Identify Candidate Genes Associated with the Weight and Egg Quality Traits in Wenshui Green Shell-Laying Chickens by the Copy Number Variation-Based Genome-Wide Association Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Population Description
2.2. Phenotyping
2.3. Sequence Alignment to Reference Genome
2.4. CNV Detection
2.5. CNV-Based GWAS
2.6. Gene Annotation
3. Results
3.1. Number and Distribution of CNVR
3.2. CNVR-Based Genome-Wide Association Study
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bras, A.; Rodrigues, A.S.; Rueff, J. Copy number variations and constitutional chromothripsis (Review). Biomed. Rep. 2020, 13, 11. [Google Scholar] [CrossRef] [PubMed]
- Stafuzza, N.B.; Silva, R.M.D.; Fragomeni, B.D.; Masuda, Y.; Huang, Y.; Gray, K.; Lourenco, D.A.L. A genome-wide single nucleotide polymorphism and copy number variation analysis for number of piglets born alive. Bmc Genom. 2019, 20, 321. [Google Scholar] [CrossRef] [PubMed]
- Stranger, B.E.; Forrest, M.S.; Dunning, M.; Ingle, C.E.; Beazley, C.; Thorne, N.; Redon, R.; Bird, C.P.; de Grassi, A.; Lee, C.; et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 2007, 315, 848–853. [Google Scholar] [CrossRef] [PubMed]
- Gamazon, E.R.; Stranger, B.E. The impact of human copy number variation on gene expression. Brief. Funct. Genom. 2015, 14, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Lemos, B.; Dopman, E.B.; Hartl, D.L. Copy-Number Variation: The Balance between Gene Dosage and Expression in Drosophila melanogaster. Genome Biol. Evol. 2011, 3, 1014–1024. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Fang, L.Z.; Liu, S.L.; Pan, M.G.; Seroussi, E.; Cole, J.B.; Ma, L.; Chen, H.; Liu, G.E. Array CGH-based detection of CNV regions and their potential association with reproduction and other economic traits in Holsteins. BMC Genom. 2019, 20, 181. [Google Scholar] [CrossRef]
- Geistlinger, L.; da Silva, V.H.; Cesar, A.S.M.; Tizioto, P.C.; Waldron, L.; Zimmer, R.; Regitano, L.C.A.; Coutinho, L.L. Widespread modulation of gene expression by copy number variation in skeletal muscle. Sci. Rep. 2018, 8, 1399. [Google Scholar] [CrossRef]
- Fernandes, A.C.; da Silva, V.H.; Goes, C.P.; Moreira, G.C.M.; Godoy, T.F.; Ibelli, A.M.G.; Peixoto, J.D.; Cantao, M.E.; Ledur, M.C.; de Rezende, F.M.; et al. Genome-wide detection of CNVs and their association with performance traits in broilers. BMC Genom. 2021, 22, 354. [Google Scholar] [CrossRef]
- Zhang, H.; Du, Z.Q.; Dong, J.Q.; Wang, H.X.; Shi, H.Y.; Wang, N.; Wang, S.Z.; Li, H. Detection of genome-wide copy number variations in two chicken lines divergently selected for abdominal fat content. BMC Genom. 2014, 15, 517. [Google Scholar] [CrossRef]
