Characterization, Quantification, and Molecular Identification of Co-Infection of Canine Parvovirus (CPV-2) Variants in Dogs Affected by Gastroenteritis in Ecuador During 2022–2023
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. DNA Extraction
2.3. qPCR Assay for CPV-2 Detection
2.4. Multiplex qPCR for CPV-2 Genotyping
2.5. Sequencing of VP1 Gene
2.6. Phylogenetic Analyses
2.7. GenBank Accession Numbers
2.8. VP2 Analysis
2.9. Statistical Analysis
3. Results
3.1. CPV-2 Detection and Genotyping
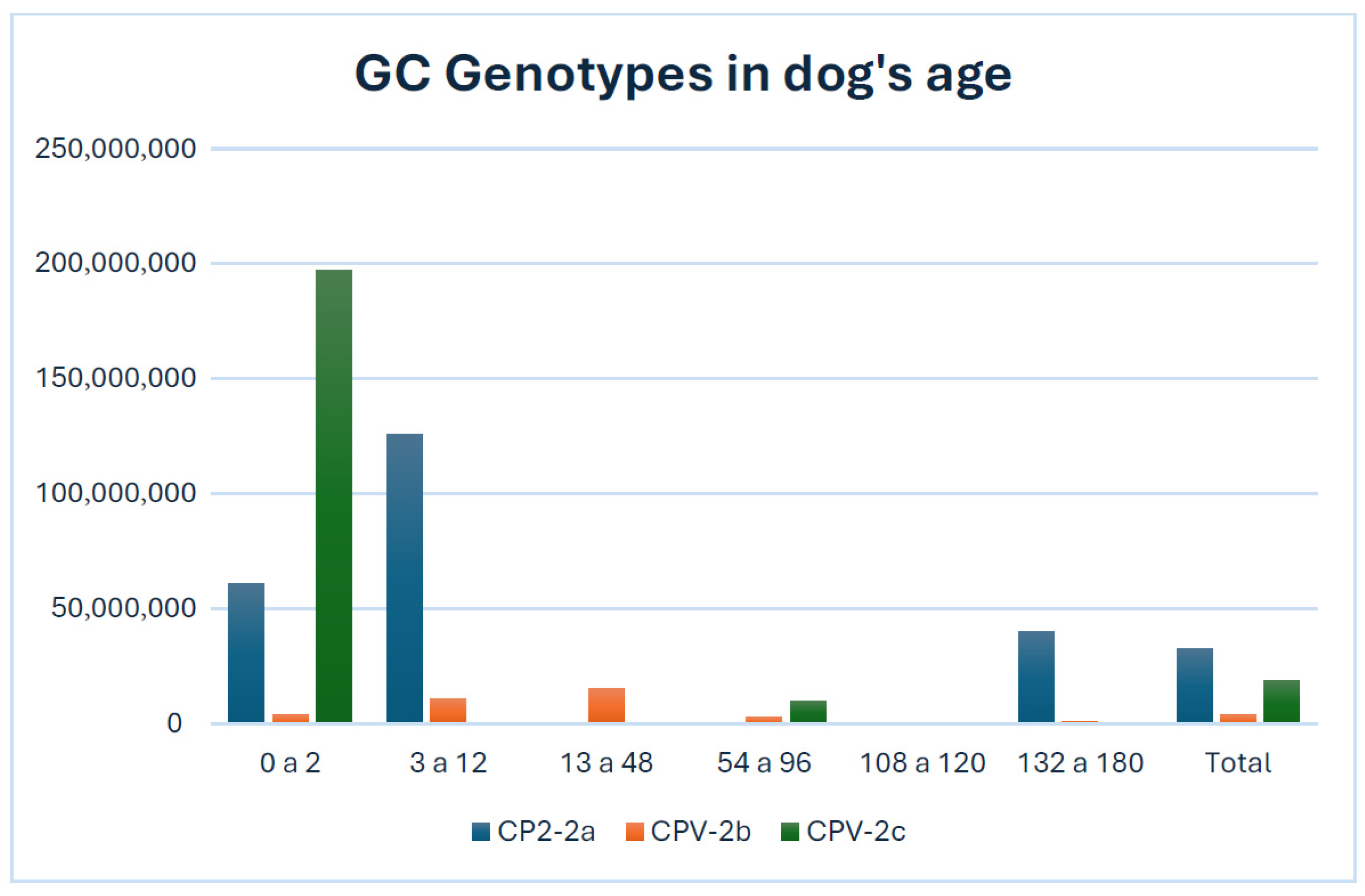
3.2. Quantification of CPV-2 Variants
3.3. Analysis by Location
3.4. Sequencing of VP1 and Phylogenetic Analysis
3.5. VP2 Protein Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goddard, A.; Leisewitz, A.L. Canine Parvovirus. Vet. Clin. N. Am. Small Anim. Pract. 2010, 40, 1041–1053. [Google Scholar] [CrossRef] [PubMed]
- Carrai, M.; Decaro, N.; Van Brusell, K.; Dall’Ara, P.; Desario, C.; Fracasso, M.; Šlapeta, J.; Colombo, E.; Bo, S.; Beatty, J.A.; et al. Canine Parvovirus Is Shed Infrequently by Cats Without Diarrhoea in Multi-Cat Environments. Vet. Microbiol. 2021, 261, 109204. [Google Scholar] [CrossRef] [PubMed]
- Williams, F.P. Astrovirus-like, Coronavirus-like, and Parvovirus-like Particles Detected in the Diarrheal Stools of Beagle Pups. Arch. Virol. 1980, 66, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Binn, L.N.; Lazar, E.C.; Eddy, G.A.; Kajima, M. Recovery and Characterization of a Minute Virus of Canines. Infect. Immun. 1970, 1, 503–508. [Google Scholar] [CrossRef]
- Rez, R.P.; Calleros, L.; Marandino, A.; Sarute, N.; Iraola, G.; Grecco, S.; Blanc, H.; Vignuzzi, M.; Isakov, O.; Shomron, N.; et al. Phylogenetic and Genome-Wide Deep-Sequencing Analyses of Canine Parvovirus Reveal Co-Infection with Field Variants and Emergence of a Recent Recombinant Strain. PLoS ONE 2014, 9, e111779. [Google Scholar] [CrossRef]
