Streptococcus suis Research Update: Serotype Prevalence and Antimicrobial Resistance Distribution in Swine Isolates Recovered in Spain from 2020 to 2022
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolate Collection and DNA Extraction
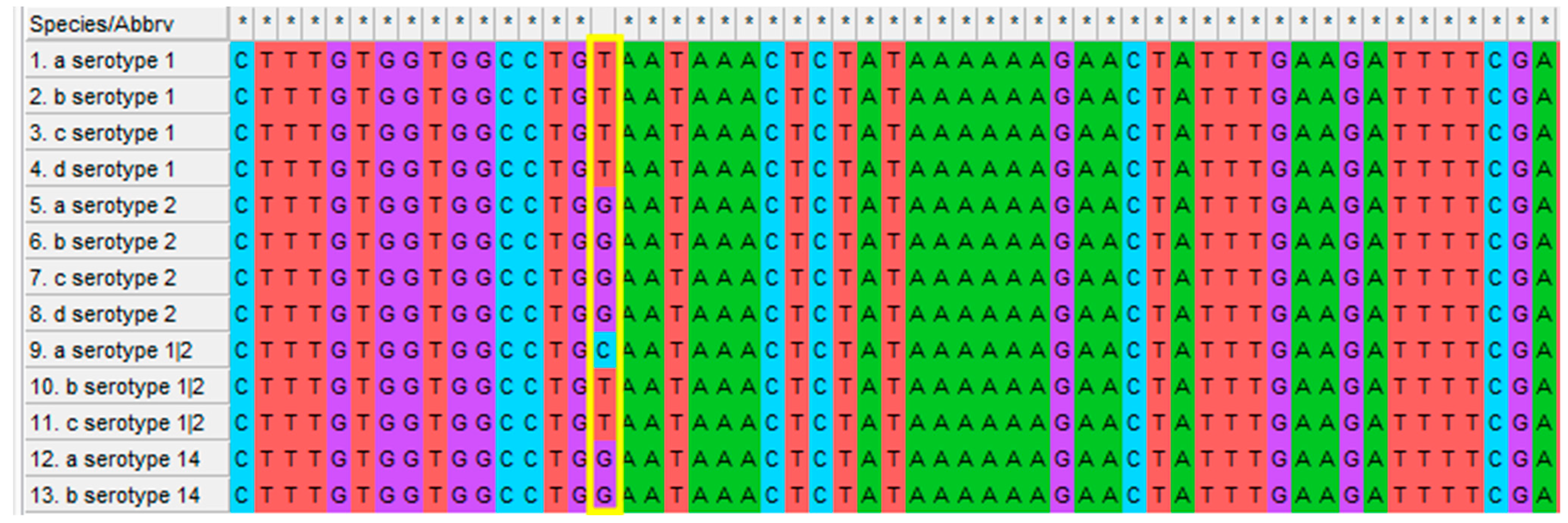
2.2. Molecular Typing
2.3. Detection of Antimicrobial Resistance Genes
2.4. Antimicrobial Susceptibility Testing
2.5. Isolate Classification in AMR Clusters
2.6. Statistical Analysis
3. Results
3.1. Molecular Typing
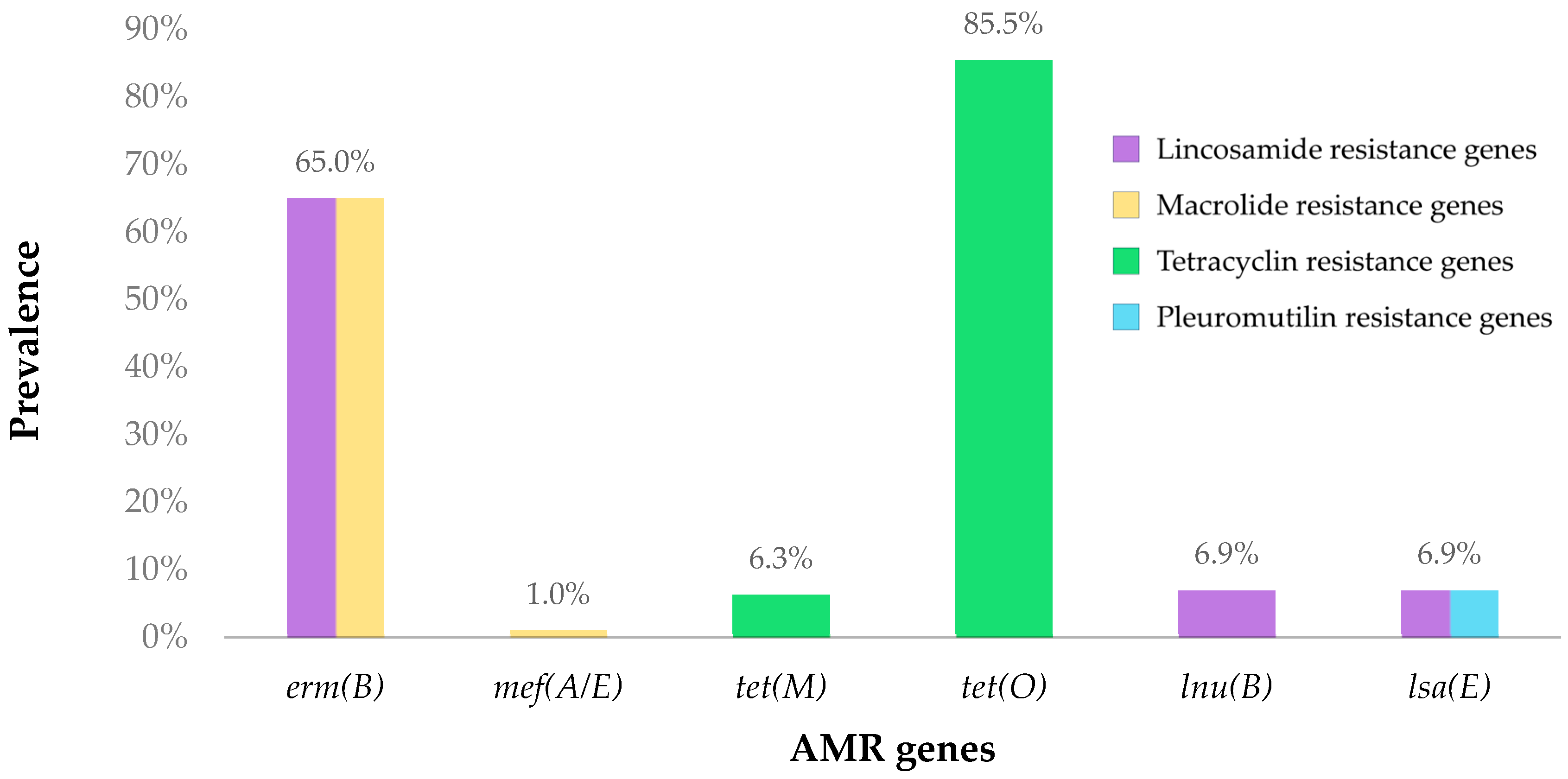
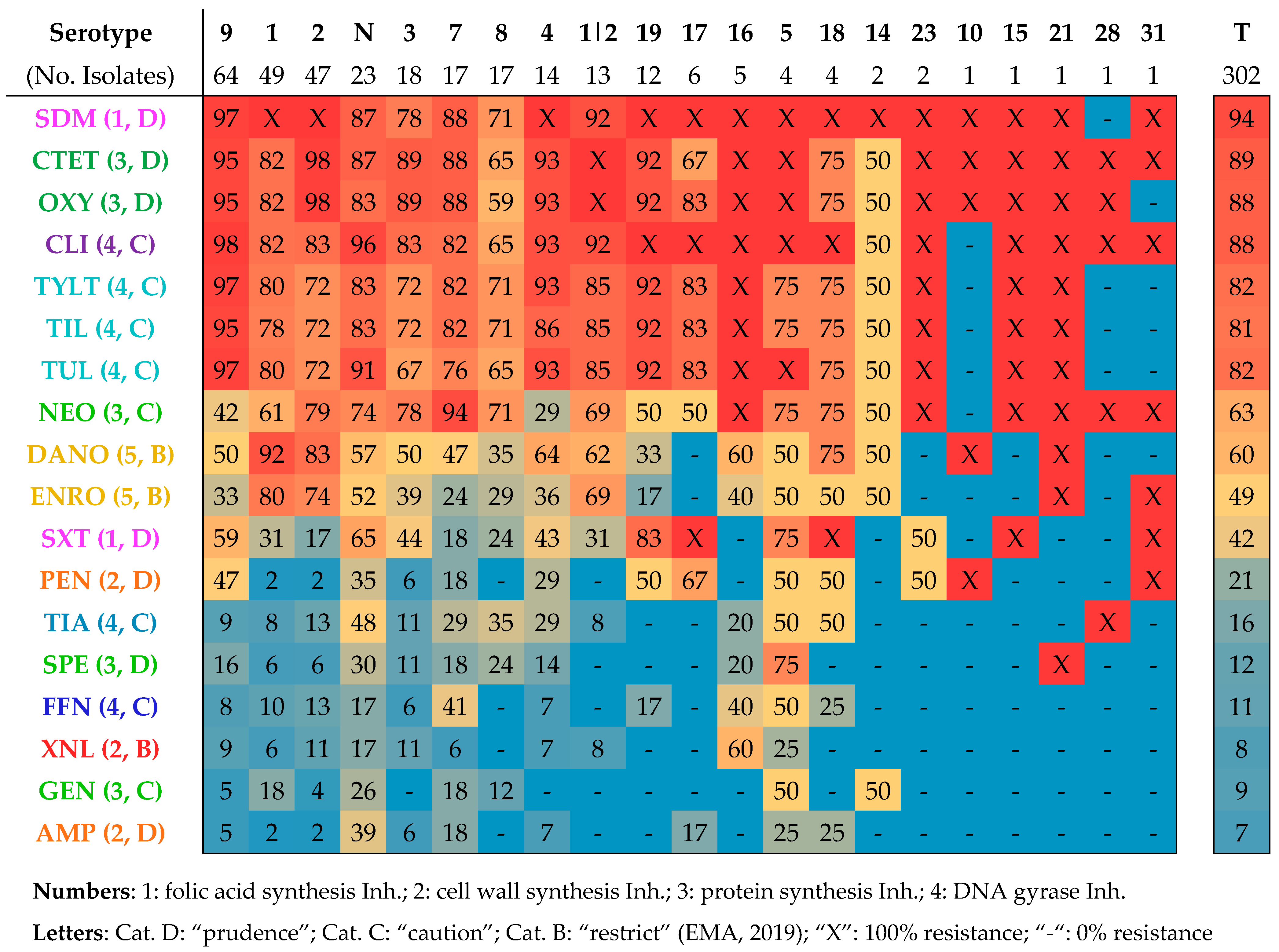
3.2. Detection of Antimicrobial Resistance Genes
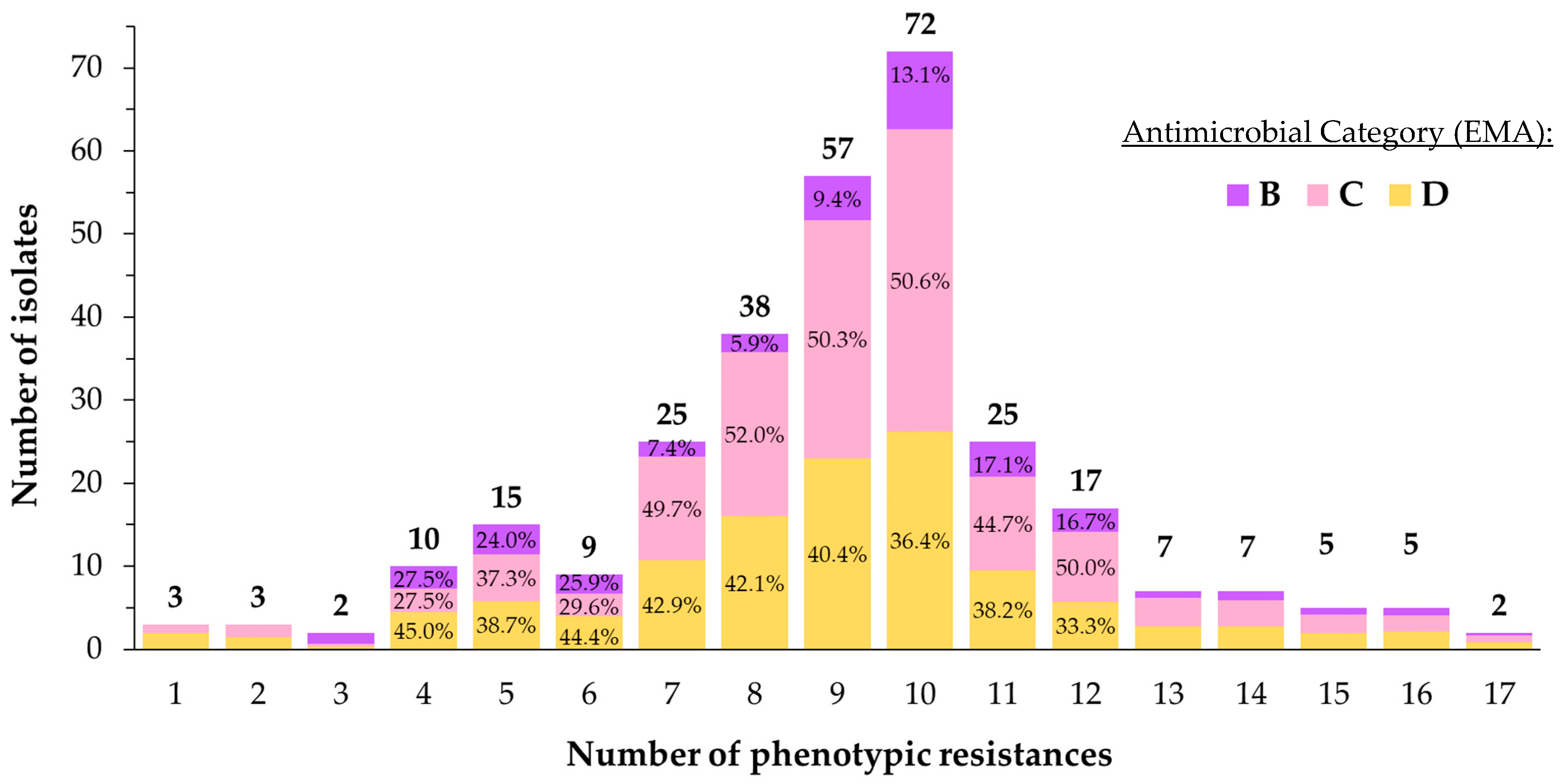
3.3. Antimicrobial Susceptibility Testing
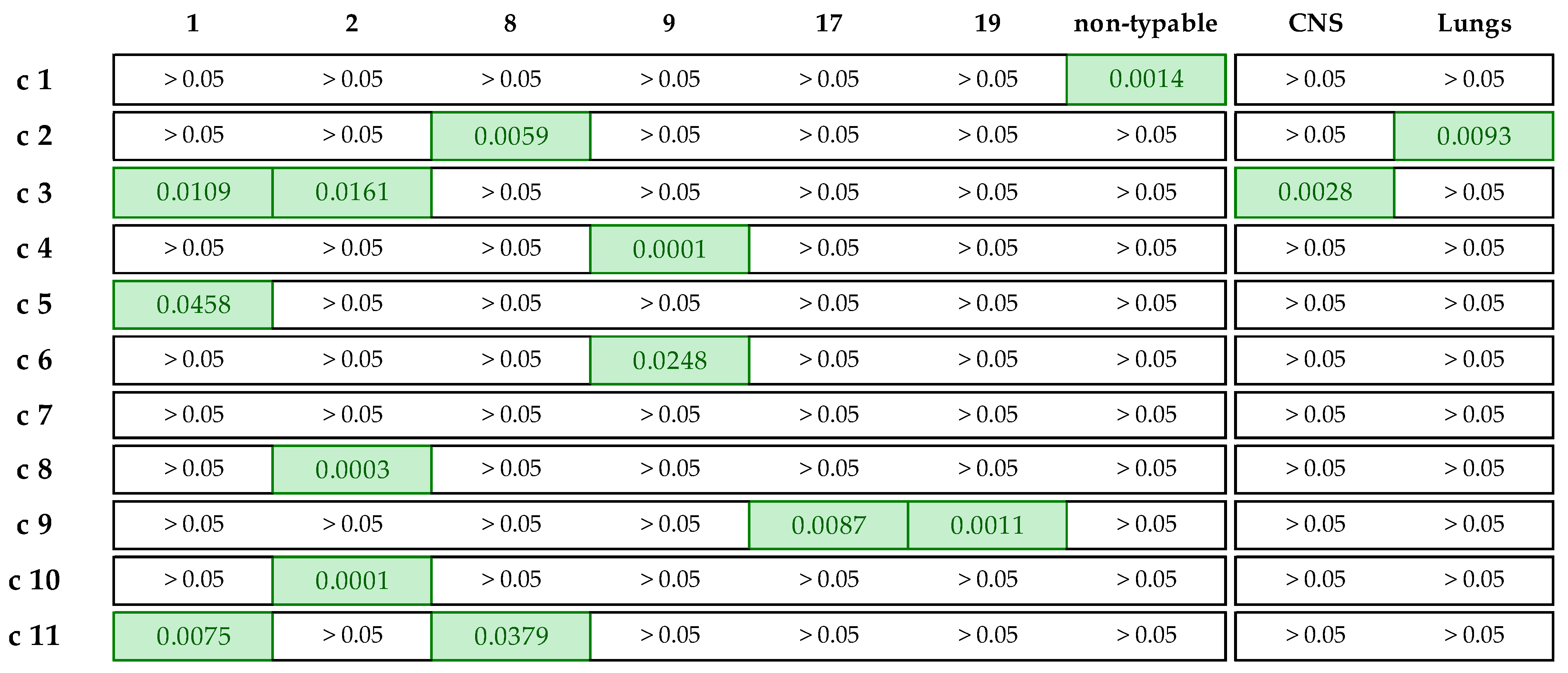
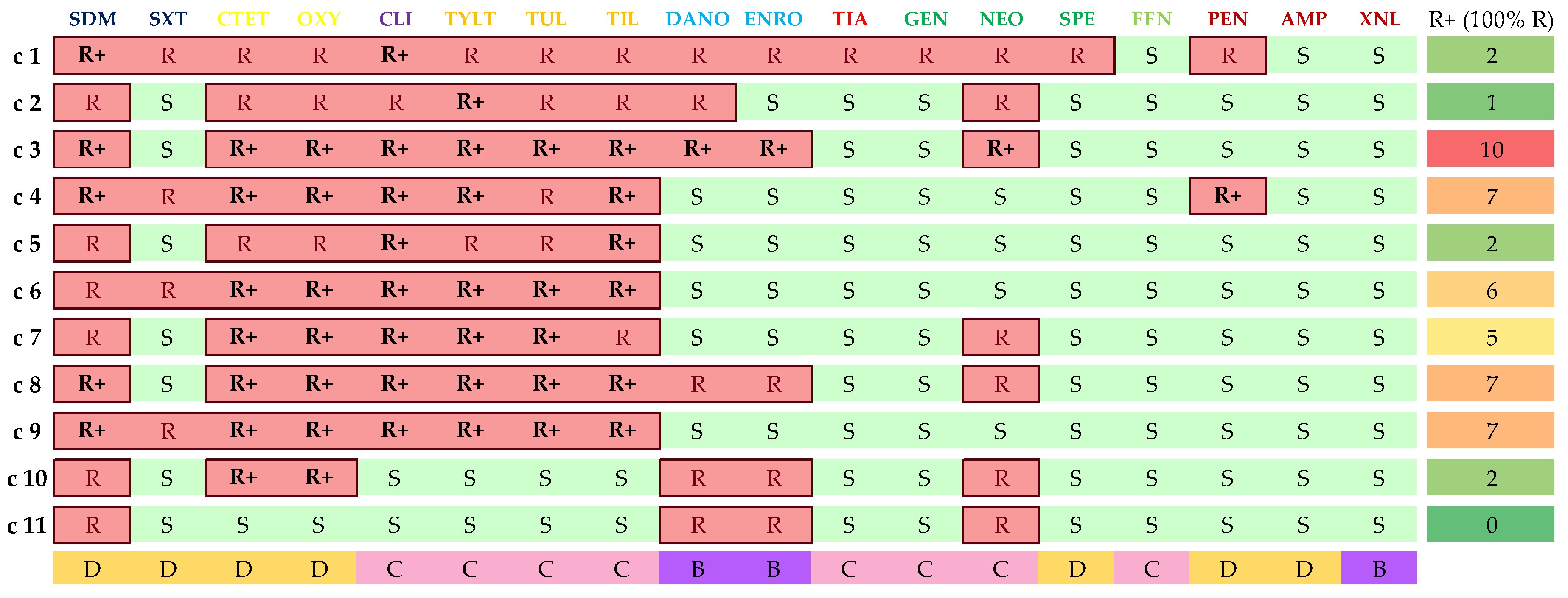
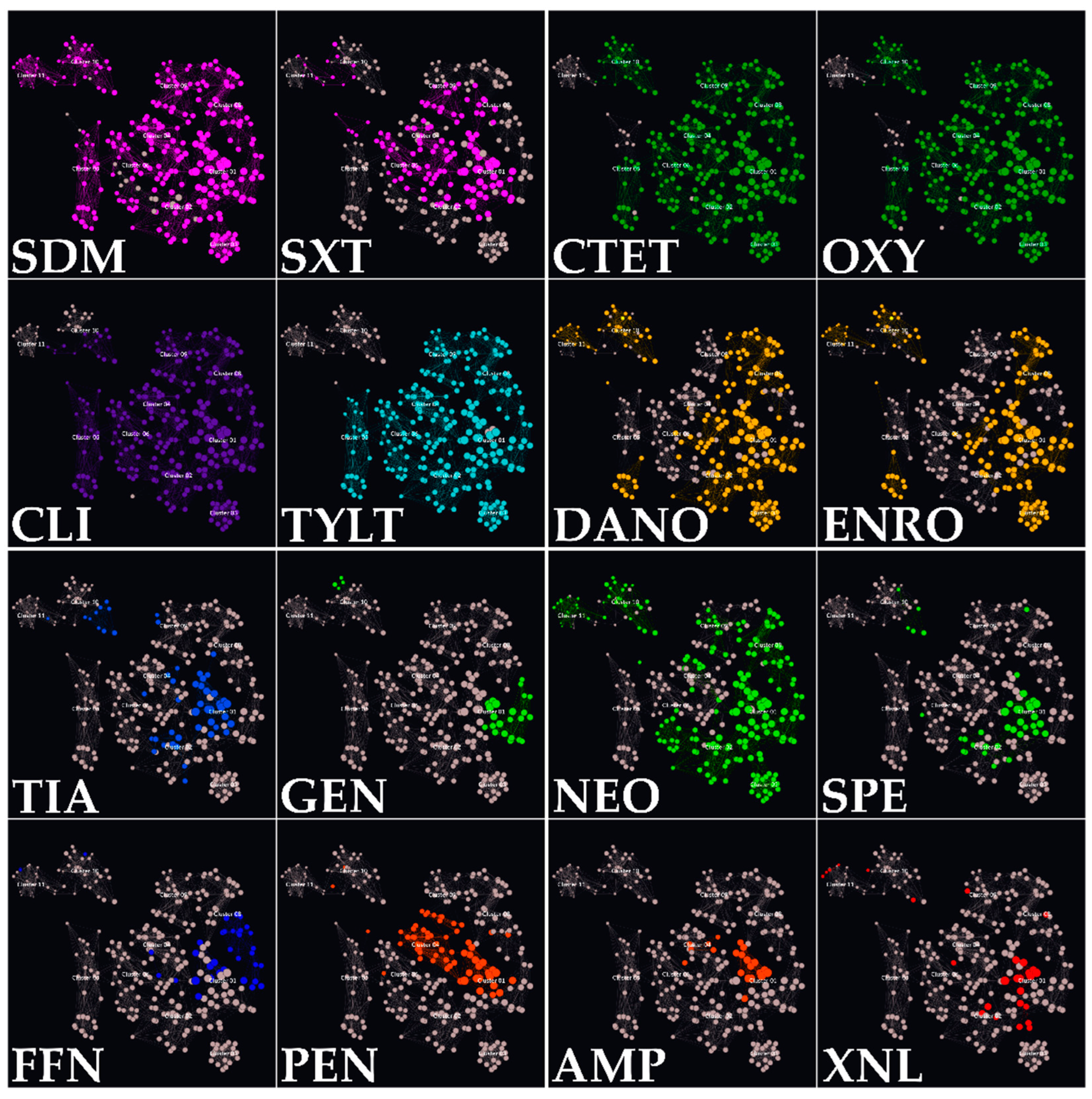
3.4. Isolate Classification in AMR Clusters
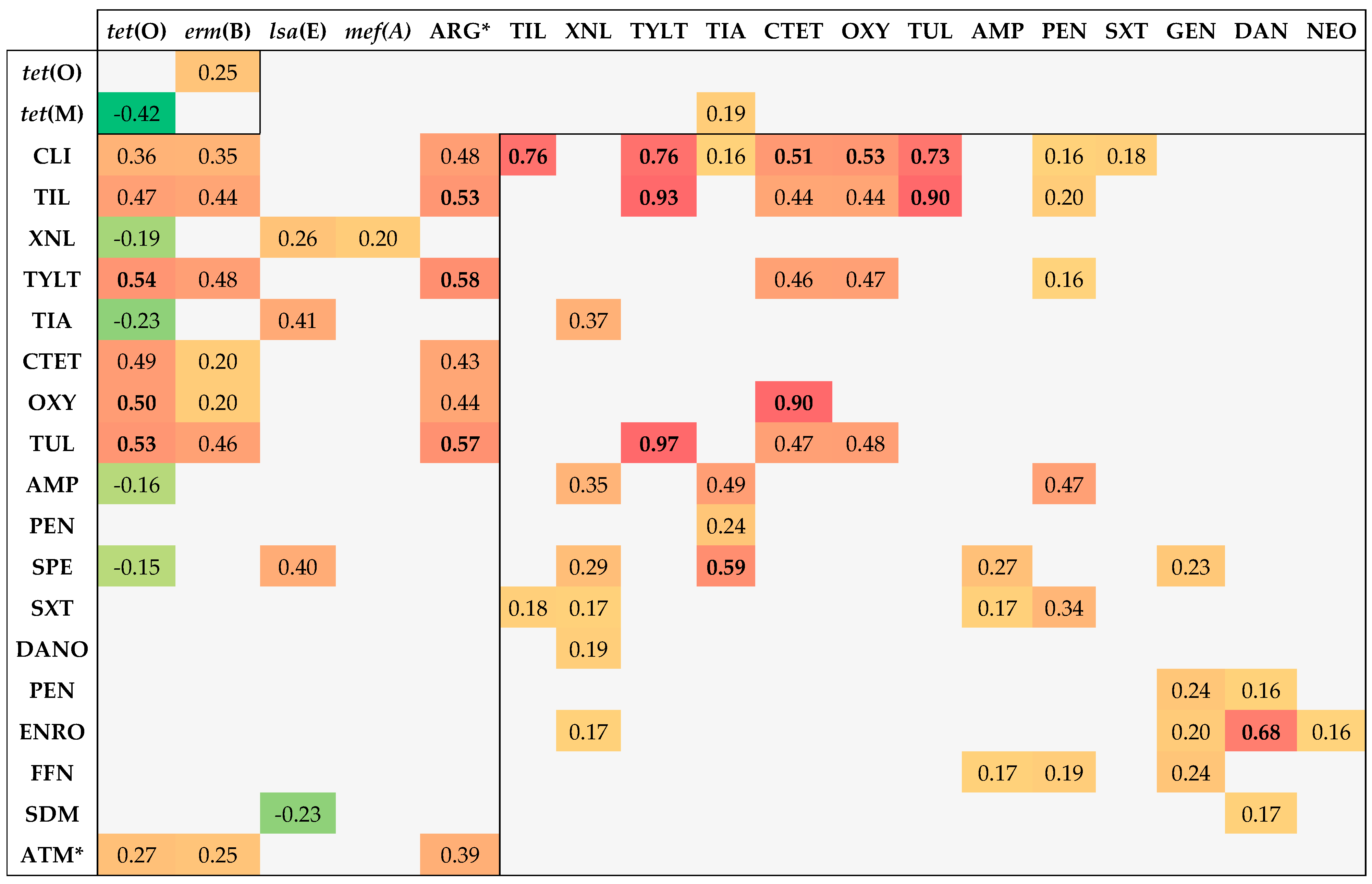
3.5. Associations between Genotypic and Phenotypic AMR
- Ten or more associations: tet(O) (13), CLI (11), and TIL (10);
- Five to ten associations: XNL (9), TYLT (9), erm(B) (8), TIA (8), CTET (8), OXY (8), TUL (8), no. of ARGs (7), AMP (7), PEN (6), SPE (6), and SXT (5);
- One to five associations: lsa(E)-lnu(B), GEN (4), DANO (4), NEO (4), ENRO (4), FFN (3), no. of phenotypic resistances (3), tet(M) (3), SDM (2), and mef(A/E) (1).
- Tetracycline–ARG correlation: tet(O)–CTET and tet(O)–OXY;
- Macrolide–ARG correlation: erm(B)–TIL, erm(B)–TUL, and erm(B)–TYLT;
- Lincosamide–ARG correlation: erm(B)–CLI;
- Pleuromutilin–ARG correlation: lsa(E)–TIA.
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Cluster | Resist. D-Cat. (7 ab.) | Resist. C-Cat. (8 ab.) | Resist. B-Cat. (3 ab.) | Total Resist./Cluster |
|---|---|---|---|---|
| c1 | 5.2 | 6.4 | 2.0 | 13.6 |
| c2 | 3.5 | 5.2 | 1.5 | 10.2 |
| c3 | 3.0 | 5.0 | 2.0 | 10.0 |