- Han, R.L.; Yang, P.K.; Tian, Y.D.; Wang, D.D.; Zhang, Z.X.; Wang, L.L.; Li, Z.J.; Jiang, R.R.; Kang, X.T. Identification and functional characterization of copy number variations in diverse chicken breeds. BMC Genom. 2014, 15, 934. [Google Scholar] [CrossRef]
- Seol, D.; Ko, B.J.; Kim, B.; Chai, H.H.; Lim, D.; Kim, H. Identification of Copy Number Variation in Domestic Chicken Using Whole-Genome Sequencing Reveals Evidence of Selection in the Genome. Animals 2019, 9, 809. [Google Scholar] [CrossRef] [PubMed]
- Pinkel, D.; Segraves, R.; Sudar, D.; Clark, S.; Poole, I.; Kowbel, D.; Collins, C.; Kuo, W.L.; Chen, C.; Zhai, Y.; et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 1998, 20, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Yau, C.; Holmes, C.C. CNV discovery using SNP genotyping arrays. Cytogenet. Genome Res. 2008, 123, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Gabrielaite, M.; Torp, M.H.; Rasmussen, M.S.; Andreu-Sanchez, S.; Vieira, F.G.; Pedersen, C.B.; Kinalis, S.; Madsen, M.B.; Kodama, M.; Demircan, G.S.; et al. A Comparison of Tools for Copy-Number Variation Detection in Germline Whole Exome and Whole Genome Sequencing Data. Cancers 2021, 13, 6283. [Google Scholar] [CrossRef] [PubMed]
- Escaramis, G.; Docampo, E.; Rabionet, R. A decade of structural variants: Description, history and methods to detect structural variation. Brief. Funct. Genom. 2015, 14, 305–314. [Google Scholar] [CrossRef]
- Xi, R.; Lee, S.; Park, P.J. A survey of copy-number variation detection tools based on high-throughput sequencing data. Curr. Protoc. Hum. Genet. 2012, 75, 7.19.1–7.19.15. [Google Scholar] [CrossRef]
- Pirooznia, M.; Goes, F.S.; Zandi, P.P. Whole-genome CNV analysis: Advances in computational approaches. Front. Genet. 2015, 6, 138. [Google Scholar] [CrossRef]
- Qiu, Y.B.; Ding, R.R.; Zhuang, Z.W.; Wu, J.; Yang, M.; Zhou, S.P.; Ye, Y.; Geng, Q.; Xu, Z.; Huang, S.X.; et al. Genome-wide detection of CNV regions and their potential association with growth and fatness traits in Duroc pigs. BMC Genom. 2021, 22, 332. [Google Scholar] [CrossRef]
- Ladeira, G.C.; Pilonetto, F.; Fernandes, A.C.; Boscollo, P.P.; Dauria, B.D.; Titto, C.G.; Coutinho, L.L.; Silva, F.F.E.; Pinto, L.F.B.; Mourao, G.B. CNV detection and their association with growth, efficiency and carcass traits in Santa Ines sheep. J. Anim. Breed. Genet. 2022, 139, 476–487. [Google Scholar] [CrossRef]
