Nonstructural Protein A238L of the African Swine Fever Virus (ASFV) Enhances Antiviral Immune Responses by Activating the TBK1-IRF3 Pathway
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Viruses
2.3. Cells
2.4. RT-PCR Analyses
2.5. Immunoblotting
2.6. Luciferase Assay
2.7. Immunofluorescence Staining
2.8. Flow Cytometry
2.9. Statistical Analysis
3. Results
3.1. A238L Inhibits the Expression of Proinflammatory Cytokine Genes
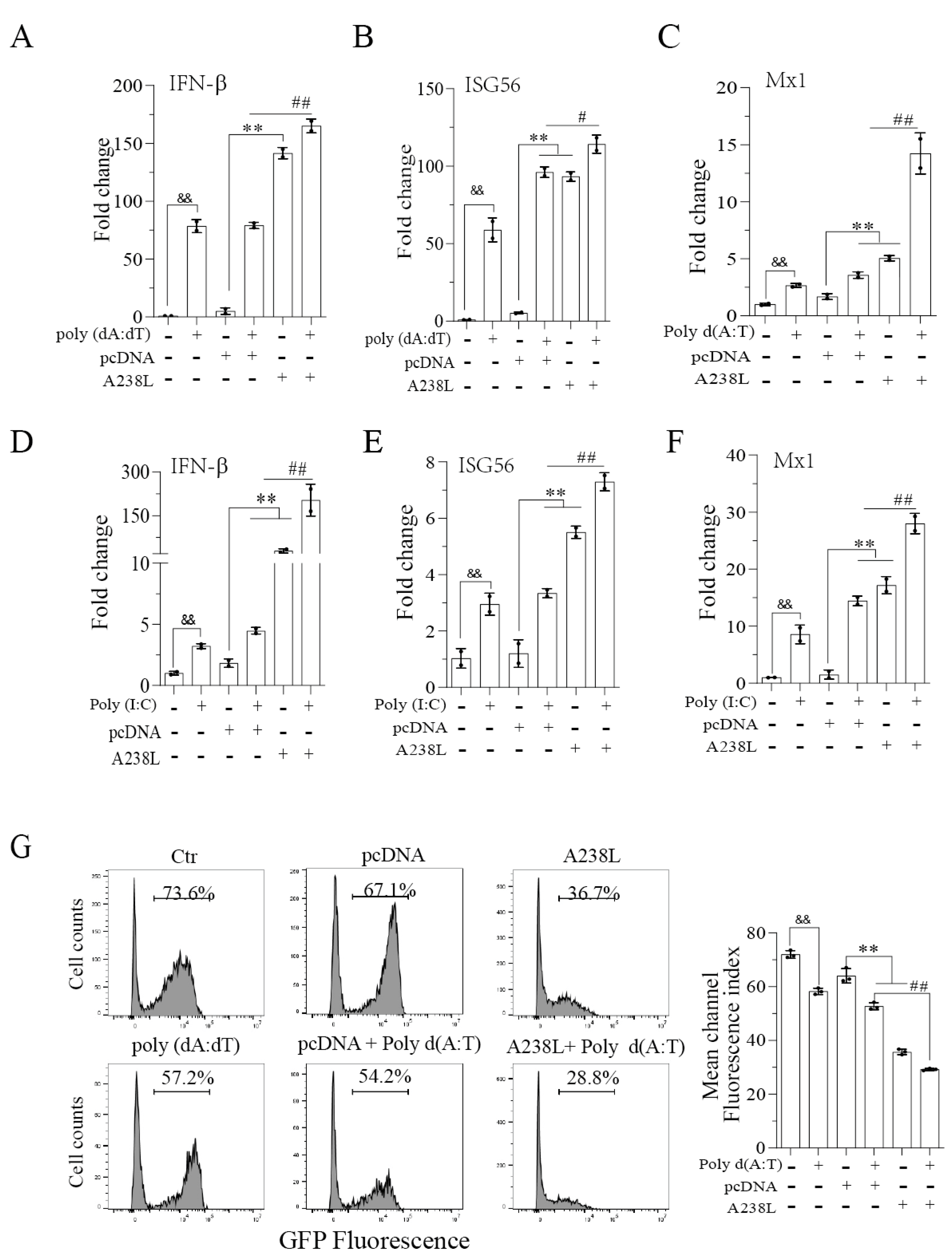
3.2. A238L Enhances the Antiviral Immune Response
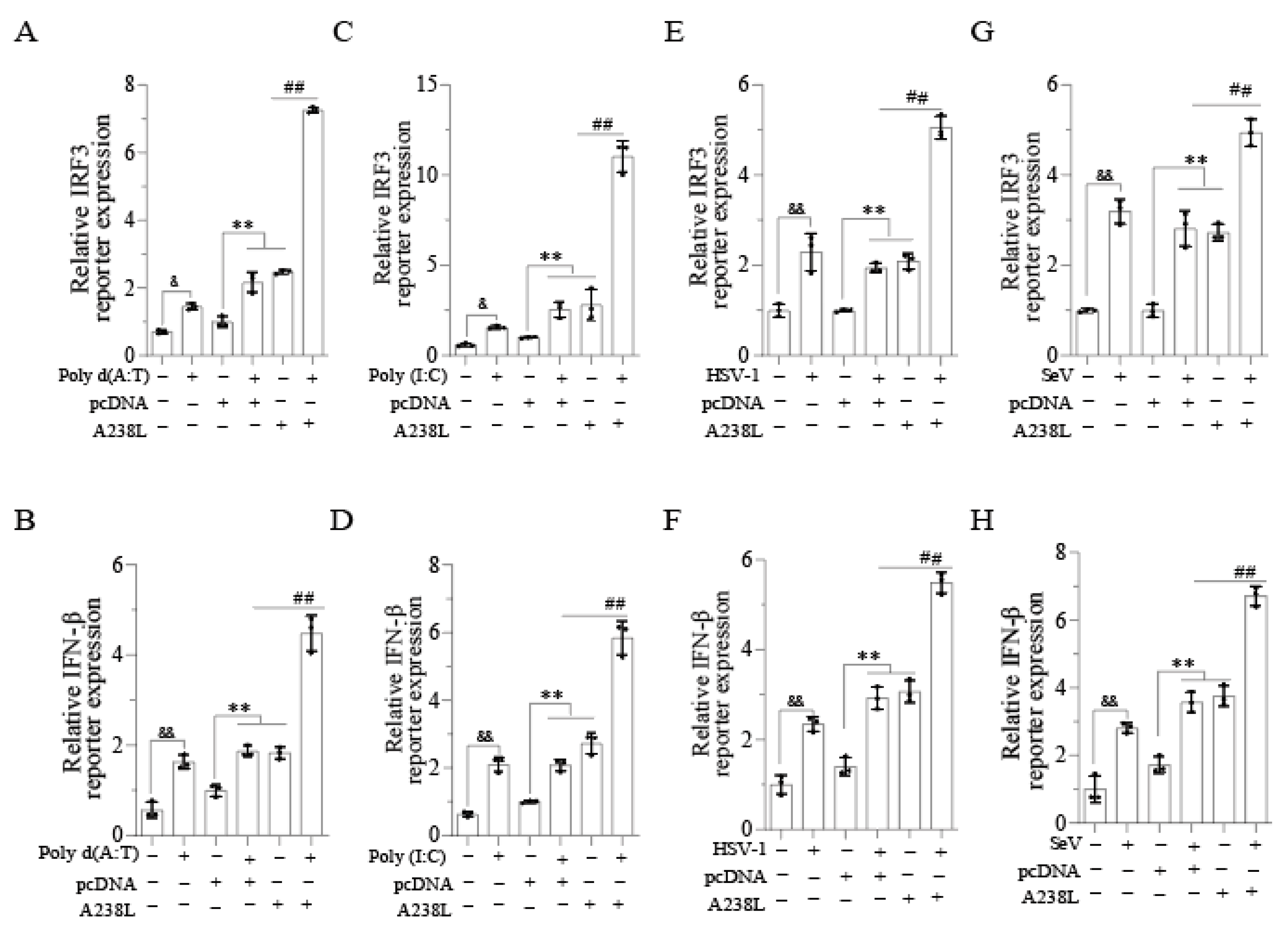
3.3. A238L Increases the IRF3-Driven Promoter Activity
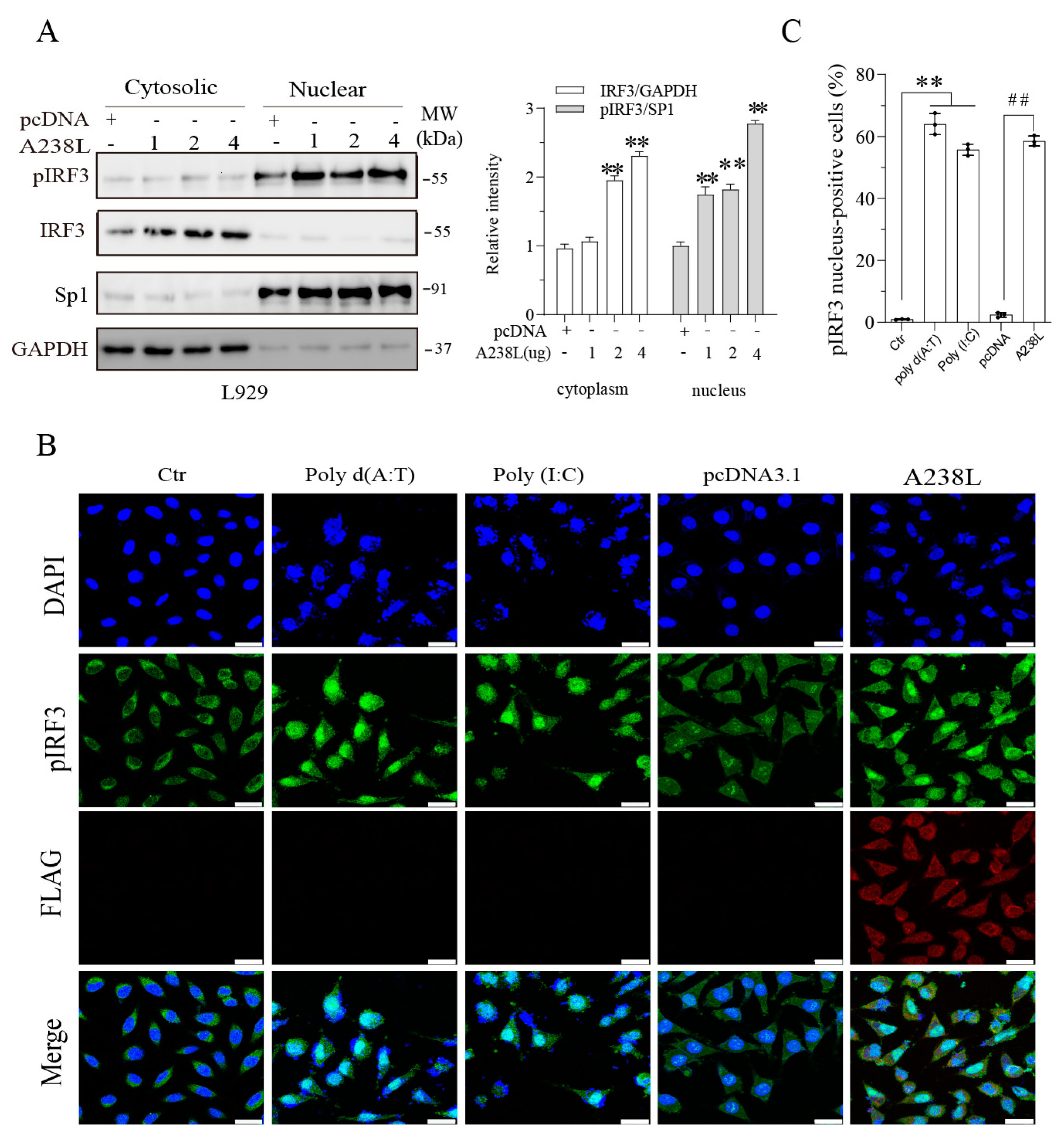
3.4. A238L Enhances TBK1 and IRF3 Phosphorylation
3.5. A238L Induces IRF3 Nuclear Translocation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- VanderWaal, K.; Deen, J. Global trends in infectious diseases of swine. Proc. Natl. Acad. Sci. USA 2018, 115, 11495–11500. [Google Scholar] [CrossRef] [PubMed]
- Zakaryan, H.; Karalova, E.; Voskanyan, H.; Ter-Pogossyan, Z.; Nersisyan, N.; Hakobyan, A.; Saroyan, D.; Karalyan, Z. Evaluation of hemostaseological status of pigs experimentally infected with African swine fever virus. Vet. Microbiol. 2014, 174, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Pikalo, J.; Zani, L.; Hühr, J.; Beer, M.; Biome, S. Pathogenesis of African swine fever in domestic pigs and European wild boar—Lessons learned from recent animal trials. Virus Res. 2019, 271, 197614. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.Q.; Li, J.M.; Fan, X.X.; Liu, F.X.; Li, L.; Wang, Q.H.; Ren, W.J.; Bao, J.Y.; Liu, C.J.; Wang, H.; et al. Molecular Characterization of African Swine Fever Virus, China, 2018. Emerg. Infect Dis. 2018, 24, 2131–2133. [Google Scholar] [CrossRef] [PubMed]
- Karger, A.; Pérez-Núñez, D.; Urquiza, J.; Hinojar, P.; Alonso, C.; Freitas, F.B.; Revilla, Y.; Le Potier, M.F.; Montoya, M. An Update on African Swine Fever Virology. Viruses 2019, 11, 864. [Google Scholar] [CrossRef] [PubMed]
