Examination of the Virulence of Actinobacillus pleuropneumoniae Serovar 16 in Pigs
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Experimental Design
2.3. Clinical and Postmortem Examinations
2.4. Serological and Bacteriological Examinations
2.5. Statistical Evaluation
3. Results
3.1. Clinical Signs
3.2. Body Mass and Mass Gain
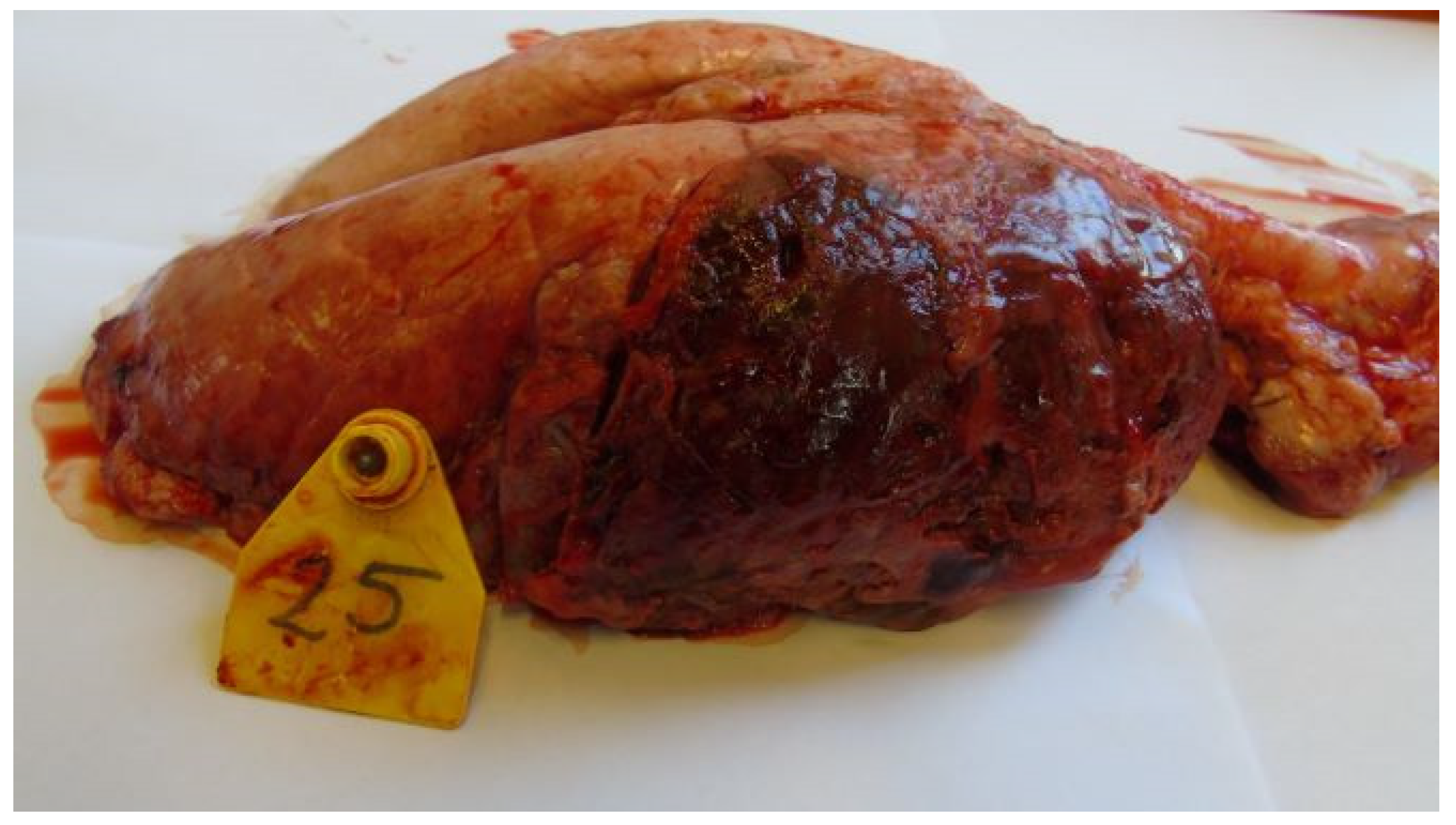
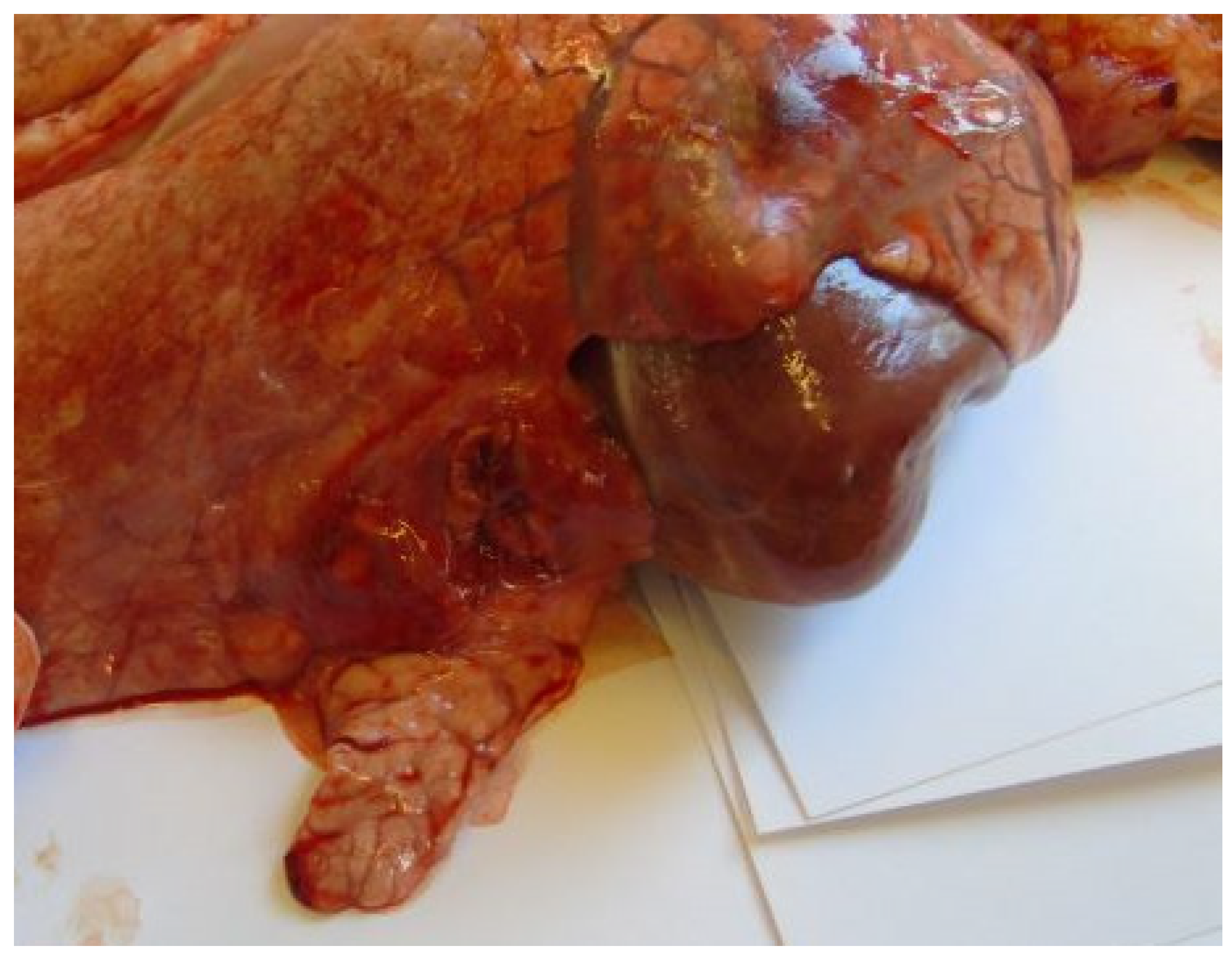
3.3. Postmortem Lesions
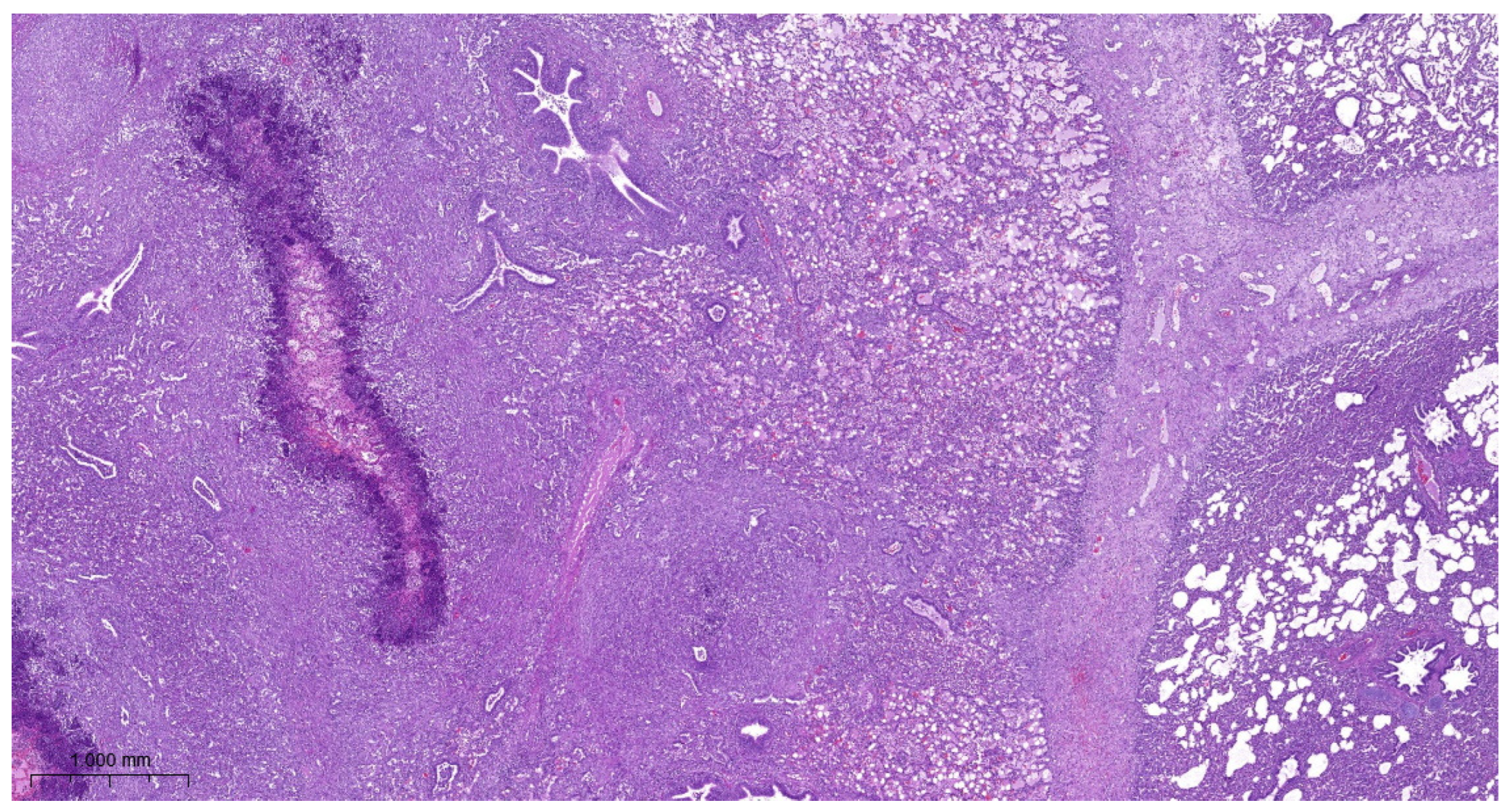
3.4. Histopathology
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hansen, M.S.; Pors, S.E.; Jensen, H.E.; Bille-Hansen, V.; Bisgaard, M.; Flachs, E.M.; Nielsen, O.L. An investigation of the pathology and pathogens associated with porcine respiratory disease complex in Denmark. J. Com. Pathol. 2010, 143, 120–131. [Google Scholar] [CrossRef]
- Gottschalk, M.; Broes, A. Actinobacillosis. In Diseases of Swine, 11th ed.; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2019; pp. 749–766. [Google Scholar]
- Cohen, L.M.; Grontvedt, C.A.; Klem, T.B.; Gulliksen, S.M.; Ranheim, B.; Nielsen, J.P.; Valheim, M.; Kielland, C. A descriptive study of acute outbreaks of respiratory disease in Norwegian fattening pig herds. Acta Vet. Scand. 2020, 62, 35. [Google Scholar] [CrossRef]
- Ruggeri, J.; Salogni, C.; Giovannini, S.; Vitale, N.; Boniotti, M.B.; Corradi, A.; Pozzi, P.; Pasquali, P.; Alborali, G.L. Association between infectious agents and lesions in post-weaned piglets and fattening heavy pigs with porcine respiratory disease complex (PRDC). Front. Vet. Sci. 2020, 7, 636. [Google Scholar] [CrossRef]
- Pattison, I.H.; Howell, D.G.; Elliot, J. A Haemophilus-like organism isolated from pig lung and the associated pneumonic lesions. J. Comp. Pathol. 1957, 67, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Százados, I.; Kádas, I. Occurrence of pleuropneumonia caused by Haemophilus parahaemolyticus in Hungary. Magy. Allatorvosok 1979, 30, 681–685. (In Hungarian) [Google Scholar]
- Molnár, L. Pleuropneumonia of swine caused by Haemophilus parahaemolyticus. Magy. Allatorvosok 1981, 36, 86–98. (In Hungarian) [Google Scholar]