| c4 | 4.8 | 4.5 | 0.5 | 9.8 |
| c5 | 2.5 | 3.9 | 0.9 | 7.3 |
| c6 | 4.0 | 4.1 | 0.8 | 8.9 |
| c7 | 3.1 | 4.8 | 0.1 | 8.0 |
| c8 | 3.4 | 5.3 | 1.5 | 10.2 |
| c9 | 3.9 | 4.6 | 0.4 | 8.9 |
| c10 | 3.4 | 1.7 | 1.4 | 6.5 |
| c11 | 1.3 | 1.0 | 1.4 | 3.7 |
References
- Petri, F.A.M.; Ferreira, G.C.; Arruda, L.P.; Malcher, C.S.; Storino, G.Y.; Almeida, H.M.d.S.; Sonalio, K.; Silva, D.G.d.; Oliveira, L.G.d. Associations between Pleurisy and the Main Bacterial Pathogens of the Porcine Respiratory Diseases Complex (PRDC). Animals 2023, 13, 1493. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Giménez-Lirola, L.G.; Halbur, P.G. Polymicrobial respiratory disease in pigs. Anim. Health Res. Rev. 2011, 12, 133–148. [Google Scholar] [CrossRef]
- Brockmeier, S.L.; Halbur, P.G.; Thacker, E.L. Porcine Respiratory Disease Complex. In Polymicrobial Diseases; Brodgen, K., Guthmiller, J., Eds.; ASM Press: Washington, DC, USA, 2002; pp. 1–24. [Google Scholar]
- Gottschalk, M.; Segura, M. Streptococcosis. In Diseases of Swine; John Wiley & Sons Ltd.: Chichester, UK, 2019; Volume 9, pp. 934–950. ISBN 9781119350927. [Google Scholar]
- Staats, J.J.; Feder, I.; Okwumabua, O.; Chengappa, M.M. Streptococcus suis: Past and present. Vet. Res. Commun. 1997, 21, 381–407. [Google Scholar] [CrossRef] [PubMed]
- Estrada, A.A.; Gottschalk, M.; Rossow, S.; Rendahl, A.; Gebhart, C.; Marthaler, D.G. Serotype and Genotype (Multilocus Sequence Type) of Streptococcus suis Isolates from the United States Serve as Predictors of Pathotype. J. Clin. Microbiol. 2019, 57, e00377-19. [Google Scholar] [CrossRef]
- Kerdsin, A.; Akeda, Y.; Hatrongjit, R.; Detchawna, U.; Sekizaki, T.; Hamada, S.; Gottschalk, M.; Oishi, K. Streptococcus suis serotyping by a new multiplex PCR. J. Med. Microbiol. 2014, 63, 824–830. [Google Scholar] [CrossRef] [PubMed]
