Differential Expression of Key Immune Markers in the Intestinal Tract of Developing Chick Embryos
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. RNA Extraction and Complementary DNA (cDNA) Synthesis
2.3. Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
2.4. Statistical Analysis
3. Results
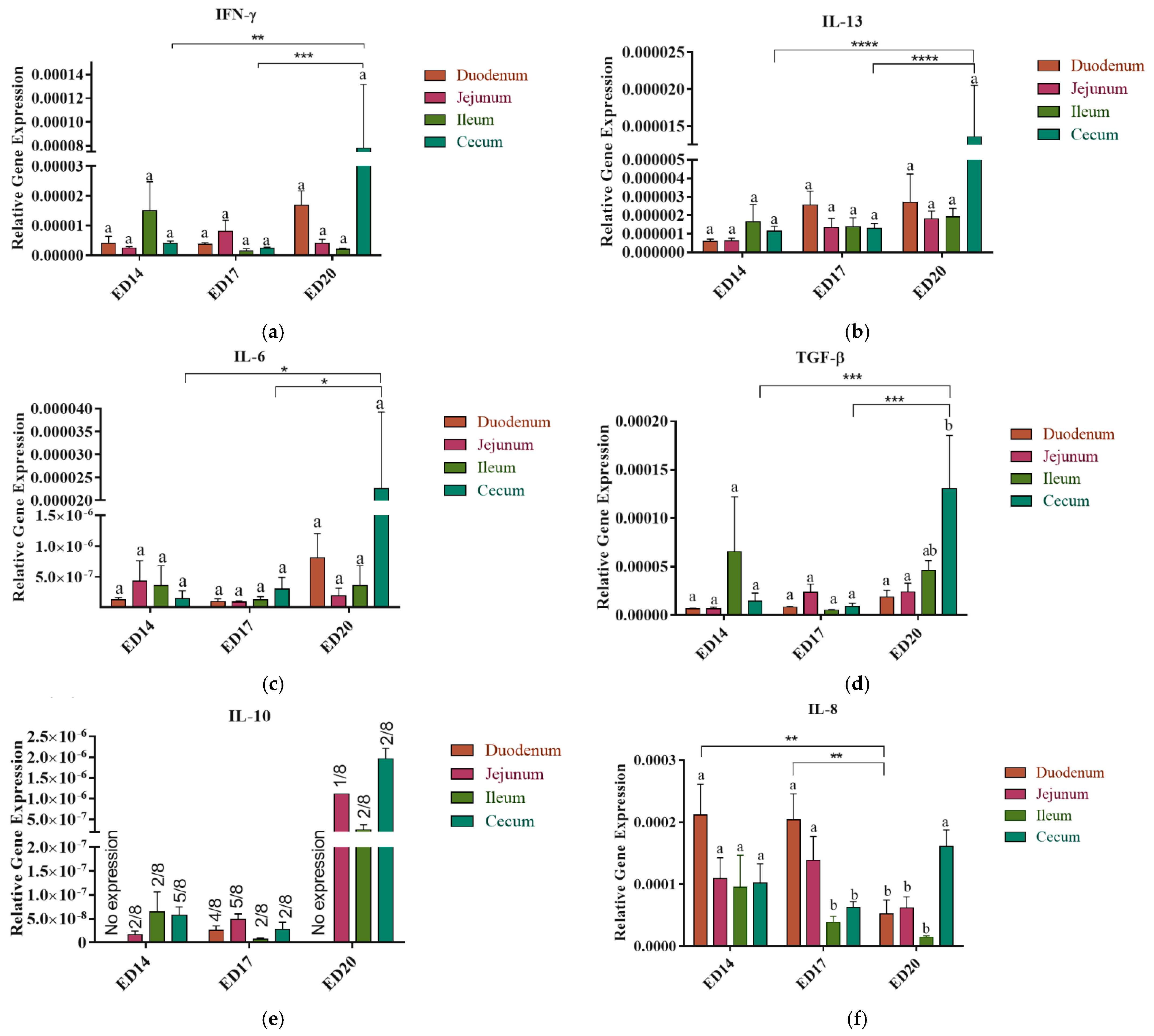
3.1. Cytokine and Chemokine Gene Expression
3.1.1. Embryonic Day 14
3.1.2. Embryonic Day 17
3.1.3. Embryonic Day 20
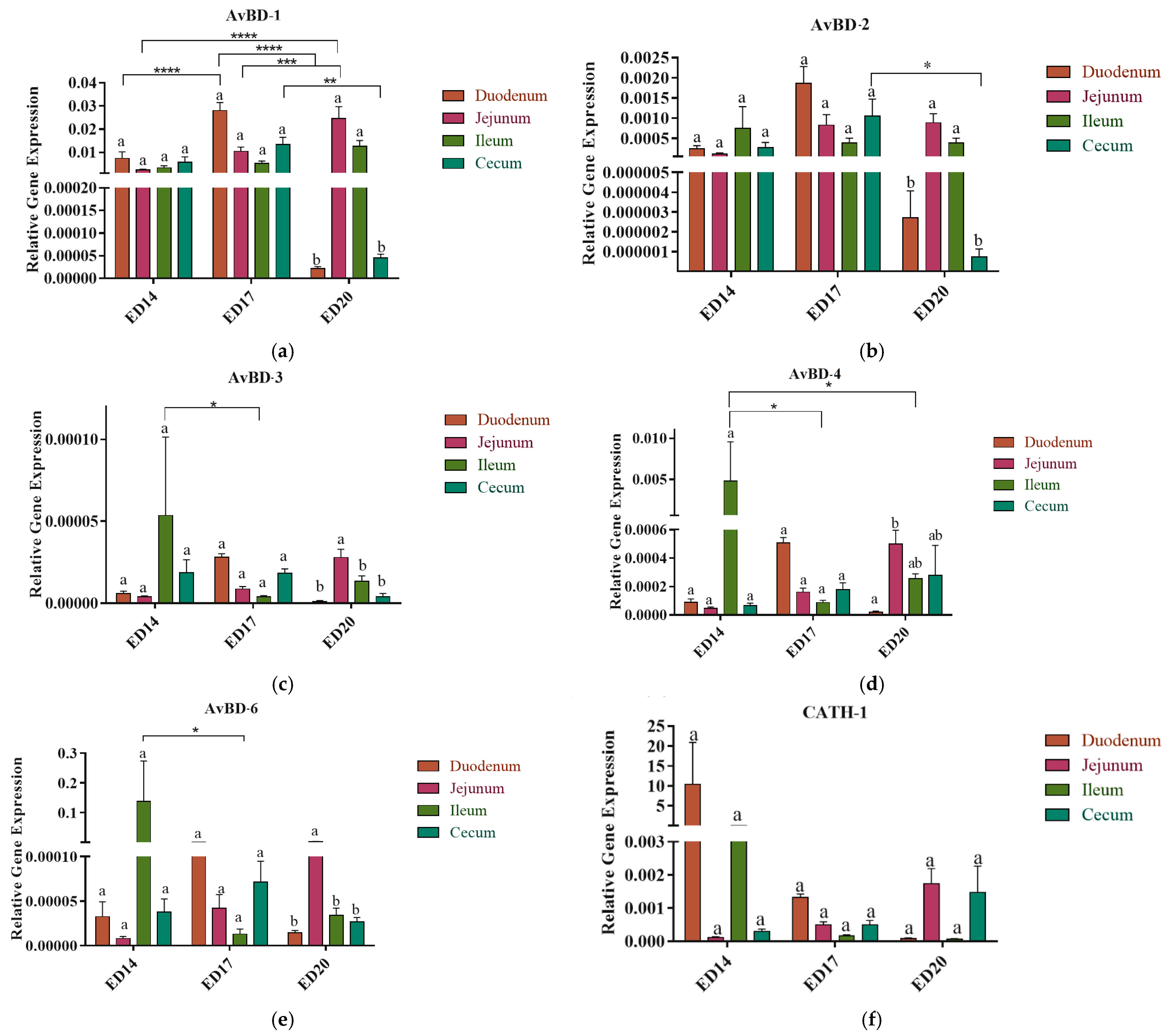
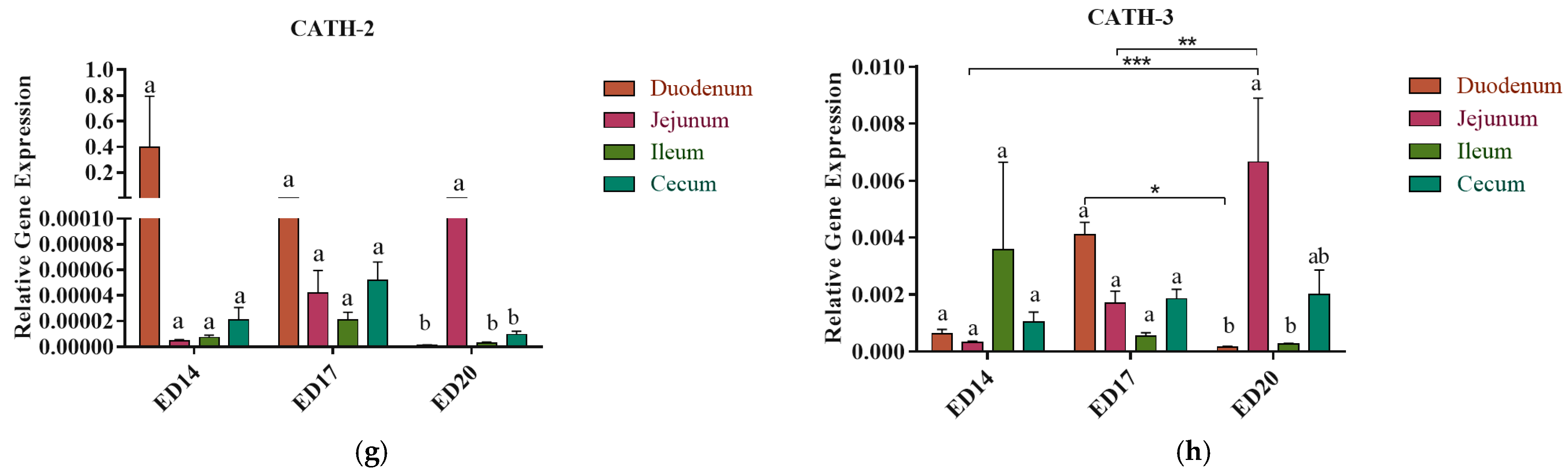
3.2. Antimicrobial Peptides (AMPs) Gene Expression
3.2.1. Embryonic Day 14
3.2.2. Embryonic Day 17
3.2.3. Embryonic Day 20
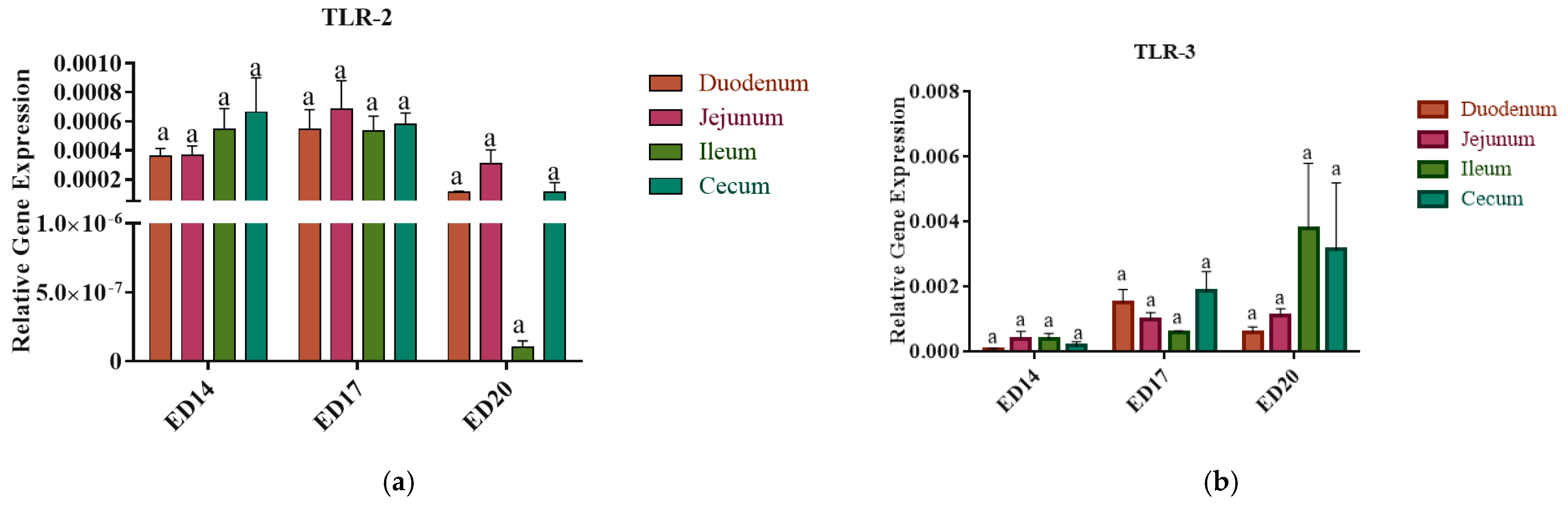