- Zhou, Y.; Connor, E.E.; Wiggans, G.R.; Lu, Y.; Tempelman, R.J.; Schroeder, S.G.; Chen, H.; Liu, G.E. Genome-wide copy number variant analysis reveals variants associated with 10 diverse production traits in Holstein cattle. BMC Genom. 2018, 19, 314. [Google Scholar] [CrossRef]
- He, X.; Tian, M.; Wang, W.; Feng, Y.; Li, Z.; Wang, J.; Song, Y.; Zhang, J.; Liu, D. Identification of Candidate Genes for Min Pig Villi Hair Traits by Genome-Wide Association of Copy Number Variation. Vet. Sci. 2023, 10, 307. [Google Scholar] [CrossRef] [PubMed]
- Risch, N.; Merikangas, K. The future of genetic studies of complex human diseases. Science 1996, 273, 1516–1517. [Google Scholar] [CrossRef]
- Liang, B.; Ding, H.; Huang, L.; Luo, H.; Zhu, X. GWAS in cancer: Progress and challenges. Mol. Genet. Genom. 2020, 295, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Strillacci, M.G.; Gorla, E.; Cozzi, M.C.; Vevey, M.; Genova, F.; Scienski, K.; Longeri, M.; Bagnato, A. A copy number variant scan in the autochthonous Valdostana Red Pied cattle breed and comparison with specialized dairy populations. PLoS ONE 2018, 13, e0204669. [Google Scholar] [CrossRef]
- Xu, Y.X.; Hu, J.; Fan, W.L.; Liu, H.H.; Zhang, Y.S.; Guo, Z.B.; Huang, W.; Liu, X.L.; Hou, S.S. Genome-wide association analysis reveals 6 copy number variations associated with the number of cervical vertebrae in Pekin ducks. Front. Cell Dev. Biol. 2022, 10, 1041088. [Google Scholar] [CrossRef] [PubMed]
- Strillacci, M.G.; Cozzi, M.C.; Gorla, E.; Mosca, F.; Schiavini, F.; Roman-Ponce, S.I.; Lopez, F.J.R.; Schiavone, A.; Marzoni, M.; Cerolini, S.; et al. Genomic and genetic variability of six chicken populations using single nucleotide polymorphism and copy number variants as markers. Animal 2017, 11, 737–745. [Google Scholar] [CrossRef]
- Fontanesi, L.; Beretti, F.; Martelli, P.L.; Colombo, M.; Dall’Olio, S.; Occidente, M.; Portolano, B.; Casadio, R.; Matassino, D.; Russo, V. A first comparative map of copy number variations in the sheep genome. Genomics 2011, 97, 158–165. [Google Scholar] [CrossRef]
- Gao, Y.; Jiang, J.; Yang, S.; Hou, Y.; Liu, G.E.; Zhang, S.; Zhang, Q.; Sun, D. CNV discovery for milk composition traits in dairy cattle using whole genome resequencing. BMC Genom. 2017, 18, 265. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- De Summa, S.; Malerba, G.; Pinto, R.; Mori, A.; Mijatovic, V.; Tommasi, S. GATK hard filtering: Tunable parameters to improve variant calling for next generation sequencing targeted gene panel data. BMC Bioinform. 2017, 18, 119. [Google Scholar] [CrossRef]
- Rausch, T.; Zichner, T.; Schlattl, A.; Stutz, A.M.; Benes, V.; Korbel, J.O. DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012, 28, i333–i339. [Google Scholar] [CrossRef] [PubMed]