- Segura, M. Streptococcus suis Research: Progress and Challenges. Pathogens 2020, 9, 707. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Chaiden, C.; Jaresitthikunchai, J.; Kerdsin, A.; Meekhanon, N.; Roytrakul, S.; Nuanualsuwan, S. Streptococcus suis serotyping by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. PLoS ONE 2021, 16, e0249682. [Google Scholar] [CrossRef]
- Scherrer, S.; Rademacher, F.; Spoerry Serrano, N.; Schrenzel, J.; Gottschalk, M.; Stephan, R.; Landolt, P. Rapid high resolution melting assay to differentiate Streptococcus suis serotypes 2, 1/2, 1, and 14. Microbiologyopen 2020, 9, e995. [Google Scholar] [CrossRef]
- Matiasovic, J.; Zouharova, M.; Nedbalcova, K.; Kralova, N.; Matiaskova, K.; Simek, B.; Kucharovicova, I.; Gottschalk, M. Resolution of Streptococcus suis Serotypes 1/2 versus 2 and 1 versus 14 by PCR-Restriction Fragment Length Polymorphism Method. J. Clin. Microbiol. 2020, 58, e00480-20. [Google Scholar] [CrossRef]
- Lacouture, S.; Okura, M.; Takamatsu, D.; Corsaut, L.; Gottschalk, M. Development of a mismatch amplification mutation assay to correctly serotype isolates of Streptococcus suis serotypes 1, 2, 1/2, and 14. J. Vet. Diagn. Investig. 2020, 32, 490–494. [Google Scholar] [CrossRef] [PubMed]
- Thu, I.S.L.; Tragoolpua, K.; Intorasoot, S.; Anukool, U.; Khamnoi, P.; Kerdsin, A.; Tharinjaroen, C.S. Direct Detection of Streptococcus suis from Cerebrospinal Fluid, Positive Hemoculture, and Simultaneous Differentiation of Serotypes 1, 1/2, 2, and 14 within Single Reaction. Pathogens 2021, 10, 996. [Google Scholar] [CrossRef] [PubMed]
- Hatrongjit, R.; Fittipaldi, N.; Gottschalk, M.; Kerdsin, A. Tools for Molecular Epidemiology of Streptococcus suis. Pathogens 2020, 9, 81. [Google Scholar] [CrossRef] [PubMed]