- Kelman, M.; Norris, J.M.; Barrs, V.R.; Ward, M.P. A History of Canine Parvovirus in Australia: What Can We Learn? Aust. Vet. J. 2020, 98, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, M.; Kalli, I.; Rallis, T. Canine Parvoviral Enteritis: An Update on the Clinical Diagnosis, Treatment, and Prevention. Vet. Med. Res. Rep. 2016, 7, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Miranda, C.; Thompson, G. Canine Parvovirus: The Worldwide Occurrence of Antigenic Variants. J. General. Virol. 2016, 97, 2043–2057. [Google Scholar] [CrossRef] [PubMed]
- Takano, T.; Hamaguchi, S.; Hasegawa, N.; Doki, T.; Soma, T. Predominance of Canine Parvovirus 2b in Japan: An Epidemiological Study during 2014–2019. Arch. Virol. 2021, 166, 3151–3156. [Google Scholar] [CrossRef] [PubMed]
- Giraldo-Ramirez, S.; Rendon-Marin, S.; Vargas-Bermudez, D.S.; Jaime, J.; Ruiz-Saenz, J. First Detection and Full Genomic Analysis of Canine Circovirus in CPV-2 Infected Dogs in Colombia, South America. Sci. Rep. 2020, 10, 17579. [Google Scholar] [CrossRef]
- Thomas, J.; Singh, M.; Goswami, T.K.; Glora, P.; Chakravarti, S.; Chander, V.; Upmanyu, V.; Verma, S.; Sharma, C.; Mahendran, K. Determination of Immune Status in Dogs against CPV-2 by Recombinant Protein Based Latex Agglutination Test. Biologicals 2017, 49, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Isolation of High-Molecular-Weight DNA Using Organic Solvents. Cold Spring Harb. Protoc. 2017, 2017, 356–359. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhang, W.; Ye, R.; Pan, Z.; Li, G.; Su, S. One-Step Multiplex TaqMan Probe-Based Method for Real-Time PCR Detection of Four Canine Diarrhea Viruses. Mol. Cell Probes 2020, 53, 101618. [Google Scholar] [CrossRef]
- Sun, Y.; Cheng, Y.; Lin, P.; Yi, L.; Tong, M.; Cao, Z.; Wang, G.; Li, S.; Cheng, S.; Yuan, W.; et al. A Multiplex TaqMan Real-Time PCR for Detection and Differentiation of Four Antigenic Types of Canine Parvovirus in China. Mol. Cell Probes 2018, 38, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Dotmatics Geneious. Available online: https://www.geneious.com (accessed on 9 January 2024).