- Blome, S.; Franzke, K.; Beer, M. African swine fever—A review of current knowledge. Virus Res. 2020, 287, 198099. [Google Scholar] [CrossRef] [PubMed]
- Dixon, L.K.; Islam, M.; Nash, R.; Reis, A.L. African swine fever virus evasion of host defences. Virus Res. 2019, 266, 25–33. [Google Scholar] [CrossRef]
- Galindo, I.; Alonso, C. African Swine Fever Virus: A Review. Viruses 2017, 9, 103. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef]
- Danilchanka, O.; Mekalanos, J.J. Cyclic Dinucleotides and the Innate Immune Response. Cell 2013, 154, 962–970. [Google Scholar] [CrossRef]
- Kleiboeker, S.B.; Scoles, G.A.; Burrage, T.G.; Sur, J. African swine fever virus replication in the midgut epithelium is required for infection of Ornithodoros ticks. J. Virol. 1999, 73, 8587–8598. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Chiu, Y.H.; Chen, Z.J.J. The cGAS-cGAMP-STING Pathway of Cytosolic DNA Sensing and Signaling. Mol. Cell 2014, 54, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Konno, H.; Konno, K.; Barber, G.N. Cyclic Dinucleotides Trigger ULK1 (ATG1) Phosphorylation of STING to Prevent Sustained Innate Immune Signaling. Cell 2013, 155, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Revilla, Y.; Granja, A.G. Viral Mechanisms Involved in the Transcriptional CBP/p300 Regulation of Inflammatory and Immune Responses. Crit. Rev. Immunol. 2009, 29, 131–154. [Google Scholar] [CrossRef] [PubMed]
- Revilla, Y.; Callejo, M.; Rodríguez, J.M.; Culebras, E.; Nogal, M.L.; Salas, M.L.; Viñuela, E.; Fresno, M. Inhibition of nuclear factor kappaB activation by a virus-encoded IkappaB-like protein. J. Biol. Chem. 1998, 273, 5405–5411. [Google Scholar] [CrossRef] [PubMed]
- Tait, S.W.G.; Reid, E.B.; Greaves, D.R.; Wileman, T.E.; Powell, P.P. Mechanism of Inactivation of NF-κB by a Viral Homologue of IκBα: Signal-induced release of Iκbα results in binding of the viral homologue to Nf-κB. J. Biol. Chem. 2000, 275, 34656–34664. [Google Scholar] [CrossRef] [PubMed]