- Segura, M.; De, A.; Gebhart, C.; Aragon, V.; Mark, A.; Kerdsin, A.; Saléry, M.; Schultsz, L.A.; Wells, J.M.; Gottschalk, M. Update on Streptococcus suis Research and Prevention in the Era of Antimicrobial Restriction: 4th International Workshop on S. suis. Pathogens 2020, 9, 374. [Google Scholar]
- Harbarth, S.; Balkhy, H.H.; Goossens, H.; Jarlier, V.; Kluytmans, J.; Laxminarayan, R.; Saam, M.; Van Belkum, A.; Pittet, D. Antimicrobial resistance: One world, one fight! Antimicrob. Resist. Infect. Control 2015, 4, 49. [Google Scholar] [CrossRef]
- Huang, J.; Ma, J.; Shang, K.; Hu, X.; Liang, Y.; Li, D.; Wu, Z.; Dai, L.; Chen, L.; Wang, L. Evolution and Diversity of the Antimicrobial Resistance Associated Mobilome in Streptococcus suis: A Probable Mobile Genetic Elements Reservoir for Other Streptococci. Front. Cell. Infect. Microbiol. 2016, 6, 118. [Google Scholar] [CrossRef] [PubMed]
- Goyette-Desjardins, G.; Auger, J.-P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 2014, 3, e45. [Google Scholar] [CrossRef]
- Segura, M. Streptococcus suis vaccines: Candidate antigens and progress. Expert Rev. Vaccines 2015, 14, 1587–1608. [Google Scholar] [CrossRef]
- Okwumabua, O.; O’Connor, M.; Shull, E. A polymerase chain reaction (PCR) assay specific for Streptococcus suis based on the gene encoding the glutamate dehydrogenase. FEMS Microbiol. Lett. 2003, 218, 79–84. [Google Scholar] [CrossRef]
- Malhotra-Kumar, S.; Lammens, C.; Piessens, J.; Goossens, H. Multiplex PCR for simultaneous detection of macrolide and tetracycline resistance determinants in streptococci. Antimicrob. Agents Chemother. 2005, 49, 4798–4800. [Google Scholar] [CrossRef]
- Bojarska, A.; Molska, E.; Janas, K.; Skoczyńska, A.; Stefaniuk, E.; Hryniewicz, W.; Sadowy, E. Streptococcus suis in invasive human infections in Poland: Clonality and determinants of virulence and antimicrobial resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 917–925. [Google Scholar] [CrossRef]