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Aldaz, J.; García-Díaz, J.; Calleros, L.; Sosa, K.; Iraola, G.; Marandino, A.; Hernández, M.; Panzera, Y.; Pérez, R. High Local Genetic Diversity of Canine Parvovirus from Ecuador. Vet. Microbiol. 2013, 166, 214–219. [Google Scholar] [CrossRef]
- Carrino, M.; Tassoni, L.; Campalto, M.; Cavicchio, L.; Mion, M.; Corrò, M.; Natale, A.; Beato, M.S. Molecular Investigation of Recent Canine Parvovirus-2 (CPV-2) in Italy Revealed Distinct Clustering. Viruses 2022, 14, 917. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, R.; Liang, J.; Zhao, N.; Li, G.; Gao, Q.; Su, S. Epidemiology, Genetic Diversity and Evolution of Canine Astrovirus. Transbound. Emerg. Dis. 2020, 67, 2901–2910. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Zhao, J.; Guo, D.; Sun, D. A Mini-Review on the Epidemiology of Canine Parvovirus in China. Front. Vet. Sci. 2020, 7, 5. [Google Scholar] [CrossRef]
- Decaro, N.; Buonavoglia, C. Canine Parvovirus—A Review of Epidemiological and Diagnostic Aspects, with Emphasis on Type 2c. Vet. Microbiol. 2012, 155, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Dema, A.; Tallapally, M.R.; Ganji, V.K.; Buddala, B.; Kodi, H.; Ramidi, A.; Yella, N.R.; Putty, K. A Comprehensive Molecular Survey of Viral Pathogens Associated with Canine Gastroenteritis. Arch. Virol. 2023, 168, 36. [Google Scholar] [CrossRef] [PubMed]
- Dandrieux, J.R.S. Inflammatory Bowel Disease versus Chronic Enteropathy in Dogs: Are They One and the Same? J. Small Anim. Pract. 2016, 57, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Unterer, S.; Busch, K. Acute Hemorrhagic Diarrhea Syndrome in Dogs. Vet. Clin. North America: Small Anim. Pract. 2021, 51, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Saltık, H.S. Concomitant Virus-Induced Gastrointestinal Infection in Dogs. Pol. J. Vet. Sci. 2023, 26, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Ojeda-Chi, M.M.; Rodriguez-Vivas, R.I.; Esteve-Gasent, M.D.; Pérez de León, A.A.; Modarelli, J.J.; Villegas-Perez, S.L. Ehrlichia Canis in Dogs of Mexico: Prevalence, Incidence, Co–Infection and Factors Associated. Comp. Immunol. Microbiol. Infect. Dis. 2019, 67, 101351. [Google Scholar] [CrossRef] [PubMed]
- Cevidanes, A.; Di Cataldo, S.; Muñoz-San Martín, C.; Latrofa, M.S.; Hernández, C.; Cattan, P.E.; Otranto, D.; Millán, J. Co-Infection Patterns of Vector-Borne Zoonotic Pathogens in Owned Free-Ranging Dogs in Central Chile. Vet. Res. Commun. 2023, 47, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Gizzi, A.B.D.R.; Oliveira, S.T.; Leutenegger, C.M.; Estrada, M.; Kozemjakin, D.A.; Stedile, R.; Marcondes, M.; Biondo, A.W. Presence of Infectious Agents and Co-Infections in Diarrheic Dogs Determined with a Real-Time Polymerase Chain Reaction-Based Panel. BMC Vet. Res. 2014, 10, 23. [Google Scholar] [CrossRef]
- Elbaz, E.; El-Tholoth, M.; Elfadl, E.A.A.; Mosad, S.M. Molecular Investigation on the Presence of Canine Parvovirus in Egypt. Comp. Immunol. Microbiol. Infect. Dis. 2021, 74, 101576. [Google Scholar] [CrossRef] [PubMed]