- Granja, A.G.; Sabina, P.; Salas, M.L.; Fresno, M.; Revilla, Y. Regulation of inducible nitric oxide synthase expression by viral A238L-mediated inhibition of p65/RelA acetylation and p300 transactivation. J. Virol. 2006, 80, 10487–10496. [Google Scholar] [CrossRef] [PubMed]
- Salguero, F.J.; Gil, S.; Revilla, Y.; Gallardo, C.; Arias, M.; Martins, C. Cytokine mRNA expression and pathological findings in pigs inoculated with African swine fever virus (E-70) deleted on A238L. Vet. Immunol. Immunop. 2008, 124, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Abkallo, H.M.; Hemmink, J.D.; Oduor, B.; Khazalwa, E.M.; Svitek, N.; Assad-Garcia, N.; Khayumbi, J.; Fuchs, W.; Vashee, S.; Steinaa, L. Co-Deletion of A238L and EP402R Genes from a Genotype IX African Swine Fever Virus Results in Partial Attenuation and Protection in Swine. Viruses 2022, 14, 2024. [Google Scholar] [CrossRef]
- Shi, J.; Liu, W.; Zhang, M.; Sun, J.; Xu, X.L. The A179L Gene of African Swine Fever Virus Suppresses Virus-Induced Apoptosis but Enhances Necroptosis. Viruses 2021, 13, 2490. [Google Scholar] [CrossRef]
- Roy, M.; Singh, R. TRIMs: Selective recruitment at different steps of the NF-κB pathway-determinant of activation or resolution of inflammation. Cell. Mol. Life Sci. 2021, 78, 6069–6086. [Google Scholar] [CrossRef] [PubMed]
- Al Hamrashdi, M.; Brady, G. Regulation of IRF3 activation in human antiviral signaling pathways. Biochem. Pharmacol. 2022, 200, 115026. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Puertas, P.; Rodríguez, F.; Oviedo, J.M.; Ramiro-Ibáñez, F.; Ruiz-Gonzalvo, F.; Alonso, C.; Escribano, J.M. Neutralizing antibodies to different proteins of African swine fever virus inhibit both virus attachment and internalization. J. Virol. 1996, 70, 5689–5694. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Matsuda, M.; Kobayashi, H.; Miyata, Y.; Nakayama, Y.; Komuro, R.; Fukuhara, A.; Shimomura, I. Effects of statins on adipose tissue inflammation: Their inhibitory effect on MyD88-independent IRF3/IFN-beta pathway in macrophages. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 871–877. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Fu, J.Y.; Li, J.X.; Guo, S.B.; Chen, Q.C.; Zhang, Y.B.; Liu, Z.K.; Tan, C.; Chen, H.C.; Wang, X.R. African Swine Fever Virus pI215L Inhibits Type I Interferon Signaling by Targeting Interferon Regulatory Factor 9 for Autophagic Degradation. J. Virol. 2022, 96, e0094422. [Google Scholar] [CrossRef] [PubMed]
- Correia, S.; Ventura, S.; Parkhouse, R.M. Identification and utility of innate immune system evasion mechanisms of ASFV. Virus Res. 2013, 173, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Dodantenna, N.; Ranathunga, L.; Chathuranga, W.A.G.; Weerawardhana, A.; Cha, J.W.; Subasinghe, A.; Gamage, N.; Haluwana, D.K.; Kim, Y.; Jheong, W.; et al. African Swine Fever Virus EP364R and C129R Target Cyclic GMP-AMP to Inhibit the cGAS-STING Signaling Pathway. J. Virol. 2022, 96, e0102222. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.W.; Yu, S.X.; Ge, H.L.; Wang, T.; Li, Y.F.; Zhou, P.P.; Pan, L.; Han, Y.; Yang, Y.Y.; Sun, Y.; et al. The A137R Protein of African Swine Fever Virus Inhibits Type I Interferon Production via the Autophagy-Mediated Lysosomal Degradation of TBK1. J. Virol. 2022, 96, e0195721. [Google Scholar] [CrossRef] [PubMed]
- Rathakrishnan, A.; Moffat, K.; Reis, A.L.; Dixon, L.K. Production of Recombinant African Swine Fever Viruses: Speeding up the Process. Viruses 2020, 12, 615. [Google Scholar] [CrossRef]
- Gladue, D.P.; O’Donnell, V.; Ramirez-Medina, E.; Rai, A.; Pruitt, S.; Vuono, E.A.; Silva, E.; Velazquez-Salinas, L.; Borca, M.V.J.V. Deletion of CD2-like (CD2v) and C-type lectin-like (EP153R) genes from African swine fever virus Georgia-Δ 9GL abrogates its effectiveness as an experimental vaccine. Viruses 2020, 12, 1185. [Google Scholar] [CrossRef]