- Sárközi, R.; Makrai, L.; Fodor, L. Actinobacillus pleuropneumoniae serotypes in Hungary. Acta Vet. Hung. 2018, 66, 343–349. [Google Scholar] [CrossRef] [PubMed]
- To, H.; Konnai, M.; Teshima, K.; Tsutsumi, N.; Ito, S.; Sato, M.; Shibuya, K.; Nagai, S. Pulmonary lesions with asteroid bodies in a pig experimentally infected with Actinobacillus pleuropneumoniae serovar 15. J. Vet. Med. Sci. 2023, 85, 1131–1135. [Google Scholar] [CrossRef] [PubMed]
- Sárközi, R.; Makrai, L.; Fodor, L. Isolation of biotype 1 serotype and detection of Actinobacillus pleuropneumoniae from wild boars. Pathogens 2022, 11, 505. [Google Scholar] [CrossRef]
- Hoeltig, D.; Rohde, J.; Frase, R.; Nietfeld, F.; Waldmann, K.-H.; Valentin-Weigand, P.; Meens, J. Multi-organ spreading of Actinobacillus pleuropneumoniae serovar 7 in weaned pigs during the first week after experimental infection. Vet. Res. 2018, 49, 97. [Google Scholar] [CrossRef]
- Hata, A.; Hiwatashi, S.; Miura, N.; Matsuo, Y.; Shibahara, T.; Ito, H. Verrucous endocarditis and pulmonary arteritis caused by Actinobacillus pleuropneumoniae serovar 8 in a slaughtered pig. J. Jpn. Vet. Med. Assoc. 2019, 72, 555–559. [Google Scholar] [CrossRef]
- Madsen, L.W.; Boyle, M.; Jensen, T.K.; Svensmark, B. Actinobacillus pleuropneumoniae demonstrated in situ in exudative meningitis and nephritis. Vet. Rec. 2001, 149, 746–747. [Google Scholar] [CrossRef]
- Ohba, T.; Shibahara, T.; Kobayashi, H.; Takashima, A.; Nagoshi, M.; Osanai, R.; Kubo, M. Multifocal granulomatous hepatitis caused by Actinobacillus pleuropneumoniae serotype 2 in slaughter pigs. J. Comp. Pathol. 2008, 139, 61–66. [Google Scholar] [CrossRef]
- Jensen, T.K.; Boyle, M.; Hagedorn-Olsen, T.; Riising, H.J.; Angen, O. Actinobacillus pleuropneumoniae osteomyelitis in pigs demonstrated by fluorescent in situ hybridization. Vet. Pathol. 1999, 36, 258–261. [Google Scholar] [CrossRef]