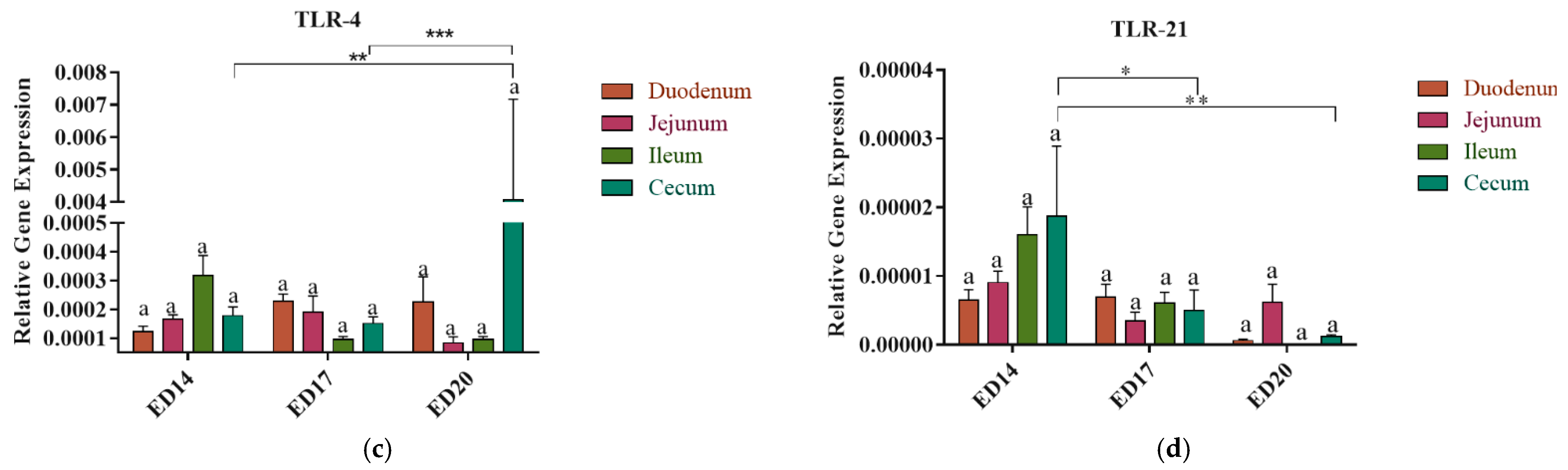
3.3. Toll-like Receptors Gene Expression
3.4. Heat Map Visualization
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lowenthal, J.; Connick, T.; McWaters, P.G.; York, J. Development of T Cell Immune Responsiveness in the Chicken. Immunol. Cell Biol. 1994, 72, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Sun, X.; Wang, T.; Chen, B.; Huang, Y.; Chen, H.; Chen, Q. The Postembryonic Development of the Immunological Barrier in the Chicken Spleens. J. Immunol. Res. 2019, 2019, 6279360. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, R.; Chan, J.; Prabakaran, M. Vaccines against Major Poultry Viral Diseases: Strategies to Improve the Breadth and Protective Efficacy. Viruses 2022, 14, 1195. [Google Scholar] [CrossRef]