- Wu, J.X.; Sun, L.J.; Chen, X.; Du, F.H.; Shi, H.P.; Chen, C.; Chen, Z.J.J. Cyclic GMP-AMP Is an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Science 2013, 339, 826–830. [Google Scholar] [CrossRef] [PubMed]
- Meylan, E.; Curran, J.; Hofmann, K.; Moradpour, D.; Binder, M.; Bartenschlager, R.; Tschopp, R. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 2005, 437, 1167–1172. [Google Scholar] [CrossRef]
- Hou, F.J.; Sun, L.J.; Zheng, H.; Skaug, B.; Jiang, Q.X.; Chen, Z.J.J. MAVS Forms Functional Prion-like Aggregates to Activate and Propagate Antiviral Innate Immune Response. Cell 2011, 146, 448–461. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.; Li, X.; Li, P.W. The mechanism of double-stranded DNA sensing through the cGAS-STING pathway. Cytokine Growth F R. 2014, 25, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.X.; Wu, J.; Wu, Y.T.; Chen, H.J.; Zhang, S.F.; Li, J.X.; Xin, T.; Jia, H.; Hou, S.H.; Jiang, Y.T.; et al. Inhibition of cGAS-STING-TBK1 signaling pathway by DP96R of ASFV China 2018/1. Biochem. Bioph. Res. Commun. 2018, 506, 437–443. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, V.L.; Almeida, S.C.P.; Soares, H.R.; Crespo, A.; Marshall-Clarke, S.; Parkhouse, R.M.E. A novel TLR3 inhibitor encoded by African swine fever virus (ASFV). Arch. Virol. 2011, 156, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhu, Z.; Feng, T.; Ma, Z.; Xue, Q.; Wu, P.; Li, P.; Li, S.; Yang, F.; Cao, W.; et al. African Swine Fever Virus E120R Protein Inhibits Interferon Beta Production by Interacting with IRF3 To Block Its Activation. J. Virol. 2021, 95, e0082421. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yang, W.P.; Li, L.L.; Li, P.; Ma, Z.; Zhang, J.; Qi, X.L.; Ren, J.J.; Ru, Y.; Niu, Q.L.; et al. African Swine Fever Virus MGF-505-7R Negatively Regulates cGAS-STING-Mediated Signaling Pathway. J. Immunol. 2021, 206, 1844–1857. [Google Scholar] [CrossRef]