- Sassu, E.L.; Bossé, J.T.; Tobias, T.J.; Gottschalk, M.; Langford, P.R.; Hennig-Pauka, I. Update on Actinobacillus pleuropneumoniae —Knowledge, gaps and challenges. Transbound. Emerg. Dis. 2018, 65 (Suppl. S1), 72–90. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zheng, B.; Wang, J.; Li, G.; Cao, S.; Wen, Y.; Huang, X.; Zuo, Z.; Zhong, Z.; Gu, Y. Quinolone resistance of Actinobacillus pleuropneumoniae revealed through genome and transcriptome analyses. Int. J. Mol. Sci. 2021, 22, 10036. [Google Scholar] [CrossRef] [PubMed]
- Vilaró, A.; Novell, E.; Enrique-Tarancon, V.; Baliellas, J.; Fraile, L. Susceptibility trends of swine respiratory pathogens from 2019 to 2022 to antimicrobials commonly used in Spain. Porc. Health Manag. 2023, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Schaller, A.; Kuhn, R.; Nicolet, J.; Anderson, T.J.; Macinnes, J.I.; Segers, R.P.A.M. Characterization of apxIVA, a new RTX determinant of Actinobacillus pleuropneumoniae. Microbiology 1999, 145, 2105–2116. [Google Scholar] [CrossRef] [PubMed]
- Frey, J. RTX toxins of animal pathogens and their role as antigens in vaccines and diagnostics. Toxins 2019, 11, 719. [Google Scholar] [CrossRef]
- Chiers, K.; de Waele, T.; Pasmans, F.; Ducatelle, R.; Haesebrouck, F. Virulence factors of Actinobacillus pleurropneumoniae involved in colonization, persistence and induction of lesions in its porcine host. Vet. Res. 2010, 41, 65–81. [Google Scholar] [CrossRef]
- Grasteau, A.; Tremblay, Y.D.N.; Labrie, J.; Jacques, M. Novel genes associated with biofilm formation of Actinobacillus pleuropneumoniae. Vet. Microbiol. 2011, 153, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Nahar, N.; Turni, C.; Tram, G.; Blackall, P.J.; Artack, J.M. Actinobacillus pleuropneumoniae: The molecular determinants of virulence and pathogenesis. In Advances in Medical Physiology; Poole, R.K., Kelly, D.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; Volume 78, pp. 179–216. [Google Scholar]
