36, 2282–2454. [CrossRef] [PubMed]
- Werner, M.; Ishii, P.E.; Pilla, R.; Lidbury, J.A.; Steiner, J.M.; Busch-Hahn, K.; Unterer, S.; Suchodolski, J.S. Prevalence of Clostridioides difficile in Canine Feces and Its Association with Intestinal Dysbiosis. Animals 2023, 13, 2441. [Google Scholar] [CrossRef] [PubMed]
- Theriot, C.M.; Bowman, A.A.; Young, V.B. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere 2016, 1, e00045-15. [Google Scholar] [CrossRef] [PubMed]
- Watson, V.E.; Jacob, M.E.; Bruno-Barcena, J.M.; Amirsultan, S.; Stauffer, S.H.; Piqueras, V.O.; Frias, R.; Gookin, J.L. Influence of the intestinal microbiota on disease susceptibility in kittens with experimentally-induced carriage of atypical enteropathogenic Escherichia coli. Vet. Microbiol. 2019, 231, 197–206. [Google Scholar] [CrossRef]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Debelius, J.; Morton, J.T.; Hyde, E.; Robbins-Pianka, A.; Knight, R. Correcting for microbial blooms in fecal samples during room-temperature shipping. mSystems 2017, 2, e00199-16. [Google Scholar] [CrossRef]
- Tal, M.; Verbrugghe, A.; Gomez, D.E.; Chau, C.; Weese, J.S. The effect of storage at ambient temperature on the feline fecal microbiota. BMC Vet. Res. 2017, 13, 256. [Google Scholar] [CrossRef]