- CLSI. M100: Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed.; Wayne, P., Ed.; Clinical Lab Standards Institute: Temecula, CA, USA, 2023; ISBN 9781684401703. [Google Scholar]
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing, Version 13.0; Breakpoint Tables for Interpretation of MICs and Zone Diameters. Available online: http://www.eucast.org (accessed on 4 March 2023).
- European Medicines Agency. Categorisation of antibiotics in the European Union. Eur. Med. Agence 2019, 31, 1–73. [Google Scholar]
- Vela, A.I.; Goyache, J.; Tarradas, C.; Luque, I.; Mateos, A.; Moreno, M.A.; Borge, C.; Perea, J.A.; Domínguez, L.; Fernández-Garayzábal, J.F. Analysis of genetic diversity of Streptococcus suis clinical isolates from pigs in Spain by pulsed-field gel electrophoresis. J. Clin. Microbiol. 2003, 41, 2498–2502. [Google Scholar] [CrossRef] [PubMed]
- Büttner, N.; Beineke, A.; de Buhr, N.; Lilienthal, S.; Merkel, J.; Waldmann, K.H.; Valentin-Weigand, P.; Baums, C.G. Streptococcus suis serotype 9 bacterin immunogenicity and protective efficacy. Vet. Immunol. Immunopathol. 2012, 146, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Du, P.; Qiu, X.; Kerdsin, A.; Roy, D.; Bai, X.; Xu, J.; Vela, A.I.; Gottschalk, M. Genomic comparisons of Streptococcus suis serotype 9 strains recovered from diseased pigs in Spain and Canada. Vet. Res. 2018, 49, 1. [Google Scholar] [CrossRef] [PubMed]
- Auger, J.P.; Payen, S.; Roy, D.; Dumesnil, A.; Segura, M.; Gottschalk, M. Interactions of Streptococcus suis serotype 9 with host cells and role of the capsular polysaccharide: Comparison with serotypes 2 and 14. PLoS ONE 2019, 14, e0223864. [Google Scholar] [CrossRef] [PubMed]
- Exopol, La Etiología Porcina en Estadísticas. Available online: https://www.exopol.com/es/servicios/estadisticas.php (accessed on 10 April 2023).