- Stancheva, S.G.; Frömbling, J.; Sassu, E.L.; Hennig-Pauka, I.; Lasinig, A.; Gerner, W.; Grunert, T.; Ehling-Schulz, E. Proteomic and immunoproteomic insights into the exoproteome of Actinobacillus pleuropneumoniae, the causative agent of porcine pleuropneumonia. Microb. Pathogen. 2022, 172, 105759. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Cuellar, E.; Guerrero-Barrera, A.L.; Avelar-González, F.J.; Díaz, J.M.; Chávez-Reyes, J.; de Santiago, A.S. Adhesion mechanisms of Actinobacillus pleuropneumoniae to porcine respiratory system and biofilm formation. Austral. J. Vet. Sci. 2022, 54, 93–102. [Google Scholar] [CrossRef]
- Liu, F.; Yao, Q.; Huang, J.; Wan, J.; Xie, T.; Gao, X.; Sun, D.; Zhang, F.; Bei, W.; Lei, L. The two-component system CpxA/CpxR is critical for full virulence of Actinobacillus pleuropneumoniae. Front. Microbiol. 2022, 13, 1029426. [Google Scholar] [CrossRef] [PubMed]
- Perezchica, M.M.S.; Barrera, A.L.G.; Gonzalez, F.J.A.; Tristan, T.Q.; Marin, O.M. Actinobacillus pleuropneumoniae, surface proteins and virulence: A review. Front. Vet. Sci. 2023, 10, 127612. [Google Scholar] [CrossRef] [PubMed]
- Sassu, E.L.; Frömbling, J.; Duvigneau, J.C.; Miller, I.; Müllebner, A.; Gutiérrez, A.M.; Grunert, T.; Patzl, M.; Saalmüller, A.; von Altrock, A.; et al. Host-pathogen interplay at primary infection sites in pigs challenged with Actinobacillus pleuropneumoniae. BMC Vet. Res. 2017, 13, 64. [Google Scholar] [CrossRef][Green Version]
- Wallgren, P.; Segall, T.; Pedersen Mörner, A.; Gunnarson, A. Experimental infections with Actinobacillus pleuropneumoniae in pigs —I. Comparison of five different parenteral antibiotic treatments. J. Vet. Med. B 1999, 46, 249–260. [Google Scholar]
- Xie, F.; Li, G.; Zhou, L.; Zhang, Y.; Cui, N.; Liu, S.; Wang, C. Attenuated Actinobacillus pleuropneumoniae double-deletion mutant S-8ΔclpP/apxIIC confers protection against homologous or heterologous strain challenge. BMC Vet. Res. 2017, 13, 14. [Google Scholar]
- Wu, H.-C.; Yeh, P.-H.; Hsueh, K.-J.; Yang, W.-J.; Chu, C.-Y. Recombinant ApxIV protein enhances protective efficacy against Actinobacillus pleuropneumoniae in mice and pigs. J. Appl. Microbiol. 2018, 124, 1366–1376. [Google Scholar] [CrossRef]
- Soutter, F.; Priestnall, S.L.; Catchpole, B.; Rycroft, A.N. An experimental dermal oedema model for Apx toxins of Actinobacillus pleuropneumoniae. J. Comp. Path. 2022, 195, 12–18. [Google Scholar] [CrossRef]