- Marsilio, S.; Ackermann, M.R.; Lidbury, J.A.; Suchodolski, J.S.; Steiner, J.M. Results of histopathology, immunohistochemistry, and molecular clonality testing of small intestinal biopsy specimens from clinically healthy client-owned cats. J. Vet. Intern. Med. 2019, 33, 551–558. [Google Scholar] [CrossRef]
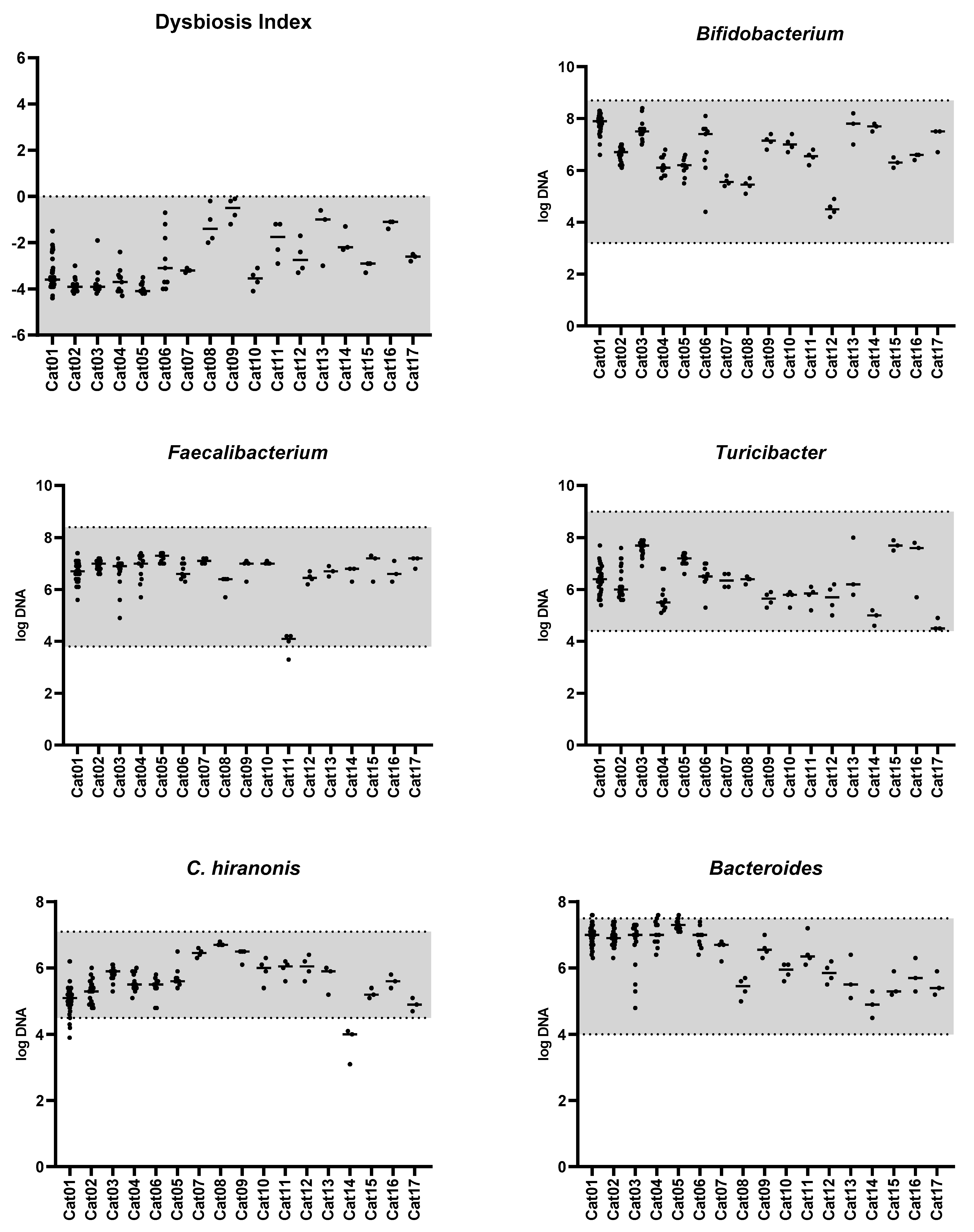
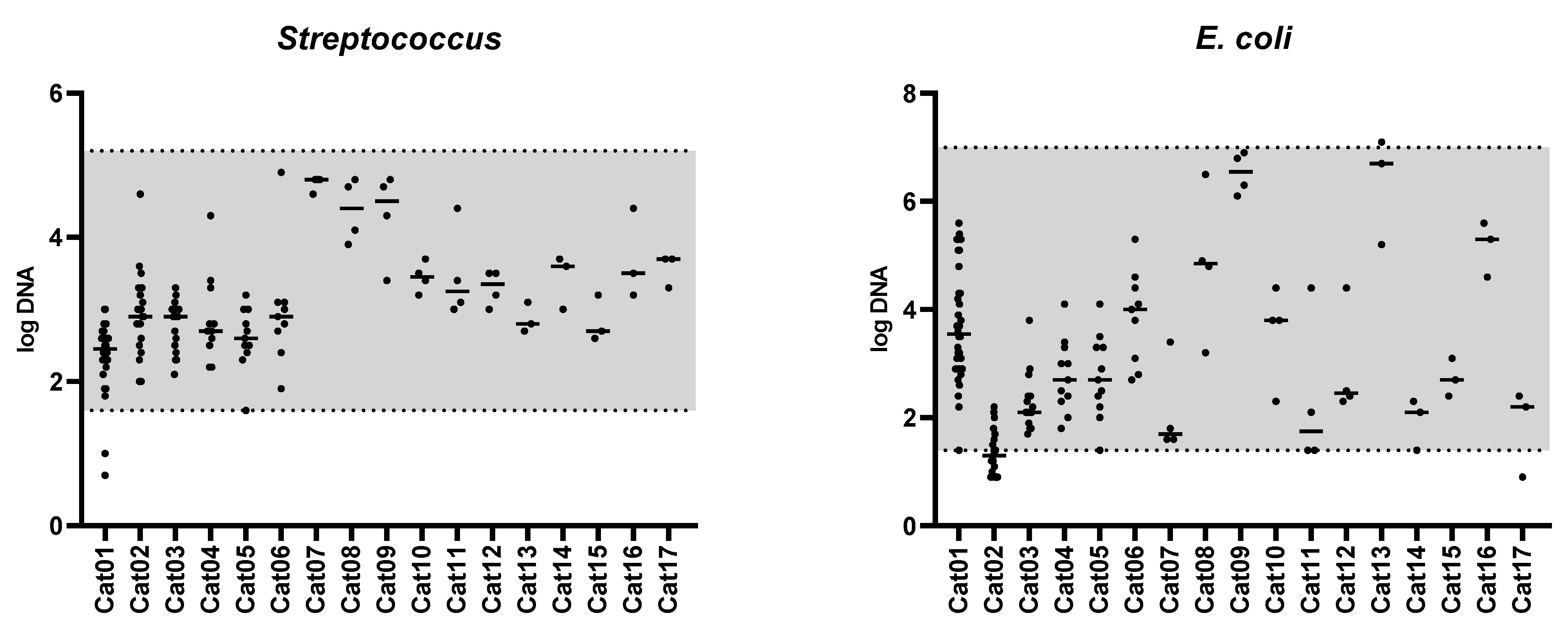
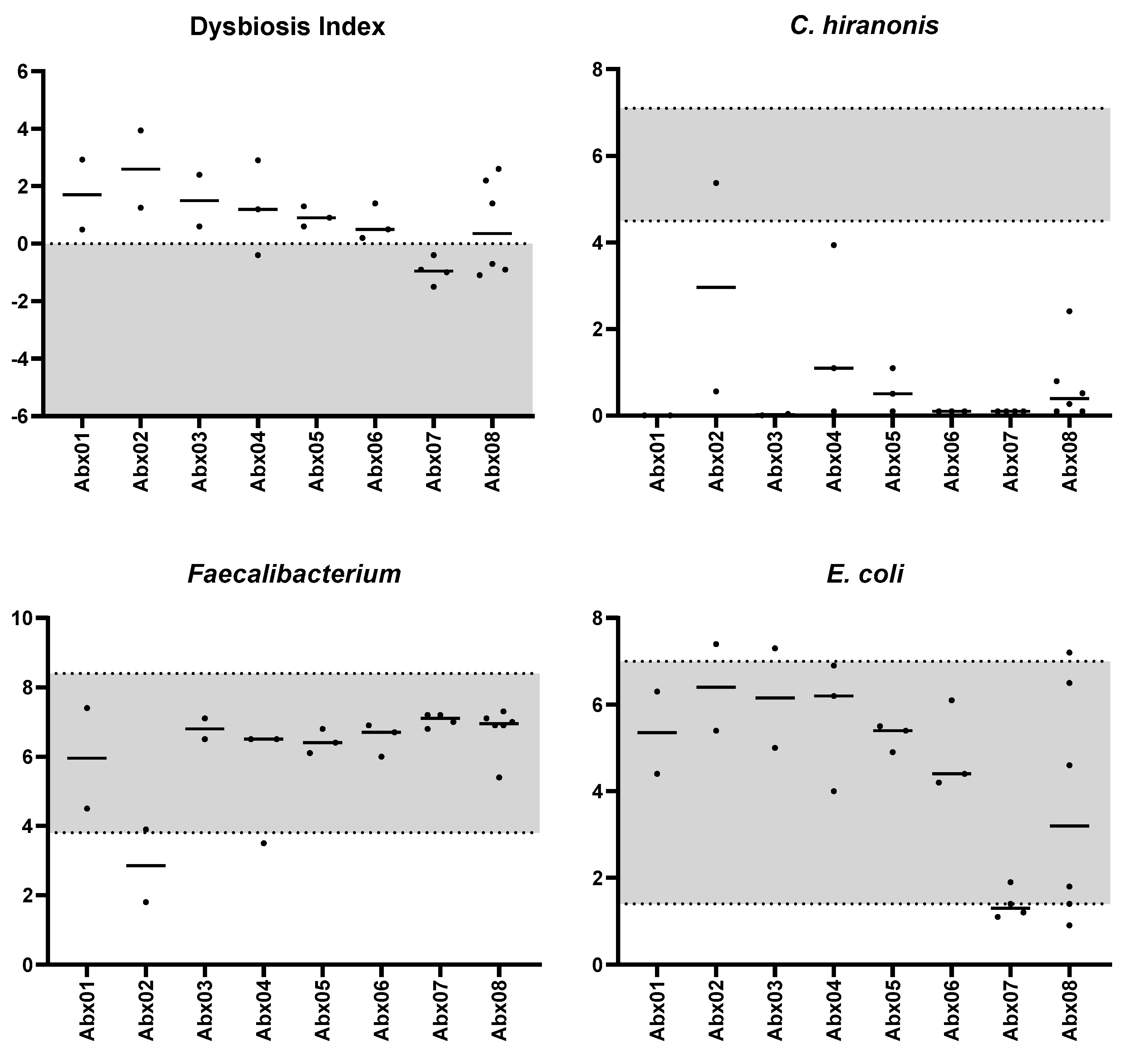
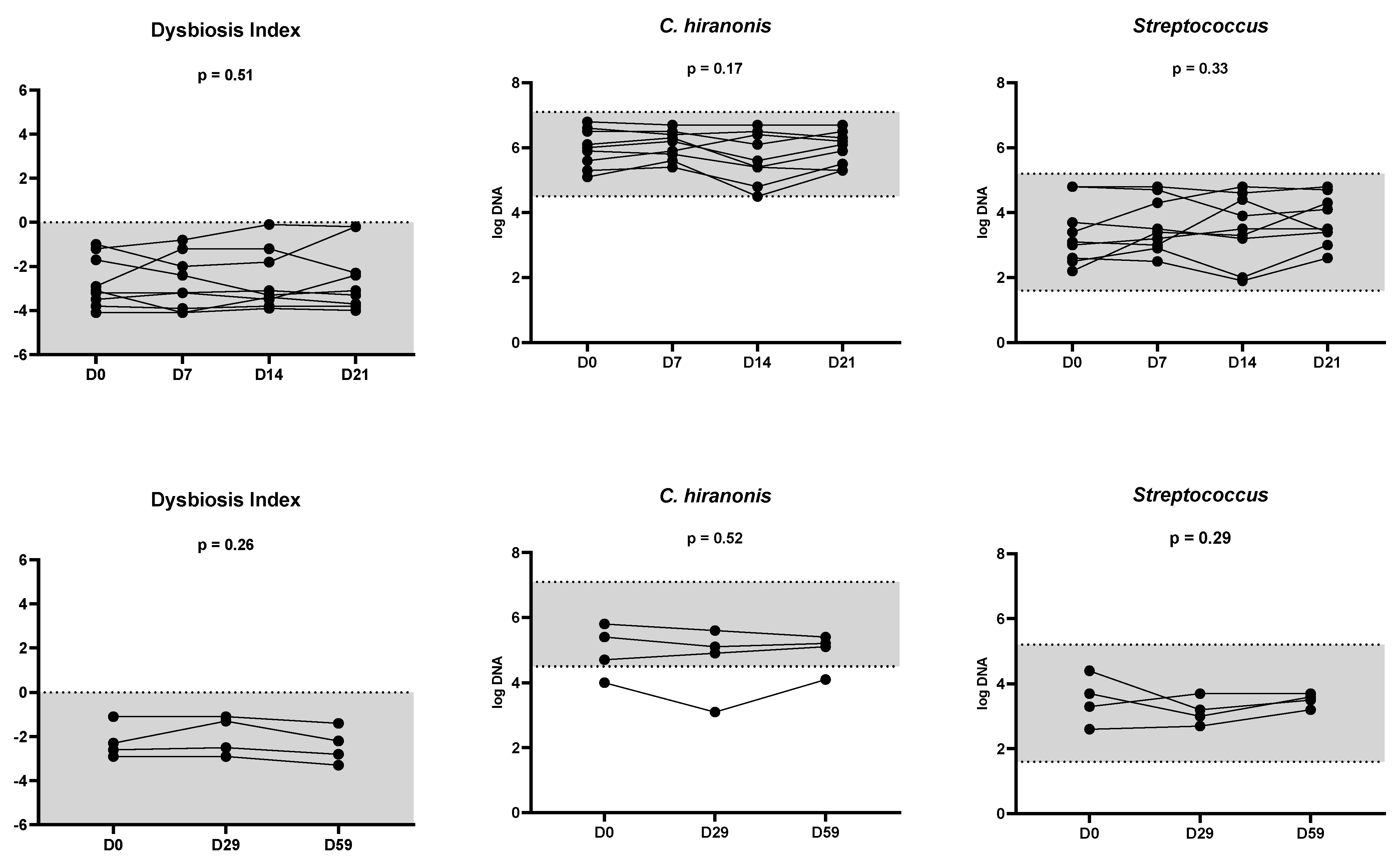



| Bacterial Group | Abundance (log DNA/g Feces) n = 142 | Coefficient of Variation (%) n = 17 | Number of Cats out of the Clinically Relevant Limit 1 n = 17 |
|---|---|---|---|
| Bacteroides | 7.6 (4.5–6.9) | 5.5 (2.2–11.8) | 0 |
| Bifidobacterium | 8.4 (4.2–6.9) | 4.6 (1.8–16.4) | 0 |