- Zhou, R.; Liu, L.; Wang, Y. Viral proteins recognized by different TLRs. J. Med. Virol. 2021, 93, 6116–6123. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Ghosh, S. The NF-κB Family of Transcription Factors and Its Regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef]
- DiDonato, J.A.; Mercurio, F.; Karin, M. NF-κB and the link between inflammation and cancer. Immunol. Rev. 2012, 246, 379–400. [Google Scholar] [CrossRef]
- Tripathi, P.; Aggarwal, A. NF-kB transcription factor: A key player in the generation of immune response. Curr. Sci. India 2006, 90, 519–531. [Google Scholar]
- Zhuo, Y.; Guo, Z.; Ba, T.; Zhang, C.; He, L.; Zeng, C.; Dai, H. African Swine Fever Virus MGF360-12L Inhibits Type I Interferon Production by Blocking the Interaction of Importin α and NF-κB Signaling Pathway. Virol. Sin. 2021, 36, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Barrado-Gil, L.; Del Puerto, A.; Galindo, I.; Cuesta-Geijo, M.; García-Dorival, I.; de Motes, C.M.; Alonso, C. African Swine Fever Virus Ubiquitin-Conjugating Enzyme Is an Immunomodulator Targeting NF-κB Activation. Viruses 2021, 13, 1160. [Google Scholar] [CrossRef] [PubMed]
- Powell, P.P.; Dixon, L.K.; Parkhouse, R.M. An IkappaB homolog encoded by African swine fever virus provides a novel mechanism for downregulation of proinflammatory cytokine responses in host macrophages. J. Virol. 1996, 70, 8527–8533. [Google Scholar] [CrossRef]
- Granja, A.G.; Perkins, N.D.; Revilla, Y. A238L inhibits NF-ATc2, NF-kappa B, and c-Jun activation through a novel mechanism involving protein kinase C-theta-mediated up-regulation of the amino-terminal transactivation domain of p300. J. Immunol. 2008, 180, 2429–2442. [Google Scholar] [CrossRef]