- Tumamao, J.Q.; Bowles, R.E.; van den Bosch, H.; Klaasen, H.L.B.M.; Fenwwick, B.W.; Storie, G.J.; Blackall, P.J. Comparison of the efficacy of a subunit and a live streptomycin-dependent porcine pleuropneumonia vaccine. Aust. Vet. J. 2004, 82, 370–374. [Google Scholar] [CrossRef]
- Mortensen, P.; Toft, N.; Kiss, I.; Palya, V.; Smits, H.; Tenk, M. Comparative efficacy in challenge dose models of a toxin expressing whole-cell vaccine against eight serovars of Actinobacillus pleuropneumoniae in pigs. Animals 2022, 12, 3244. [Google Scholar] [CrossRef] [PubMed]
- Hur, J.; Lee, J.H. Optimization of immune strategy for a construct of Salmonella-delivered ApxIA, ApxIIA, ApxIIIA and OmpA antigens of Actinobacillus pleuropneumoniae for prevention of porcine pleuropneumonia using murine model. Vet. Res. Commun. 2014, 38, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Luo, W.; Xiong, R.; Li, H.; Yao, Z.; Zhuo, W.; Zou, G.; Huang, Q.; Zhou, R. A combinatorial vaccine containing inactivated bacterin and subunits provides protection against Actinobacillus pleuropneumoniae infection in mice and pigs. Front. Vet. Sci. 2022, 9, 902497. [Google Scholar] [CrossRef]
- Pereira, M.F.; Rossi, C.C.; de Queiroz, M.V.; Martins, G.F.; Isaac, C.; Bossé, J.T.; Li, Y.; Wren, B.W.; Terra, V.S.; Cuccui, J.; et al. Galleria mellonella is an effective model to study Actinobacillus pleuropneumoniae infection. Microbiology 2015, 161, 387–400. [Google Scholar] [CrossRef]
- Stringer, O.W.; Bossé, J.T.; Lacouture, S.; Gottschalk, M.; Fodor, L.; Angen, O.; Velasquez, E.; Penny, P.; Lei, L.; Langford, P.R.; et al. Proposal of Actinobacillus pleuropneumoniae serovar 19, and reformulation of previous multiplex PCRs for capsule-specific typing of all known serovars. Vet. Microbiol. 2021, 255, 109021. [Google Scholar] [CrossRef]
- Dubreuil, J.D.; Jacques, M.; Mittal, K.R.; Gottschalk, M. Actinobacillus pleuropneumoniae surface polysaccharides: Their role in diagnosis and immunogenicity. Anim. Health Res. Rev. 2000, 1, 73–93. [Google Scholar] [CrossRef]