| C. hiranonis | 6.8 (3.1–5.5) | 5.1 (0.7–14.8) | 2 |
| Escherichia coli | 7.1 (0.9–2.8) | 25.1 (5.9–61.2) | 1 |
| Faecalibacterium | 7.4 (3.3–6.9) | 4.7 (0.7–10.9) | 1 |
| Streptococcus | 4.9 (0.7–2.8) | 12.6 (2.1–27.4) | 0 |
| Turicibacter | 8.0 (4.5–6.1) | 6.2 (2–17.6) | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sung, C.-H.; Marsilio, S.; Pilla, R.; Wu, Y.-A.; Cavasin, J.P.; Hong, M.-P.; Suchodolski, J.S. Temporal Variability of the Dominant Fecal Microbiota in Healthy Adult Cats. Vet. Sci. 2024, 11, 31. https://doi.org/10.3390/vetsci11010031
Sung C-H, Marsilio S, Pilla R, Wu Y-A, Cavasin JP, Hong M-P, Suchodolski JS. Temporal Variability of the Dominant Fecal Microbiota in Healthy Adult Cats. Veterinary Sciences. 2024; 11(1):31. https://doi.org/10.3390/vetsci11010031
Chicago/Turabian StyleSung, Chi-Hsuan, Sina Marsilio, Rachel Pilla, Yu-An Wu, Joao Pedro Cavasin, Min-Pyo Hong, and Jan S. Suchodolski. 2024. "Temporal Variability of the Dominant Fecal Microbiota in Healthy Adult Cats" Veterinary Sciences 11, no. 1: 31. https://doi.org/10.3390/vetsci11010031
APA StyleSung, C.-H., Marsilio, S., Pilla, R., Wu, Y.-A., Cavasin, J. P., Hong, M.-P., & Suchodolski, J. S. (2024). Temporal Variability of the Dominant Fecal Microbiota in Healthy Adult Cats. Veterinary Sciences, 11(1), 31. https://doi.org/10.3390/vetsci11010031