- García-Belmonte, R.; Pérez-Núñez, D.; Pittau, M.; Richt, J.A.; Revilla, Y. African Swine Fever Virus Armenia/07 Virulent Strain Controls Interferon Beta Production through the cGAS-STING Pathway. J. Virol. 2019, 93, e02298-18. [Google Scholar] [CrossRef]


| Primer | Sequence |
|---|---|
| TNF-α-F | CCCTCACACTCAGATCATCTTCT |
| TNF-α-R | GCTACGACGTGGGCTACAG |
| IL-6-F | TGAGATCTACTCGGCAAACCTAGTG |
| IL-6-R | CTTCGTAGAGAACAACATAAGTCAGATACC |
| IL-1β-F | TGGACCTTCCAGGATGAGGACA |
| IL-1β-R | TTCATCTCGGAGCCTGTAGTG |
| IFN-β-F | CAGCTCCAAGAAAGGACGAAC |
| IFN-β-R | GGCAGTGTAACTCTTCTGCAT |
| ISG56-F | TAGCCAACATGTCCTCACAGAC |
| ISG56-R | TCTTCTACCACTGGTTTCATGC |
| Mx1-F | GACCATAGGGGTCTTGACCAA |
| Mx1-R | AGACTTGCTCTTTCTGAAAAGCC |
| Actin-F | CATCCGTAAAGACCTCTATGCCAAC |
| Actin-R | ATGGAGCCACCGATCCACA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Yang, L.; Di, C.; Sun, J.; Liu, P.; Liu, H. Nonstructural Protein A238L of the African Swine Fever Virus (ASFV) Enhances Antiviral Immune Responses by Activating the TBK1-IRF3 Pathway. Vet. Sci. 2024, 11, 252. https://doi.org/10.3390/vetsci11060252
Liu W, Yang L, Di C, Sun J, Liu P, Liu H. Nonstructural Protein A238L of the African Swine Fever Virus (ASFV) Enhances Antiviral Immune Responses by Activating the TBK1-IRF3 Pathway. Veterinary Sciences. 2024; 11(6):252. https://doi.org/10.3390/vetsci11060252
Chicago/Turabian StyleLiu, Wei, Lanlan Yang, Chuanyuan Di, Jing Sun, Penggang Liu, and Huisheng Liu. 2024. "Nonstructural Protein A238L of the African Swine Fever Virus (ASFV) Enhances Antiviral Immune Responses by Activating the TBK1-IRF3 Pathway" Veterinary Sciences 11, no. 6: 252. https://doi.org/10.3390/vetsci11060252
APA StyleLiu, W., Yang, L., Di, C., Sun, J., Liu, P., & Liu, H. (2024). Nonstructural Protein A238L of the African Swine Fever Virus (ASFV) Enhances Antiviral Immune Responses by Activating the TBK1-IRF3 Pathway. Veterinary Sciences, 11(6), 252. https://doi.org/10.3390/vetsci11060252