- Maldonaldo, J.; Valls, L.; Martínez, E.; Riera, P. Isolation rates, serovars, and toxin genotypes of nicotinamide adenine dinucleotide-independent Actinobacillus pleuropneumoniae among pigs suffering from pleuropneumonia in Spain. J. Vet. Diagn. Investig. 2009, 21, 854–857. [Google Scholar] [CrossRef]
- Schuwerk, L.; Hoeltig, D.; Waldmann, K.-H.; Valentin-Weigand, P.; Rohde, J. Sero- and apx-typing of German Actinobacillus pleuropneumoniae field isolates from 2010 to 2019 reveals predominance of serovar 2 with regular apx-profile. Vet. Res. 2021, 52, 10. [Google Scholar] [CrossRef]
- Ozawa, M.; Kawano, M.; Abo, H.; Issiki, Y.; Kumakawa, M.; Kawanishi, M.; Iwamoto, S. Characterization of Actinobacillus pleuropneumoniae isolated from pigs in Japan using whole genome sequencing. Comp. Immunol. Microbiol. Inf. Dis. 2023, 102, 102062. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Lacouture, S. Distribution of Streptococcus suis (from 2012 to 2014) and Actinobacillus pleuropneumoniae (from 2011 to 2014) serotypes isolated from diseased pigs. Canad. Vet. J. 2015, 56, 1093–1094. [Google Scholar]
- Sárközi, R.; Makrai, L.; Fodor, L. Identification of a proposed new serovar of Actinobacillus pleuropneumoniae: Serovar 16. Acta Vet. Hung. 2015, 63, 444–450. [Google Scholar] [CrossRef]
- Kardos, G.; Sárközi, R.; Laczkó, L.; Marton, S.; Makrai, L.; Bányai, K.; Fodor, L. Genetic diversity of Actinobacillus pleuropneumoniae serovars in Hungary. Vet. Sci. 2022, 9, 511. [Google Scholar] [CrossRef]
- Bossé, J.T.; Li, Y.; Sárközi, R.; Gottschalk, M.; Angen, O.; Nedbalcona, K.; Rycroft, A.N.; Fodor, L.; Langford, P.R. A unique capsule locus in the newly designated Actinobacillus pleuropneumoniae serovar 16 and development of a diagnostic PCR assay. J. Clin. Microbiol. 2017, 22, 902–907. [Google Scholar] [CrossRef]
- Tenk, M.; Országh, G.; Benyeda, J.; Biksi, I.; Kis, T.; Alapi, I.; Herczeg, J.; Thevenon, J.; Kupcsulik, B.; Siklódi, B.; et al. Efficacy of COGLAPIX® in susceptible piglets against a challenge with Actinobacillus pleuropneumoniae; evaluation of post-mortem findings, clinical data and immune response. In Proceedings of the 21st International Pig Veterinary Society Congress, Vancouver, BC, Canada, 18–21 July 2010; p. 614. [Google Scholar]