- Giraldo-Ramirez, S.; Rendon-Marin, S.; Ruiz-Saenz, J.Z. Phylogenetic, Evolutionary and Structural Analysis of Canine Parvovirus (CPV-2) Antigenic Variants Circulating in Colombia. Viruses 2020, 12, 500. [Google Scholar] [CrossRef] [PubMed]
- Luna Espinoza, L.R.; Carhuaricra Huamán, D.; Quino Quispe, R.; Rosadio Alcántara, R.H.; Maturrano Hernández, A.L. Carnivore Protoparvovirus 1 in Peruvian Dogs: Temporal/Geographical and Evolutionary Dynamics of Virus. Infect. Genet. Evol. 2022, 99, 105255. [Google Scholar] [CrossRef]
- Hao, X.; He, Y.; Wang, C.; Xiao, W.; Liu, R.; Xiao, X.; Zhou, P.; Li, S. The Increasing Prevalence of CPV-2c in Domestic Dogs in China. PeerJ 2020, 8, e9869. [Google Scholar] [CrossRef]
- Busch, T.J. Canine Parvovirus Infection. N. Z. Vet. J. 1981, 29, 20. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, M.; Freisl, M.; Hartmann, K. Prophylaxe Der Kaninen Parvovirose. Tierärztliche Prax. Ausg. K Kleintiere/Heimtiere 2021, 49, 44–50. [Google Scholar] [CrossRef]
- Decaro, N.; Buonavoglia, C.; Barrs, V.R. Canine Parvovirus Vaccination and Immunisation Failures: Are We Far from Disease Eradication? Vet. Microbiol. 2020, 247, 108760. [Google Scholar] [CrossRef]
- Yip, H.Y.E.; Peaston, A.; Woolford, L.; Khuu, S.J.; Wallace, G.; Kumar, R.S.; Patel, K.; Ahani Azari, A.; Akbarzadeh, M.; Sharifian, M.; et al. Diagnostic Challenges in Canine Parvovirus 2c in Vaccine Failure Cases. Viruses 2020, 12, 980. [Google Scholar] [CrossRef] [PubMed]
- Abhiram, S.; Mondal, T.; Samanta, S.; Batabyal, K.; Joardar, S.N.; Samanta, I.; Isore, D.P.; Dey, S. Occurrence of Canine Parvovirus Type 2c in Diarrhoeic Faeces of Dogs in Kolkata, India. Virusdisease 2023, 34, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Alzuheir, I.M.; Fayyad, A.F.; Abu Helal, B.Y.; Atalla, H.A.; Jalboush, N.H. Detection of Canine Parvovirus Type 2c (CPV-2c) in Palestine. J. Infect. Dev. Ctries. 2024, 18, 809–816. [Google Scholar] [CrossRef] [PubMed]
- De la Torre, D.; Mafla, E.; Puga, B.; Erazo, L.; Astolfi-Ferreira, C.; Ferreira, A.P. Molecular Characterization of Canine Parvovirus Variants (CPV-2a, CPV-2b, and CPV-2c) Based on the VP2 Gene in Affected Domestic Dogs in Ecuador. Vet. World 2018, 11, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Mazzaferro, E.M. Update on Canine Parvoviral Enteritis. Vet. Clin. N. Am. Small Anim. Pract. 2020, 50, 1307–1325. [Google Scholar] [CrossRef] [PubMed]
- Stanley, K.K.; Szewczuk, E. Multiplexed Tandem PCR: Gene Profiling from Small Amounts of RNA Using SYBR Green Detection. Nucleic Acids Res. 2005, 33, e180. [Google Scholar] [CrossRef]
- Meggiolaro, M.N.; Ly, A.; Rysnik-Steck, B.; Silva, C.; Zhang, J.; Higgins, D.P.; Muscatello, G.; Norris, J.M.; Krockenberger, M.; Šlapeta, J. MT-PCR Panel Detection of Canine Parvovirus (CPV-2): Vaccine and Wild-Type CPV-2 Can Be Difficult to Differentiate in Canine Diagnostic Fecal Samples. Mol. Cell Probes 2017, 33, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Buonavoglia, C.; Martella, V.; Pratella, A.; Tempesta, M.; Cavalli, A.; Buonavoglia, D.; Bozzo, G.; Elia, G.; Decaro, N.; Carmichael, L. Evidence for Evolution of Canine Parvovirus Type 2 in Italy. J. Gen. Virol. 2001, 82, 3021–3025. [Google Scholar] [CrossRef] [PubMed]