- Redon, R.; Ishikawa, S.; Fitch, K.R.; Feuk, L.; Perry, G.H.; Andrews, T.D.; Fiegler, H.; Shapero, M.H.; Carson, A.R.; Chen, W.; et al. Global variation in copy number in the human genome. Nature 2006, 444, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Tang, H.; Liu, J.F.; Xu, S.Z.; Zhang, Q.; Ning, C. Rapid epistatic mixed-model association studies by controlling multiple polygenic effects. Bioinformatics 2020, 36, 4833–4837. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.S.; Li, J.; Zhang, R.; Lin, X.R.; Xu, J.G.; Xie, L.; Xu, Z.Q.; Wang, L.; Gan, J.K.; Xie, X.J.; et al. Copy number variation identification and analysis of the chicken genome using a 60K SNP BeadChip. Poult. Sci. 2016, 95, 1750–1756. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Bai, X.; Liu, H.G.; Zhao, B.B.; Yan, Z.X.; Hou, Y.L.; Chu, Q. Population Genomic Sequencing Delineates Global Landscape of Copy Number Variations that Drive Domestication and Breed Formation of in Chicken. Front. Genet. 2022, 13, 830393. [Google Scholar] [CrossRef]
- Zhang, G.X.; Fan, Q.C.; Wang, J.Y.; Zhang, T.; Xue, Q.; Shi, H.Q. Genome-wide association study on reproductive traits in Jinghai Yellow Chicken. Anim. Reprod. Sci. 2015, 163, 30–34. [Google Scholar] [CrossRef]
- Jin, C.F.; Chen, Y.J.; Yang, Z.Q.; Shi, K.; Chen, C.K. A genome-wide association study of growth trait-related single nucleotide polymorphisms in Chinese Yancheng chickens. Genet. Mol. Res. 2015, 14, 15783–15792. [Google Scholar] [CrossRef]
- Perez-Bonilla, A.; Jabbour, C.; Frikha, M.; Mirzaie, S.; Garcia, J.; Mateos, G.G. Effect of crude protein and fat content of diet on productive performance and egg quality traits of brown egg-laying hens with different initial body weight. Poult. Sci. 2012, 91, 1400–1405. [Google Scholar] [CrossRef]
- Kirikçi, K.; Günlü, A.; Cetin, O.; Garip, M. Effect of hen weight on egg production and some egg quality characteristics in the partridge (Alectoris graeca). Poult. Sci. 2007, 86, 1380–1383. [Google Scholar] [CrossRef]
- Leeson, S.; Summers, J.D. Effect of immature body weight on laying performance. Poult. Sci. 1987, 66, 1924–1928. [Google Scholar] [CrossRef]
- Santiago, G.G.; Siqueira, F.; Cardoso, F.F.; Regitano, L.C.A.; Ventura, R.; Sollero, B.P.; Souza, M.D.; Mokry, F.B.; Ferreira, A.B.R.; Torres, R.A.A. Genomewide association study for production and meat quality traits in Canchim beef cattle. J. Anim. Sci. 2017, 95, 3381–3390. [Google Scholar] [CrossRef] [PubMed]
- Anton, I.; Huth, B.; Fuller, I.; Rozsa, L.; Hollo, G.; Zsolnai, A. Effect of single nucleotide polymorphisms on intramuscular fat content in Hungarian Simmental cattle. Asian Austral J. Anim. 2018, 31, 1415–1419. [Google Scholar] [CrossRef]