- Vötsch, D.; Willenborg, M.; Weldearegay, Y.B.; Valentin-Weigand, P. Streptococcus suis-The «Two Faces» of a Pathobiont in the Porcine Respiratory Tract. Front. Microbiol. 2018, 9, 480. [Google Scholar] [CrossRef] [PubMed]
- Kerdsin, A.; Hatrongjit, R.; Gottschalk, M.; Takeuchi, D.; Hamada, S.; Akeda, Y.; Oishi, K. Emergence of Streptococcus suis serotype 9 infection in humans. J. Microbiol. Immunol. Infect. 2017, 50, 545–546. [Google Scholar] [CrossRef]
- De Greeff, A.; Wisselink, H.J.; De Bree, F.M.; Schultsz, C.; Baums, C.G.; Thi, H.N.; Stockhofe-Zurwieden, N.; Smith, H.E. Genetic diversity of Streptococcus suis isolates as determined by comparative genome hybridization. BMC Microbiol. 2011, 11, 161. [Google Scholar] [CrossRef]
- Sánchez del Rey, V.; Fernández-Garayzábal, J.F.; Bárcena, C.; Briones, V.; Domínguez, L.; Gottschalk, M.; Vela, A.I. Molecular typing of Streptococcus suis isolates from Iberian pigs: A comparison with isolates from common intensively-reared commercial pig breeds. Vet. J. 2014, 202, 597–602. [Google Scholar] [CrossRef]
- Dong, W.; Zhu, Y.; Ma, Y.; Ma, J.; Zhang, Y.; Yuan, L.; Pan, Z.; Wu, Z.; Yao, H. Multilocus sequence typing and virulence genotyping of Streptococcus suis serotype 9 isolates revealed high genetic and virulence diversity. FEMS Microbiol. Lett. 2017, 364, fnx192. [Google Scholar] [CrossRef]
- Scherrer, S.; Rosato, G.; Spoerry Serrano, N.; Stevens, M.J.A.; Rademacher, F.; Schrenzel, J.; Gottschalk, M.; Stephan, R.; Peterhans, S. Population structure, genetic diversity and pathotypes of Streptococcus suis isolated during the last 13 years from diseased pigs in Switzerland. Vet. Res. 2020, 51, 85. [Google Scholar] [CrossRef] [PubMed]
- Fittipaldi, N.; Xu, J.; Lacouture, S.; Tharavichitkul, P.; Osaki, M.; Sekizaki, T.; Takamatsu, D.; Gottschalk, M. Lineage and virulence of Streptococcus suis serotype 2 isolates from North America. Emerg. Infect. Dis. 2011, 17, 2239–2244. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Li, C.; Wang, Y.; Bian, Z.; Chu, P.; Zhai, S.; Yang, D.; Song, S.; Li, Y.; Jiang, Z.; et al. Accelerated loop-mediated isothermal amplification method for the rapid detection of Streptococcus suis serotypes 2 and 14 based on single nucleotide polymorphisms. Front. Cell. Infect. Microbiol. 2022, 12, 1034762. [Google Scholar] [CrossRef] [PubMed]
- King, S.J.; Heath, P.J.; Luque, I.; Tarradas, C.; Dowson, C.G.; Whatmore, A.M. Distribution and Genetic Diversity of Suilysin in Streptococcus suis Isolated from Different Diseases of Pigs and Characterization of the Genetic Basis of Suilysin Absence. Infect. Immun. 2001, 69, 7572–7582. [Google Scholar] [CrossRef] [PubMed]
- Segura, M.; Fittipaldi, N.; Calzas, C.; Gottschalk, M. Critical Streptococcus suis Virulence Factors: Are They All Really Critical? Trends Microbiol. 2017, 25, 585–599. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Wei, Z.; He, H.; Zhang, A.; Jin, M. Antimicrobial susceptibility, tetracycline and erythromycin resistance genes, and multilocus sequence typing of Streptococcus suis isolates from diseased pigs in China. J. Vet. Med. Sci. 2013, 75, 583–587. [Google Scholar] [CrossRef]
- Dechêne-Tempier, M.; Marois-Créhan, C.; Libante, V.; Jouy, E.; Leblond-Bourget, N.; Payot, S. Update on the mechanisms of antibiotic resistance and the mobile resistome in the emerging zoonotic pathogen Streptococcus suis. Microorganisms 2021, 9, 1765. [Google Scholar] [CrossRef]
- Vela, A.I.; Moreno, M.A.; Cebolla, J.A.; González, S.; Latre, M.V.; Domínguez, L.; Fernández-Garayzábal, J.F. Antimicrobial susceptibility of clinical strains of Streptococcus suis isolated from pigs in Spain. Vet. Microbiol. 2005, 105, 143–147. [Google Scholar] [CrossRef]
- Hayer, S.S.; Rovira, A.; Olsen, K.; Johnson, T.J.; Vannucci, F.; Rendahl, A.; Perez, A.; Alvarez, J. Prevalence and time trend analysis of antimicrobial resistance in respiratory bacterial pathogens collected from diseased pigs in USA between 2006–2016. Res. Vet. Sci. 2020, 128, 135–144. [Google Scholar] [CrossRef]
- Arndt, E.R.; Farzan, A.; MacInnes, J.I.; Friendship, R.M. Antimicrobial resistance of Streptococcus suis isolates recovered from clinically ill nursery pigs and from healthy pigs at different stages of production. Can. Vet. J. = La Rev. Vet. Can. 2019, 60, 519–522. [Google Scholar]
- Dechêne-Tempier, M.; Jouy, E.; Bayon-Auboyer, M.H.; Bougeard, S.; Chauvin, C.; Libante, V.; Payot, S.; Marois-Créhan, C. Antimicrobial resistance profiles of Streptococcus suis isolated from pigs, wild boars, and humans in France between 1994 and 2020. J. Clin. Microbiol. 2023, 61, e0016423. [Google Scholar] [CrossRef]
- Holmer, I.; Salomonsen, C.M.; Jorsal, S.E.; Astrup, L.B.; Jensen, V.F.; Høg, B.B.; Pedersen, K. Antibiotic resistance in porcine pathogenic bacteria and relation to antibiotic usage. BMC Vet. Res. 2019, 15, 449. [Google Scholar] [CrossRef] [PubMed]
- Li, L.L.; Liao, X.P.; Sun, J.; Yang, Y.R.; Liu, B.T.; Yang, S.S.; Zhao, D.H.; Liu, Y.H. Antimicrobial resistance, serotypes, and virulence factors of Streptococcus suis isolates from diseased pigs. Foodborne Pathog. Dis. 2012, 9, 583–588. [Google Scholar] [CrossRef]