- Chastant, S.; Mila, H. Passive Immune Transfer in Puppies. Anim. Reprod. Sci. 2019, 207, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, L.; Noll, L.; Stoy, C.; Porter, E.; Fu, J.; Feng, Y.; Peddireddi, L.; Liu, X.; Dodd, K.A.; et al. Development of a Real-Time PCR Assay for Detection of African Swine Fever Virus with an Endogenous Internal Control. Transbound. Emerg. Dis. 2020, 67, 2446–2454. [Google Scholar] [CrossRef] [PubMed]
- Aurnhammer, C.; Haase, M.; Muether, N.; Hausl, M.; Rauschhuber, C.; Huber, I.; Nitschko, H.; Busch, U.; Sing, A.; Ehrhardt, A.; et al. Universal Real-Time PCR for the Detection and Quantification of Adeno-Associated Virus Serotype 2-Derived Inverted Terminal Repeat Sequences. Hum. Gene Ther. Methods 2012, 23, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Allison, A.B.; Kohler, D.J.; Ortega, A.; Hoover, E.A.; Grove, D.M.; Holmes, E.C.; Parrish, C.R. Host-Specific Parvovirus Evolution in Nature Is Recapitulated by In Vitro Adaptation to Different Carnivore Species. PLoS Pathog. 2014, 10, e1004475. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Guo, D.; Li, C.; Wang, E.; Wei, S.; Wang, Z.; Yao, S.; Zhao, X.; Su, M.; Wang, X.; et al. Co-Circulation of the Rare CPV-2c with Unique Gln370Arg Substitution, New CPV-2b with Unique Thr440Ala Substitution, and New CPV-2a with High Prevalence and Variation in Heilongjiang Province, Northeast China. PLoS ONE 2015, 10, e0137288. [Google Scholar] [CrossRef]
- Chung, H.-C.; Kim, S.-J.; Nguyen, V.G.; Shin, S.; Kim, J.Y.; Lim, S.-K.; Park, Y.H.; Park, B. New Genotype Classification and Molecular Characterization of Canine and Feline Parvoviruses. J. Vet. Sci. 2020, 21, e43. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.; Chowdhury, Q.M.M.K.; Roy, S.; Chowdhury, M.S.R.; Hasan, M.; Al Mamun, M.; Uddin, M.B.; Hossain, M.M.; Rahman, M.M.; Rahman, M.M. Molecular Detection and Phylogenetic Analysis of Canine Parvovirus (CPV) in Diarrhoeic Pet Dogs in Bangladesh. Vet. Anim. Sci. 2021, 14, 100224. [Google Scholar] [CrossRef]
- Horiuchi, M.; Goto, H.; Ishiguro, N.; Shinagawa, M. Mapping of Determinants of the Host Range for Canine Cells in the Genome of Canine Parvovirus Using Canine Parvovirus/Mink Enteritis Virus Chimeric Viruses. J. Gen. Virol. 1994, 75, 1319–1328. [Google Scholar] [CrossRef]
- Kelman, M.; Barrs, V.R.; Norris, J.M.; Ward, M.P. Canine Parvovirus Prevention and Prevalence: Veterinarian Perceptions and Behaviors. Prev. Vet. Med. 2020, 174, 104817. [Google Scholar] [CrossRef] [PubMed]
- Packianathan, R.; Hodge, A.; Wright, J.; Lavidis, L.; Ameiss, K.; Yip, H.Y.E.; Akbarzadeh, M.; Sharifian, M.; Amanollahi, R.; Khabiri, A.; et al. Cross-Neutralization of Vanguard C4 Vaccine Against Australian Isolates of Canine Parvovirus Variants CPV-2a, CPV-2b, and CPV-2c. Viral Immunol. 2022, 35, 553–558. [Google Scholar] [CrossRef] [PubMed]

| Primer | Gene | Assay | Sequences (5′–3′) | Product | Reference |
|---|---|---|---|---|---|
| CPV-QF | NS | qPCR | TTCGGTAAACTTAACACCAAC | 84 bp | [13] |
| CPV-QR | CTGTATGTTAATATAGTCACCCA | ||||
| CPV-QP | 6-FAM-CTGCAATTTCTCTGAGCTTA-MGB | ||||
| 2161F | VP2 | PCR end point | TTGGCGTTACTCACAAAGACGTGC | 2662 bp | [5] |
| 4823R | ACCAACCACCCACACCATAACAAC | ||||