- Niu, Q.H.; Zhang, T.L.; Xu, L.; Wang, T.Z.; Wang, Z.Z.; Zhu, B.; Gao, X.; Chen, Y.; Zhang, L.P.; Gao, H.J.; et al. Identification of Candidate Variants Associated With Bone Weight Using Whole Genome Sequence in Beef Cattle. Front. Genet. 2021, 12, 750746. [Google Scholar] [CrossRef] [PubMed]
- Al-Mamun, H.A.; Kwan, P.; Clark, S.A.; Ferdosi, M.H.; Tellam, R.; Gondro, C. Genome-wide association study of body weight in Australian Merino sheep reveals an orthologous region on OAR6 to human and bovine genomic regions affecting height and weight. Genet. Sel. Evol. 2015, 47, 66. [Google Scholar] [CrossRef]
- Ramos, Z.; Garrick, D.J.; Blair, H.T.; Vera, B.; Ciappesoni, G.; Kenyon, P.R. Genomic Regions Associated with Wool, Growth and Reproduction Traits in Uruguayan Merino Sheep. Genes 2023, 14, 167. [Google Scholar] [CrossRef]
- Zhuang, X.N.; Xie, F.; Lin, Z.K.; Luo, J.Y.; Chen, T.; Xi, Q.Y.; Zhang, Y.L.; Sun, J.J. Effect of miR-493-5p on proliferation and differentiation of myoblast by targeting ANKRD17. Cell Tissue Res. 2023, 393, 119–132. [Google Scholar] [CrossRef]
- Zheng, X.; Ju, Z.H.; Wang, J.; Li, Q.L.; Huang, J.M.; Zhang, A.W.; Zhong, J.F.; Wang, C.F. Single nucleotide polymorphisms, haplotypes and combined genotypes of LAP3 gene in bovine and their association with milk production traits. Mol. Biol. Rep. 2011, 38, 4053–4061. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.R.; Sun, Y.F.; Zhao, G.P.; Wang, F.J.; Wu, D.; Zheng, M.Q.; Chen, J.L.; Zhang, L.; Hu, Y.D.; Wen, J. Genome-Wide Association Study Identifies Loci and Candidate Genes for Body Composition and Meat Quality Traits in Beijing-You Chickens. PLoS ONE 2013, 8, e61172. [Google Scholar] [CrossRef]
- Chen, J.C.; Dai, F.P.; Balakrishnan-Renuka, A.; Leese, F.; Schempp, W.; Schaller, F.; Hoffmann, M.M.; Morosan-Puopolo, G.; Yusuf, F.; Bisschoff, I.J.; et al. Diversification and Molecular Evolution of ATOH8, a Gene Encoding a bHLH Transcription Factor. PLoS ONE 2011, 6, e23005. [Google Scholar] [CrossRef]
- Divvela, S.S.K.; Offei, E.B.; Suerland, F.; Garcia, D.R.; Kwiatkowski, J.; Balakrishnan-Renuka, A.; Bohne, P.; Boing, M.; Morosan-Puopolo, G.; Mark, M.D.; et al. Atonal homolog 8/Math6 regulates differentiation and maintenance of skeletal muscle. Front. Cell Dev. Biol. 2022, 10, 950414. [Google Scholar] [CrossRef]
- Balakrishnan-Renuka, A.; Morosan-Puopolo, G.; Yusuf, F.; Abduelmula, A.; Chen, J.C.; Zoidl, G.; Philippi, S.; Dai, F.P.; Brand-Saberi, B. ATOH8, a regulator of skeletal myogenesis in the hypaxial myotome of the trunk. Histochem. Cell Biol. 2014, 141, 289–300. [Google Scholar] [CrossRef]