- Ichikawa, T.; Oshima, M.; Yamagishi, J.; Muramatsu, C.; Asai, T. Changes in antimicrobial resistance phenotypes and genotypes in Streptococcus suis strains isolated from pigs in the tokai area of Japan. J. Vet. Med. Sci. 2020, 82, 9–13. [Google Scholar] [CrossRef]
- Granados-Chinchilla, F.; Rodríguez, C. Tetracyclines in Food and Feedingstuffs: From Regulation to Analytical Methods, Bacterial Resistance, and Environmental and Health Implications. J. Anal. Methods Chem. 2017, 2017, 1315497. [Google Scholar] [CrossRef] [PubMed]
- Cantin, M.; Harel, J.; Higgins, R.; Gottschalk, M. Antimicrobial resistance patterns and plasmid profiles of Streptococcus suis isolates. J. Vet. Diagnostic Investig. 1992, 4, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Uruén, C.; García, C.; Fraile, L.; Tommassen, J.; Arenas, J. How Streptococcus suis escapes antibiotic treatments. Vet. Res. 2022, 53, 91. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Z.; Song, L.; Fan, X.; Wen, F.; Xu, S.; Ning, Y. Antimicrobial Resistance Profile and Genotypic Characteristics of Streptococcus suis Capsular Type 2 Isolated from Clinical Carrier Sows and Diseased Pigs in China. Biomed Res. Int. 2015, 2015, 284303. [Google Scholar] [CrossRef]
- Wang, C.-Z.; Wang, M.-G.; Chu, Y.-F.; Sun, R.-Y.; Li, J.-G.; Li, X.-A.; Sun, J.; Liu, Y.-H.; Zhou, Y.-F.; Liao, X.-P. Antibiotic Resistance Patterns and Molecular Characterization of Streptococcus suis Isolates from Swine and Humans in China. Microbiol. Spectr. 2023, 11, e0030923. [Google Scholar] [CrossRef]
- Nicholson, T.L.; Bayles, D.O. Comparative virulence and antimicrobial resistance distribution of Streptococcus suis isolates obtained from the United States. Front. Microbiol. 2022, 13, 1043529. [Google Scholar] [CrossRef] [PubMed]
- Lekagul, A.; Tangcharoensathien, V.; Yeung, S. Patterns of antibiotic use in global pig production: A systematic review. Vet. Anim. Sci. 2019, 7, 100058. [Google Scholar] [CrossRef]
- Kittiwan, N.; Calland, J.K.; Mourkas, E.; Hitchings, M.D.; Murray, S.; Tadee, P.; Tadee, P.; Duangsonk, K.; Meric, G.; Sheppard, S.K.; et al. Genetic diversity and variation in antimicrobial-resistance determinants of non-serotype 2 Streptococcus suis isolates from healthy pigs. Microb. Genom. 2022, 8, 882. [Google Scholar] [CrossRef] [PubMed]
- Matajira, C.E.C.; Moreno, L.Z.; Poor, A.P.; Gomes, V.T.M.; Dalmutt, A.C.; Parra, B.M.; de Oliveira, C.H.; Barbosa, M.R.F.; Sato, M.I.Z.; Calderaro, F.F.; et al. Streptococcus suis in Brazil: Genotypic, Virulence, and Resistance Profiling of Strains Isolated from Pigs between 2001 and 2016. Pathogens 2019, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Benford, D.; Ceccatelli, S.; Cottrill, B.; DiNovi, M.; Dogliotti, E.; Edler, L.; Farmer, P.; Fürst, P.; Hoogenboom, L.; Katrine Knutsen, H.; et al. Scientific Opinion on Chloramphenicol in food and feed. EFSA J. 2014, 12, 3907. [Google Scholar] [CrossRef]
- Trif, E.; Cerbu, C.; Olah, D.; Zăblău, S.D.; Spînu, M.; Potârniche, A.V.; Pall, E.; Brudașcă, F. Old Antibiotics Can Learn New Ways: A Systematic Review of Florfenicol Use in Veterinary Medicine and Future Perspectives Using Nanotechnology. Animals 2023, 13, 1695. [Google Scholar] [CrossRef] [PubMed]
- Haenni, M.; Lupo, A.; Madec, J.-Y. Antimicrobial Resistance in Streptococcus spp. Microbiol. Spectr. 2018, 6, 10–1128. [Google Scholar] [CrossRef]
- Gottschalk, M.; Turgeon, P.; Higgins, R.; Beaudoin, M.; Bourgault, A.M. Susceptibility of Streptococcus suis to penicillin. J. Vet. Diagn. Investig. 1991, 3, 170–172. [Google Scholar] [CrossRef]
- Lunha, K.; Chumpol, W.; Jiemsup, S.; Samngamnim, S.; Assavacheep, P.; Yongkiettrakul, S. Relationship between Penicillin-Binding Proteins Alterations and β-Lactams Non-Susceptibility of Diseased Pig-Isolated Streptococcus suis. Antibiotics 2023, 12, 158. [Google Scholar] [CrossRef]
- Yongkiettrakul, S.; Maneerat, K.; Arechanajan, B.; Malila, Y.; Srimanote, P.; Gottschalk, M.; Visessanguan, W. Antimicrobial susceptibility of Streptococcus suis isolated from diseased pigs, asymptomatic pigs, and human patients in Thailand. BMC Vet. Res. 2019, 15, 5. [Google Scholar] [CrossRef]
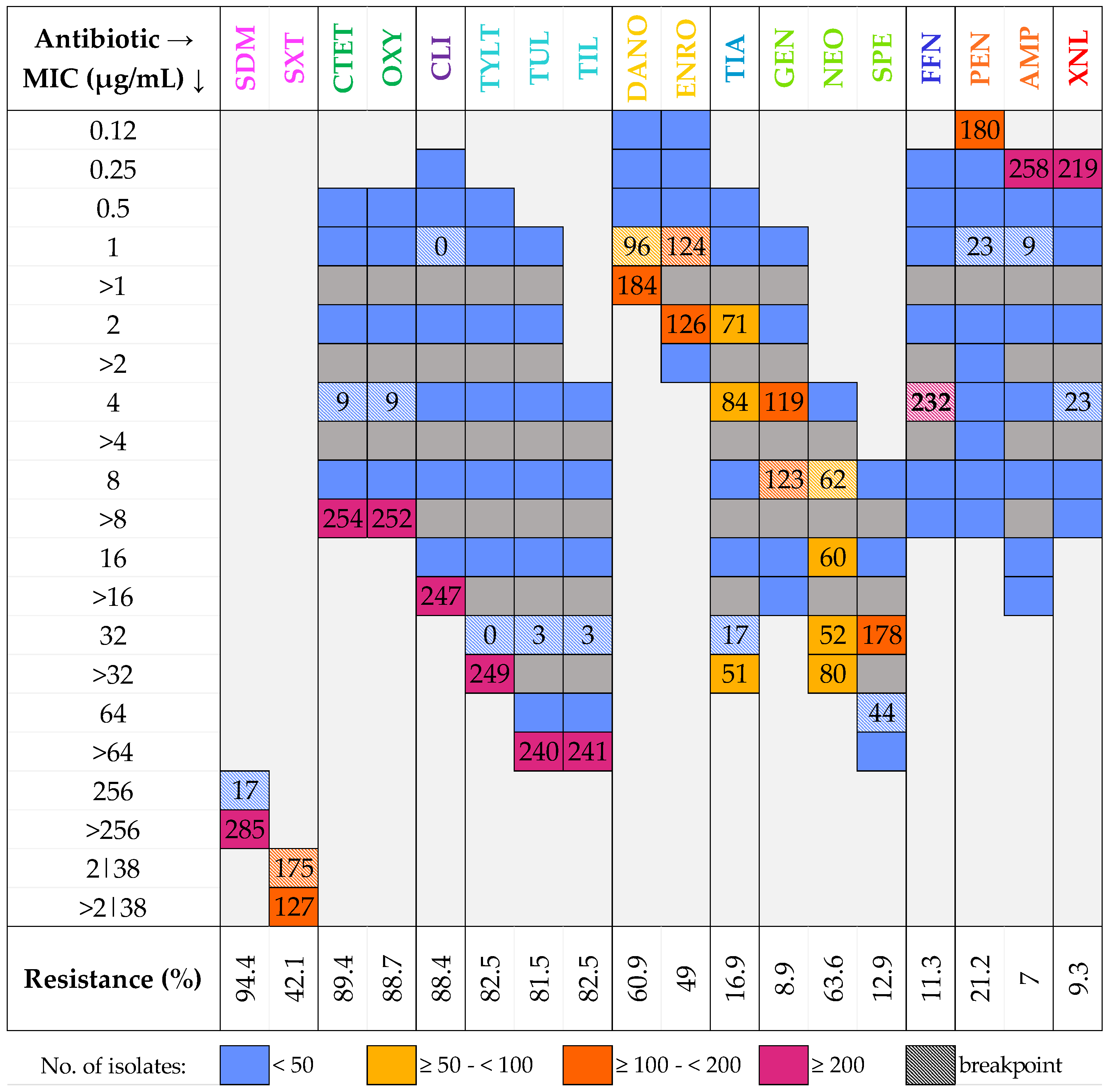

| Antimicrobial Family | Antimicrobial | Range (µg/mL) | BP (µg/mL) |
|---|---|---|---|
| Aminoglycosides | Spectinomycin (SPE) | 8–64 | 64 |
| Gentamicin (GEN) | 1–16 | 8 | |
| Neomycin (NEO) | 4–32 | 8 | |
| Cephalosporins | Ceftiofur (XNL) | 0.25–8 | 4 |
| Amphenicols | Florfenicol (FFN) | 0.25–8 | 4 |
| Lincosamides | Clindamycin (CLI) | 0.25–16 | 1 |
| Macrolides | Tylosin tartrate (TYLT) | 0.5–32 | 32 |
| Tilmicosin (TIL) | 4–64 | 32 | |
| Tulathromycin (TUL) | 1–64 | 32 | |
| Penicillins | Ampicillin (AMP) | 0.25–16 | 1 |
| Penicillin (PEN) | 0.12–8 | 1 | |
| Pleuromutilins | Tiamulin (TIA) | 0.5–32 | 32 |
| Quinolones | Danofloxacin (DANO) | 0.12–1 | 1 |
| Enrofloxacin (ENRO) | 0.12–2 | 1 | |
| Sulfonamides | Sulfadimethoxine (SDM) | 256 | 256 |
| Trimethoprim-sulfamethoxazole (SXT) | 2|38 | 2|38 | |
| Tetracyclines | Chlortetracycline (CTET) | 0.5–8 | 4 |
| Oxytetracycline (OXY) | 0.5–8 | 4 |
| Serotype | R.O.A. *1 | N° of Isolates | % of Isolates | % R.O.A. |