| CPV-305F | qPCR | CGTTGCCTCAATCTGAAGGAGCTA | 85 bp | [14] | |
| CPV-305R | TTGCCCATTTGAGTTACACCACGT | ||||
| CPV-2-305P | FAM-ACTCCTATATAACCAAAGTTAGTA-MGB | ||||
| CPV-426F | Multiplex qPCR | AGGAAGATATCCAGAAGGAGATTGGA | 93 bp | ||
| CPV-426R | CCAATTGGATCTGTTGGTAGCAATACA | ||||
| CPV-2a-426P | FAM-CCTGTAACAAATGATAATGTATTGC-MGB | ||||
| CPV-2b-426P | VIC-CTTCCTGTAACAGATGATAATGTATT-MGB | ||||
| CPV-2c-426P | TAMRA-CTTCCTGTAACAGAAGATAATGTATT-MGB |
| Age (Months) | CPV-2 Detection | Genotyping | |||||
|---|---|---|---|---|---|---|---|
| Negative | Positive | Total | CPV-2 | CPV-2a | CPV-2b | CPV-2c | |
| 0 to2 | 30 (27.3%) | 85 (21.2%) | 115(22.5%) | 51 * | 47 | 79 * | 49 * |
| 3 to 12 | 39 (35.5%) | 160 (39.9%) | 199 (38.9%) | 55 * | 86 | 148 * | 61 * |
| 13 to 48 | 34 (30.9%) | 104 (25.9%) | 138 (27.0%) | 19 | 66 | 72 | 36 |
| 54 to 96 | 4 (3.6%) | 37 (9.2%) | 38 (12.8%) | 13 | 25 | 31 | 15 |
| 108 to 120 | 3 (2.7%) | 8 (2.0%) | 11 (2.2%) | 2 | 7 | 6 | 3 |
| 132 to 180 | 0 (0.0%) | 7 (1.7%) | 7 (1.4%) | 4 | 7 | 7 | 3 |
| Total | 110 (100.0%) | 401 (100.0%) | 511 (100.0%) | 144 | 258 | 343 | 167 |
| Combination of Variants | |||||
|---|---|---|---|---|---|
| Combination | CPV-2 | CPV-2a | CPV-2b | CPV-2c | TOTAL |
| C.1 | + | + | + | + | 35 |
| C.2 | + | + | + | 55 | |
| C.3 | + | + | + | 1 | |
| C.4 | + | + | + | 36 | |
| C.5 | + | + | 1 | ||
| C.6 | + | + | 10 | ||
| C.7 | + | + | 4 | ||
| C.8 | + | + | + | 42 | |
| C.9 | + | + | 74 | ||
| C.10 | + | + | 11 | ||
| C.11 | + | + | 21 | ||
| C.12 | + | 2 | |||
| C.14 | + | 39 | |||
| C.15 | + | 70 | |||
| C.16 | + | 17 | |||
| CG by Genotypes of CPV-2 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Age (Months) | Average | Maximum | ||||||
| CPV-2 | CPV-2a | CPV-2b | CPV-2c | CPV-2 | CPV-2a | CPV-2b | CPV-2c | |
| 0 a 2 | 133 (SD = 1.33 × 104) | 5.17 × 107 (SD = 2.07 × 108) | 5588 (SD = 7.06 × 107) | 1.80 × 108 (SD = 9.01 × 108) | 1612 | 4.73 × 1010 | 1.29 × 107 | 1.58 × 1011 |
| 3 a 12 | 39 (SD = 10) | 3036 (SD = 2.10 × 108) | 300 (SD = 6.98 × 107) | 56 (SD = 9.30 × 108) | 39 | 2.14 × 1011 | 2.43 × 1010 | 2.15 × 106 |
| 13 a 48 | 28 (SD = 17) | 115 (SD = 2.07 × 108) | 133 (SD = 6.83 × 107) | 57 (SD = 8.88 × 104) | 52 | 6.19 × 108 | 4.80 × 107 | 8458 |
| 54 a 96 | 34 (SD = 40) | 345 (SD = 1.07 × 107) | 242 (SD = 4.07 × 108) | 50 (SD = 5.02 × 108) | 74 | 4.99 × 107 | 1.03 × 109 | 3.01 × 109 |
| 108 a 120 | 4 SD = 0) | 173 (SD = 2.70 × 108) | 42 (SD = 1165) | 48 (SD = 358) | 4 | 1.43 × 108 | 1186 | 400 |
| 132 a 180 | 18 (SD = 0) | 11203 (SD = 4.01 × 108) | 659 (SD = 1.07 × 108) | 28 (SD = 50) | 89 | 4.77 × 109 | 6.38 × 108 | 72 |
| CPV-2 | |||
|---|---|---|---|
| Province | Negative | Positive | Total |
| Carchi | 1 (0.9%) | 64 (16.0%) | 65 (12.7%) |
| Chimborazo | 1 (0.9%) | 3 (0.7%) | 4 (0.8%) |
| Guayas | 8 (7.3%) | 42 (10.5%) | 50 (9.8%) |
| Imbabura | 6 (5.5%) | 22 (5.5%) | 28 (5.5%) |
| * Pichincha | 73 (66.4%) | 225 (56.1%) | 298 (58.3%) |
| Santo Domingo de los Tsachillas | 21 (19.1%) | 45 (11.2%) | 66 (12.9%) |
| Total | 110 (100.0%) | 401 (100.0%) | 511 (100.0%) |
| Name | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino Acid Similarity | ||||||||||||||||||||||