- Erensoy, K.; Raynaud, E.; Petit, A.; Baumard, Y.; Metayer-Coustard, S.; Le Bihan-Duval, E. Research Note: Divergent selection for breast muscle ultimate pH affects egg quality traits in broiler breeders. Poult. Sci. 2022, 101, 102142. [Google Scholar] [CrossRef] [PubMed]
- Strekalova, T.; Veniaminova, E.; Svirin, E.; Kopeikina, E.; Veremeyko, T.; Yung, A.W.Y.; Proshin, A.; Tan, S.Z.K.; Khairuddin, S.; Lim, L.W.; et al. Sex-Specific ADHD-like Behaviour, Altered Metabolic Functions, and Altered EEG Activity in Sialyltransferase ST3GAL5-Deficient Mice. Biomolecules 2021, 11, 1759. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; van der Zon, G.; Ma, J.; Mei, H.; Cabukusta, B.; Agaser, C.C.; Madunić, K.; Wuhrer, M.; Zhang, T.; Ten Dijke, P. ST3GAL5-catalyzed gangliosides inhibit TGF-β-induced epithelial-mesenchymal transition via TβRI degradation. EMBO J. 2023, 42, e110553. [Google Scholar] [CrossRef] [PubMed]
- Kidani, S.; Kaneoka, H.; Okuzaki, Y.; Asai, S.; Kojima, Y.; Nishijima, K.; Iijima, S. Analyses of chicken sialyltransferases related to O-glycosylation. J. Biosci. Bioeng. 2016, 122, 379–384. [Google Scholar] [CrossRef]
- Gu, Y.F.; Chen, Y.P.; Jin, R.; Wang, C.; Wen, C.; Zhou, Y.M. A comparison of intestinal integrity, digestive function, and egg quality in laying hens with different ages. Poult. Sci. 2021, 100, 100949. [Google Scholar] [CrossRef]
- Javerzat, S.; Franco, M.; Herbert, J.; Platonova, N.; Peille, A.L.; Pantesco, V.; De Vos, J.; Assou, S.; Bicknell, R.; Bikfalvi, A.; et al. Correlating global gene regulation to angiogenesis in the developing chick extra-embryonic vascular system. PLoS ONE 2009, 4, e7856. [Google Scholar] [CrossRef]
- Ostendorff, H.P.; Tursun, B.; Cornils, K.; Schluter, A.; Drung, A.; Gungor, C.; Bach, I. Dynamic expression of LIM cofactors in the developing mouse neural tube. Dev. Dyn. 2006, 235, 786–791. [Google Scholar] [CrossRef]
- Gu, X.; Feng, C.; Ma, L.; Song, C.; Wang, Y.; Da, Y.; Li, H.; Chen, K.; Ye, S.; Ge, C.; et al. Genome-wide association study of body weight in chicken F2 resource population. PLoS ONE 2011, 6, e21872. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Li, Y.; Wu, J.; Wang, X.; Bian, C.; Tian, Y.; Sun, G.; Han, R.; Liu, X.; et al. Genome-wide association study reveals the genetic determinism of growth traits in a Gushi-Anka F(2) chicken population. Heredity 2021, 126, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.H.; Wang, J.Y.; Zhang, T.; Wang, Y.; Zhang, Y.; Han, K. Genome-wide association study of growth traits in Jinghai Yellow chicken hens using SLAF-seq technology. Anim. Genet. 2019, 50, 175–176. [Google Scholar] [CrossRef] [PubMed]