|---|---|---|---|---|
| 1 | First *2 | 49 | 16.2% | 71.2% |
| 14 | First *2 | 2 | 0.7% | |
| 2 | First *2 | 47 | 15.6% | |
| 1/2 | First *2 | 13 | 4.3% | |
| 3 | First | 18 | 6.0% | |
| 7 | First | 17 | 5.6% | |
| 9 | First | 64 | 21.2% | |
| 16 | First | 5 | 1.7% | |
| 4 | Second | 14 | 4.6% | 16.9% |
| 5 | Second | 4 | 1.3% | |
| 8 | Second | 17 | 5.6% | |
| 18 | Second | 4 | 1.3% | |
| 19 | Second | 12 | 4.0% | |
| 10 | Third | 1 | 0.3% | 3.6% |
| 15 | Third | 1 | 0.3% | |
| 17 | Third | 6 | 2.0% | |
| 23 | Third | 2 | 0.7% | |
| 31 | Third | 1 | 0.3% | |
| 21 | Fourth | 1 | 0.3% | 0.7% |
| 28 | Fourth | 1 | 0.3% | |
| NT | - | 23 | 7.6% | - |
| Serotype | C.N.S. | Lungs | Joints | Other * | Total |
|---|---|---|---|---|---|
| 9 | 34 | 16 | 13 | 1 | 64 |
| 1 | 20 | 9 | 19 | 1 | 49 |
| 2 | 20 | 14 | 9 | 4 | 47 |
| 3 | 2 | 15 | 0 | 1 | 18 |
| 7 | 7 | 3 | 4 | 3 | 17 |
| 8 | 1 | 14 | 1 | 1 | 17 |
| 4 | 5 | 7 | 1 | 1 | 14 |
| 1|2 | 10 | 1 | 2 | 0 | 13 |
| 19 | 8 | 2 | 1 | 1 | 12 |
| OS * | 11 | 11 | 6 | 0 | 28 |
| NT * | 10 | 7 | 4 | 2 | 23 |
| Total | 128 | 99 | 60 | 15 | 302 |
| Serotype | Antimicrobial Resistance Pattern | Isolates (%) |
|---|---|---|
| 1 | erm(B)+|mef(A/E)−|tet(M)−|tet(O)+|lsa(E)−|lnu(B)− | 61.2% |
| erm(B)−|mef(A/E)−|tet(M)−|tet(O)−|lsa(E)−|lnu(B)− | 16.3% | |
| erm(B)−|mef(A/E)−|tet(M)−|tet(O)+|lsa(E)−|lnu(B)− | 14.3% | |
| Other antimicrobial resistance patterns | 8.2% | |
| 2 | erm(B)−|mef(A/E)−|tet(M)−|tet(O)+|lsa(E)−|lnu(B)− | 48.9% |
| erm(B)+|mef(A/E)−|tet(M)−|tet(O)+|lsa(E)−|lnu(B)− | 36.2% | |
| erm(B)−|mef(A/E)−|tet(M)+|tet(O)−|lsa(E)−|lnu(B)− | 8.5% | |
| Other antimicrobial resistance patterns | 6.4% | |
| 9 | erm(B)+|mef(A/E)−|tet(M)−|tet(O)+|lsa(E)−|lnu(B)− | 73.8% |
| erm(B)−|mef(A/E)−|tet(M)−|tet(O)+|lsa(E)−|lnu(B)− | 13.8% | |
| erm(B)+|mef(A/E)−|tet(M)−|tet(O)−|lsa(E)−|lnu(B)− | 7.7% | |
| Other antimicrobial resistance patterns | 4.6% | |
| OS * (including 3, 4, 7, 8, 1|2, and 19) | erm(B)+|mef(A/E)−|tet(M)−|tet(O)+|lsa(E)−|lnu(B)− | 36.1% |
| erm(B)−|mef(A/E)−|tet(M)−|tet(O)+|lsa(E)−|lnu(B)− | 32.8% | |
| erm(B)−|mef(A/E)−|tet(M)−|tet(O)−|lsa(E)−|lnu(B)− | 12.6% | |
| Other antimicrobial resistance patterns | 18.5% | |
| NT * | erm(B)−|mef(A/E)−|tet(M)−|tet(O)+|lsa(E)−|lnu(B)− | 26.1% |
| erm(B)−|mef(A/E)−|tet(M)−|tet(O)−|lsa(E)−|lnu(B)− | 21.7% | |
| erm(B)+|mef(A/E)−|tet(M)−|tet(O)+|lsa(E)−|lnu(B)− | 13.0% | |
| Other antimicrobial resistance patterns | 39.1% | |
| Total | erm(B)+|mef(A/E)−|tet(M)−|tet(O)+|lsa(E)−|lnu(B)− | 46.5% |
| erm(B)−|mef(A/E)−|tet(M)−|tet(O)+|lsa(E)−|lnu(B)− | 27.7% | |
| erm(B)−|mef(A/E)−|tet(M)−|tet(O)−|lsa(E)−|lnu(B)− | 9.9% | |
| Other antimicrobial resistance patterns | 15.8% |
| AMR Cluster | No. of Isolates | Main Two Serotypes | Main Isolat. Site | erm(B) | mef(A/E) | tet(M) | tet(O) | lsa(E)/ lnu(B) | Phenotypic AMRs |
|---|---|---|---|---|---|---|---|---|---|
| c1 | 36 | 9, NT | CNS | + | − | − | + | − | 12 (10–17) |
| c2 | 35 | 1, 8 | Lungs | + | − | − | + | − | 11 (7–13) |
| c3 | 20 | 2, 1 | CNS | + | − | − | + | − | 10 |
| c4 | 28 | 9, 4 | CNS | + | − | − | + | − | 10 (8–12) |
| c5 | 28 | 9, 1 | CNS | + | − | − | + | − | 9 (4–9) |
| c6 | 24 | 9, 1 | CNS | + | − | − | + | − | 8 (7–10) |
| c7 | 15 | 3, 7 | Lungs | + | − | − | + | − | 8 (7–9) |
| c8 | 37 | 2, 1 | CNS | − | − | − | + | − | 10 (9–13) |
| c9 | 30 | 2, 19 | CNS | − | − | − | + | − | 9 (7–11) |
| c10 | 27 | 2, 3 | Lungs | − | − | − | + | − | 7 (4–9) |
| c11 | 23 | 1, 8 | Lungs | − | − | − | − | − | 4 (1–7) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrocchi Rilo, M.; Gutiérrez Martín, C.B.; Acebes Fernández, V.; Aguarón Turrientes, Á.; González Fernández, A.; Miguélez Pérez, R.; Martínez Martínez, S. Streptococcus suis Research Update: Serotype Prevalence and Antimicrobial Resistance Distribution in Swine Isolates Recovered in Spain from 2020 to 2022. Vet. Sci. 2024, 11, 40. https://doi.org/10.3390/vetsci11010040
Petrocchi Rilo M, Gutiérrez Martín CB, Acebes Fernández V, Aguarón Turrientes Á, González Fernández A, Miguélez Pérez R, Martínez Martínez S. Streptococcus suis Research Update: Serotype Prevalence and Antimicrobial Resistance Distribution in Swine Isolates Recovered in Spain from 2020 to 2022. Veterinary Sciences. 2024; 11(1):40. https://doi.org/10.3390/vetsci11010040
Chicago/Turabian StylePetrocchi Rilo, Máximo, César Bernardo Gutiérrez Martín, Vanessa Acebes Fernández, Álvaro Aguarón Turrientes, Alba González Fernández, Rubén Miguélez Pérez, and Sonia Martínez Martínez. 2024. "Streptococcus suis Research Update: Serotype Prevalence and Antimicrobial Resistance Distribution in Swine Isolates Recovered in Spain from 2020 to 2022" Veterinary Sciences 11, no. 1: 40. https://doi.org/10.3390/vetsci11010040
APA StylePetrocchi Rilo, M., Gutiérrez Martín, C. B., Acebes Fernández, V., Aguarón Turrientes, Á., González Fernández, A., Miguélez Pérez, R., & Martínez Martínez, S. (2024). Streptococcus suis Research Update: Serotype Prevalence and Antimicrobial Resistance Distribution in Swine Isolates Recovered in Spain from 2020 to 2022. Veterinary Sciences, 11(1), 40. https://doi.org/10.3390/vetsci11010040