- Porcine actinobacillosis vaccine (inactivated). In European Pharmacopoeia, 10th ed.; Council of Europe: London, UK, 2013; Volume 10, pp. 1139–1140.
- Exbrayat, J.M. Microscopy: Light Microscopy and Histochemical Methods. In Encyclopedia of Food and Health; Caballero, B., Finglas, P.M., Toldrá, F., Eds.; Academic Press: Oxford, UK, 2016; pp. 715–723. [Google Scholar]
- Markey, B.; Leonard, F.; Archambault, M.; Cullinane, A.; Maguire, D. Clinical Veterinary Microbiology, 2nd ed.; Mosby Elsevier: Edinburgh, UK, 2013; p. 901. [Google Scholar]
- Bossé, J.T.; Janson, H.; Sheehan, B.J.; Beddek, A.J.; Rycroft, A.N.; Kroll, J.S.; Langford, P.R. Actinobacillus pleuropneumoniae: Pathobiology and pathogenesis of infection. Microbes Infect. 2002, 4, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Idris, U.E.A.; Harmon, B.G.; Udeze, F.A.; Kadis, S. Pulmonary lesions in mice inoculated with Actinobacillus pleuropneumoniae hemolysin and lipopolysaccharide. Vet. Pathol. 1993, 30, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Montaraz, J.A.; Rosales, M.E.; Bautista, E.; Barcenas, G.; Lara, V. A novel experimental model for the study and evaluation of experimental vaccines to Actinobacillus pleuropneumoniae. Vet. Immunol. Immunpathol. 1994, 41, 375–380. [Google Scholar] [CrossRef]
- Dao, H.T.; Truong, Q.L.; Do, V.T.; Hahn, T.-W. Construction and immunization with double mutant ΔapxIBD Δpnp forms of Actinobacillus pleuropneumoniae serotypes 1 and 5. J. Vet. Sci. 2020, 21, 20. [Google Scholar] [CrossRef] [PubMed]



| Scores | |||
|---|---|---|---|
| Clinical Signs | 0 | 1 | 2 |
| Weakness | No | Moderate | Intensive |
| Dyspnea | No | Moderate | Intensive |
| Respiratory rattle | No | Moderate | Intensive |
| Cough | No | Yes | - |
| Vomit | No | Yes | - |
| Anorexia | No | Yes | - |
| Groups | Diseased Animals | Average Clinical Score | Febrile Animals | Average Length of Clinical Signs | Average Number of Febrile Days |