| 1 | MT152350 | - | 100.0 | 99.8 | 98.8 | 100.0 | 99.3 | 98.1 | 99.1 | 99.6 | 99.8 | 99.6 | 99.6 | 98.9 | 100.0 | 99.1 | 99.8 | 100.0 | 98.8 | 99.1 | 99.1 | 100.0 |
| 2 | MT152373 | 99.0 | - | 99.8 | 98.8 | 100.0 | 99.3 | 98.1 | 99.1 | 99.6 | 99.8 | 99.6 | 99.6 | 98.9 | 100.0 | 99.1 | 99.8 | 100.0 | 98.8 | 99.1 | 99.1 | 100.0 |
| 3 | UDLA 425D | 99.0 | 99.0 | - | 98.8 | 99.8 | 99.3 | 98.1 | 99.1 | 99.9 | 99.8 | 99.6 | 99.6 | 98.9 | 99.8 | 99.1 | 99.8 | 99.8 | 98.8 | 99.1 | 99.1 | 99.8 |
| 4 | UDLA 197D | 99.0 | 99.0 | 99.0 | - | 98.8 | 98.2 | 96.9 | 97.9 | 98.4 | 98.6 | 98.6 | 98.4 | 97.7 | 98.8 | 97.9 | 99.8 | 98.8 | 97.9 | 97.9 | 97.9 | 98.8 |
| 5 | MG264075 | 99.0 | 99.0 | 99.0 | 99.0 | - | 99.3 | 98.1 | 99.1 | 99.6 | 99.8 | 99.6 | 99.6 | 98.9 | 100.0 | 99.1 | 99.8 | 100.0 | 98.8 | 99.1 | 99.1 | 100.0 |
| 6 | UDLA 234D | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | - | 97.4 | 98.4 | 98.2 | 99.1 | 99.3 | 98.9 | 98.2 | 99.3 | 98.4 | 99.8 | 99.3 | 99.1 | 98.4 | 98.4 | 99.3 |
| 7 | MW815499 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | - | 97.2 | 97.7 | 97.9 | 97.7 | 98.1 | 97.9 | 98.1 | 98.1 | 99.8 | 98.1 | 97.9 | 98.1 | 98.1 | 98.1 |
| 8 | UDLA 151D | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | - | 98.8 | 98.9 | 98.8 | 98.8 | 98.1 | 99.1 | 98.8 | 99.8 | 99.1 | 98.1 | 98.8 | 98.8 | 99.1 |
| 9 | UDLA 223D | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | - | 99.4 | 99.3 | 99.6 | 98.6 | 99.6 | 98.8 | 99.8 | 99.6 | 98.6 | 98.8 | 98.8 | 99.6 |
| 10 | MT585713 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | - | 98.8 | 99.4 | 99.1 | 99.8 | 99.3 | 99.8 | 99.8 | 98.8 | 99.1 | 99.1 | 99.8 |
| 11 | UDLA 429D | 98.5 | 98.5 | 98.5 | 98.5 | 98.5 | 98.5 | 98.5 | 98.5 | 98.5 | 98.5 | - | 99.3 | 98.9 | 99.6 | 99.1 | 99.8 | 99.6 | 98.6 | 98.9 | 98.9 | 99.6 |
| 12 | UDLA 195D | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | - | 98.6 | 99.6 | 98.8 | 99.8 | 99.6 | 98.6 | 98.8 | 98.8 | 99.6 |
| 13 | UDLA 15D | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | - | 98.9 | 99.8 | 99.1 | 98.9 | 98.9 | 99.3 | 99.1 | 98.9 |
| 14 | MF177269 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 99.0 | - | 99.1 | 99.8 | 100.0 | 98.8 | 99.1 | 99.1 | 100.0 |
| 15 | KF149985 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 99.0 | 99.0 | - | 99.8 | 99.1 | 99.1 | 99.4 | 99.4 | 99.1 |
| 16 | MT585711 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | - | 99.8 | 98.8 | 99.1 | 99.1 | 99.8 |
| 17 | KY921604 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 99.0 | - | 98.8 | 99.1 | 99.1 | 100.0 |
| 18 | UDLA 82D | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 99.0 | 99.0 | - | 99.6 | 99.6 | 98.8 |
| 19 | MG26077 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 99.0 | 99.0 | 99.0 | - | 99.9 | 99.1 |
| 20 | UDLA 88D | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 99.0 | 99.0 | 99.0 | 99.0 | - | 99.1 |
| 21 | MZ36281 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 98.0 | 99.0 | 99.0 | 99.0 | 99.0 | 99.0 | - |
| Nucleotide similarity | ||||||||||||||||||||||
| CPV-2a | CPV-2b | CPV-2c | ||||||||||||||||||||