- Dou, D.; Shen, L.; Zhou, J.; Cao, Z.; Luan, P.; Li, Y.; Xiao, F.; Guo, H.; Li, H.; Zhang, H. Genome-wide association studies for growth traits in broilers. BMC Genom. Data 2022, 23, 1. [Google Scholar] [CrossRef]
- Li, Z.; Zheng, M.; Abdalla, B.A.; Zhang, Z.; Xu, Z.; Ye, Q.; Xu, H.; Luo, W.; Nie, Q.; Zhang, X. Genome-wide association study of aggressive behaviour in chicken. Sci. Rep. 2016, 6, 30981. [Google Scholar] [CrossRef]
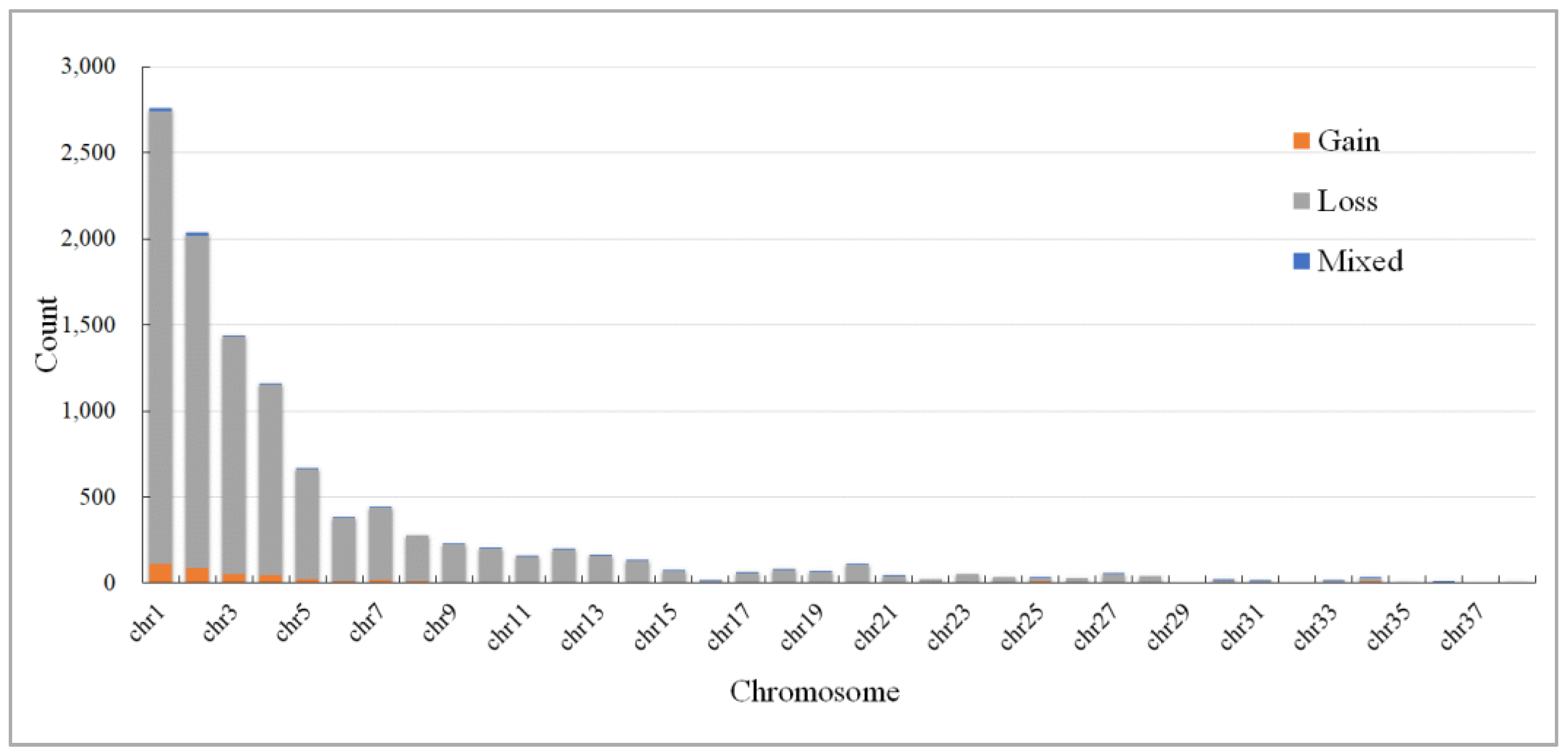



| Chr | Chr Length (bp) | CNVR Count | Length of CNVR (bp) | Coverage (%) | Max Size (bp) | Average (bp) | Min Size (bp) |
|---|---|---|---|---|---|---|---|
| 1 | 197,778,178 | 2761 | 4,102,658 | 2.1 | 642,753 | 1485.9 | 51 |
| 2 | 149,541,958 | 2038 | 1,795,942 | 1.2 | 76,049 | 881.2 | 51 |
| 3 | 110,815,227 | 1435 | 1,011,600 | 0.9 | 37,549 | 704.9 | 51 |
| 4 | 91,021,375 | 1162 | 894,849 | 1.0 | 104,295 | 770.1 | 51 |
| 5 | 59,471,259 | 664 | 581,860 | 1.0 | 38,456 | 876.3 | 51 |
| 6 | 35,339,061 | 382 | 241,170 | 0.7 | 21,044 | 631.3 | 51 |
| 7 | 36,318,844 | 437 | 353,483 | 1.0 | 36,899 | 808.9 | 51 |
| 8 | 29,613,760 | 276 | 143,465 | 0.5 | 20,456 | 519.8 | 52 |
| 9 | 23,556,363 | 224 | 177,489 | 0.8 | 15,124 | 792.4 | 51 |
| 10 | 20,214,400 | 204 | 122,553 | 0.6 | 12,825 | 600.8 | 51 |
| 11 | 19,755,808 | 157 | 81,894 | 0.4 | 16,494 | 521.6 | 51 |
| 12 | 20,438,972 | 194 | 124,811 | 0.6 | 19,162 | 643.4 | 51 |
| 13 | 18,437,548 | 161 | 285,249 | 1.5 | 50,758 | 1771.7 | 51 |
| 14 | 15,523,295 | 135 | 89,883 | 0.6 | 18,875 | 665.8 | 51 |
| 15 | 12,662,000 | 71 | 80,617 | 0.6 | 14,389 | 1135.5 | 51 |
| 16 | 1,595,800 | 11 | 615,070 | 38.5 | 542,075 | 55,915.5 | 63 |
| 17 | 10,229,956 | 57 | 22,414 | 0.2 | 4896 | 393.2 | 51 |
| 18 | 11,472,971 | 78 | 101,581 | 0.9 | 35,771 | 1302.3 | 51 |
| 19 | 10,411,340 | 66 | 65,454 | 0.6 | 12,765 | 991.7 | 51 |
| 20 | 14,040,156 | 110 | 78,193 | 0.6 | 16,021 | 710.8 | 51 |
| 21 | 6,776,000 | 42 | 36,126 | 0.5 | 27,274 | 860.1 | 52 |
| 22 | 4,690,381 | 23 | 15,414 | 0.3 | 4663 | 670.2 | 53 |
| 23 | 5,830,993 | 53 | 17,315 | 0.3 | 6364 | 326.7 | 52 |
| 24 | 6,352,200 | 37 | 16,508 | 0.3 | 6088 | 446.2 | 53 |
| 25 | 2,575,857 | 34 | 44,705 | 1.7 | 10,788 | 1314.9 | 52 |
| 26 | 5,288,600 | 30 | 23,279 | 0.4 | 13,044 | 776.0 | 51 |
| 27 | 5,930,361 | 56 | 425,849 | 7.2 | 311,709 | 7604.4 | 53 |
| 28 | 5,407,282 | 42 | 29,718 | 0.5 | 12,414 | 707.6 | 52 |
| 29 | 1,064,585 | 0 | 0 | 0 | 0 | 0 | 0 |
| 30 | 979,082 | 21 | 8930 | 0.9 | 1139 | 425.2 | 54 |