|---|---|---|---|---|---|
| Group 1 (108 cfu) * | 10/10 | 12.60 ± 10.16 a | 5/10 | 4.20 ± 1.55 a | 0.90 ± 1.29 a |
| Group 2 (107 cfu) | 9/10 | 8.70 ± 6.88 a,c | 4/10 | 4.00 ± 2.05 a | 0.50 ± 0.71 a |
| Group 3 (106 cfu) | 7/9 | 3.89 ± 6.51 b,c | 2/9 | 1.89 ± 1.83 b | 0.33 ± 0.71 a |
| Group | Body Mass (Day −1, kg) | Body Mass (Day 6, kg) | Mass Gain (kg) |
|---|---|---|---|
| Group 1 (108 cfu) * | 23.20 ± 2.54 a | 25.55 ± 3.58 a | 2.35 ± 1.92 a |
| Group 2 (107 cfu) | 23.55 ± 1.83 a | 27.30 ± 2.87 a,c | 3.75 ± 2.69 a |
| Group 3 (106 cfu) | 24.83 ± 2.44 a | 29.11 ± 2.93 b,c | 4.28 ± 2.20 a |
| Group | Lung Lesions | Postmortem Score | Re-isolation of A. pleuropneumoniae |
|---|---|---|---|
| Group 1 (108 cfu) | 9/10 | 5.60 ± 6.08 | 8/9 |
| Group 2 (107 cfu) | 8/10 | 4.50 ± 4.50 | 7/8 |
| Group 3 (106 cfu) | 8/9 | 5.78 ± 6.85 | 7/8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tenk, M.; Tóth, G.; Márton, Z.; Sárközi, R.; Szórádi, A.; Makrai, L.; Pálmai, N.; Szalai, T.; Albert, M.; Fodor, L. Examination of the Virulence of Actinobacillus pleuropneumoniae Serovar 16 in Pigs. Vet. Sci. 2024, 11, 62. https://doi.org/10.3390/vetsci11020062
Tenk M, Tóth G, Márton Z, Sárközi R, Szórádi A, Makrai L, Pálmai N, Szalai T, Albert M, Fodor L. Examination of the Virulence of Actinobacillus pleuropneumoniae Serovar 16 in Pigs. Veterinary Sciences. 2024; 11(2):62. https://doi.org/10.3390/vetsci11020062
Chicago/Turabian StyleTenk, Miklós, Gergely Tóth, Zsuzsanna Márton, Rita Sárközi, Alejandra Szórádi, László Makrai, Nimród Pálmai, Tamás Szalai, Mihály Albert, and László Fodor. 2024. "Examination of the Virulence of Actinobacillus pleuropneumoniae Serovar 16 in Pigs" Veterinary Sciences 11, no. 2: 62. https://doi.org/10.3390/vetsci11020062
APA StyleTenk, M., Tóth, G., Márton, Z., Sárközi, R., Szórádi, A., Makrai, L., Pálmai, N., Szalai, T., Albert, M., & Fodor, L. (2024). Examination of the Virulence of Actinobacillus pleuropneumoniae Serovar 16 in Pigs. Veterinary Sciences, 11(2), 62. https://doi.org/10.3390/vetsci11020062