| Sample | Amino Acid Residue Position | Genotype | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 20 | 28 | 139 | 144 | 159 | 166 | 174 | 184 | 191 | 199 | 201 | 297 | 311 | 313 | 322 | 324 | 426 | 440 | 514 | 571 | 572 | 577 | 581 | 582 | ||
| Consensus | M | A | G | V | E | Q | N | M | P | R | P | K | N | D | R | T | I | N | T | S | I | V | Q | R | K | |
| 82D | - | - | - | I | - | - | - | - | - | - | - | - | A | - | - | - | Y | E | - | A | - | - | - | - | - | CPV-2c |
| 88D | - | - | - | - | - | - | - | - | - | - | - | - | A | - | - | - | Y | E | - | A | - | - | - | - | - | |
| 15D | - | - | - | - | - | - | - | - | - | - | - | - | A | - | - | S | Y | D | S | A | - | - | - | - | - | CPV-2b |
| 429D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | - | - | - | - | - | - | - | |
| 151D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | CPV-2a |
| 246D | - | - | - | - | - | H | - | - | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| 245D | - | - | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | F | G | - | S | - | |
| 223D | - | - | - | - | - | - | - | - | - | - | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| 224D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S | - | - | - | - | - | - | - | - | - | |
| 90D | - | - | - | - | - | - | - | - | - | K | - | E | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| 6D | - | - | - | - | - | - | - | - | - | - | - | E | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| 282D | - | - | - | - | - | - | - | - | - | - | - | - | - | E | k | - | - | - | - | - | - | - | - | - | - | |
| 425D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | N | |
| 195D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | H | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loor-Giler, A.; Santander-Parra, S.; Castillo-Reyes, S.; Campos, M.; Mena-Pérez, R.; Prado-Chiriboga, S.; Nuñez, L. Characterization, Quantification, and Molecular Identification of Co-Infection of Canine Parvovirus (CPV-2) Variants in Dogs Affected by Gastroenteritis in Ecuador During 2022–2023. Vet. Sci. 2025, 12, 46. https://doi.org/10.3390/vetsci12010046
Loor-Giler A, Santander-Parra S, Castillo-Reyes S, Campos M, Mena-Pérez R, Prado-Chiriboga S, Nuñez L. Characterization, Quantification, and Molecular Identification of Co-Infection of Canine Parvovirus (CPV-2) Variants in Dogs Affected by Gastroenteritis in Ecuador During 2022–2023. Veterinary Sciences. 2025; 12(1):46. https://doi.org/10.3390/vetsci12010046
Chicago/Turabian StyleLoor-Giler, Anthony, Silvana Santander-Parra, Sara Castillo-Reyes, Martin Campos, Renán Mena-Pérez, Santiago Prado-Chiriboga, and Luis Nuñez. 2025. "Characterization, Quantification, and Molecular Identification of Co-Infection of Canine Parvovirus (CPV-2) Variants in Dogs Affected by Gastroenteritis in Ecuador During 2022–2023" Veterinary Sciences 12, no. 1: 46. https://doi.org/10.3390/vetsci12010046
APA StyleLoor-Giler, A., Santander-Parra, S., Castillo-Reyes, S., Campos, M., Mena-Pérez, R., Prado-Chiriboga, S., & Nuñez, L. (2025). Characterization, Quantification, and Molecular Identification of Co-Infection of Canine Parvovirus (CPV-2) Variants in Dogs Affected by Gastroenteritis in Ecuador During 2022–2023. Veterinary Sciences, 12(1), 46. https://doi.org/10.3390/vetsci12010046