| 31 | 2,139,823 | 13 | 308,985 | 14.4 | 193,165 | 23,768.1 | 58 |
| 32 | 454,000 | 0 | 0 | 0 | 0 | 0 | 0 |
| 33 | 3,524,363 | 19 | 1,142,884 | 32.4 | 504,390 | 60,151.8 | 52 |
| 34 | 2,223,258 | 30 | 28,054 | 1.3 | 16,716 | 935.1 | 73 |
| 35 | 327,777 | 2 | 17,147 | 5.2 | 15,351 | 8573.5 | 1796 |
| 36 | 493,600 | 7 | 7529 | 1.5 | 3094 | 1075.6 | 226 |
| 37 | 316,000 | 0 | 0 | 0 | 0 | 0 | 0 |
| 38 | 298,400 | 3 | 1258 | 0.4 | 477 | 419.3 | 365 |
| overall | 6,247,088 | 11,035 | 13,093,936 | 1.4 | 642,753 | 1186.6 | 51 |
| Trait 1 | CNVR ID | Type 2 | Chromosome | CNVR Position (bp) 3 | p-Value 4 | Proximal Gene 5 |
|---|---|---|---|---|---|---|
| 30-EW | DUP00035918 | Gain | 4 | 85,203,669–85,205,350 | 2.52 × 10−7 | ATOH8 ST3GAL5 |
| 30-EW 40-EW 40-EWW | DEL00035336 | Loss | 4 | 75,882,077–75,882,619 | 2.22 × 10−6 3.93 × 10−6 0.65 × 10−6 | FAM184B MED28 LAP3 |
| 8-W | DEL00035339 | Loss | 4 | 76,019,681–76,020,779 | 2.35 × 10−6 | LOC112532307 LDB2 |
| 8-W 38-W | DEL00035404 | Loss | 4 | 77,124,281–77,124,469 | 5.01 × 10−8 2.02 × 10−7 | LOC107053295 |
| 8-W | DEL00035425 | Loss | 4 | 77,492,948–77,493,042 | 3.16 × 10−6 | LOC121110716 |
| 8-W | DEL00035578 | Loss | 4 | 80,144,379–80,145,908 | 1.35 × 10−6 | SORCS2 LOC121110591 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Ning, C.; Yang, C.; Li, W.; Zhang, Q.; Wang, D.; Tang, H. Identify Candidate Genes Associated with the Weight and Egg Quality Traits in Wenshui Green Shell-Laying Chickens by the Copy Number Variation-Based Genome-Wide Association Study. Vet. Sci. 2024, 11, 76. https://doi.org/10.3390/vetsci11020076
Yang S, Ning C, Yang C, Li W, Zhang Q, Wang D, Tang H. Identify Candidate Genes Associated with the Weight and Egg Quality Traits in Wenshui Green Shell-Laying Chickens by the Copy Number Variation-Based Genome-Wide Association Study. Veterinary Sciences. 2024; 11(2):76. https://doi.org/10.3390/vetsci11020076
Chicago/Turabian StyleYang, Suozhou, Chao Ning, Cheng Yang, Wenqiang Li, Qin Zhang, Dan Wang, and Hui Tang. 2024. "Identify Candidate Genes Associated with the Weight and Egg Quality Traits in Wenshui Green Shell-Laying Chickens by the Copy Number Variation-Based Genome-Wide Association Study" Veterinary Sciences 11, no. 2: 76. https://doi.org/10.3390/vetsci11020076
APA StyleYang, S., Ning, C., Yang, C., Li, W., Zhang, Q., Wang, D., & Tang, H. (2024). Identify Candidate Genes Associated with the Weight and Egg Quality Traits in Wenshui Green Shell-Laying Chickens by the Copy Number Variation-Based Genome-Wide Association Study. Veterinary Sciences, 11(2), 76. https://doi.org/10.3390/vetsci11020076