- Abaidullah, M.; Peng, S.; Kamran, M.; Song, X.; Yin, Z. Current Findings on Gut Microbiota Mediated Immune Modulation against Viral Diseases in Chicken. Viruses 2019, 11, 681. [Google Scholar] [CrossRef]
- Peralta, M.F.; Magnoli, A.; Alustiza, F.; Nilson, A.; Miazzo, R.; Vivas, A. Gut-Associated Lymphoid Tissue: A Key Tissue Inside the Mucosal Immune System of Hens Immunized with Escherichia coli F4. Front. Immunol. 2017, 8, 568. [Google Scholar] [CrossRef]
- Sharma, S.; Kulkarni, R.R.; Sharif, S.; Hassan, H.; Alizadeh, M.; Pratt, S.; Abdelaziz, K. In Ovo Feeding of Probiotic Lactobacilli Differentially Alters Expression of Genes Involved in the Development and Immunological Maturation of Bursa of Fabricius in Pre-Hatched Chicks. Poult. Sci. 2024, 103, 103237. [Google Scholar] [CrossRef]
- Taha-Abdelaziz, K.; Hodgins, D.C.; Lammers, A.; Alkie, T.N.; Sharif, S. Effects of Early Feeding and Dietary Interventions on Development of Lymphoid Organs and Immune Competence in Neonatal Chickens: A Review. Vet. Immunol. Immunopathol. 2018, 201, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.W.; Artis, D. Intestinal Epithelial Cells: Regulators of Barrier Function and Immune Homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef]
- Takiishi, T.; Fenero, C.I.M.; Câmara, N.O.S. Intestinal Barrier and Gut Microbiota: Shaping Our Immune Responses throughout Life. Tissue Barriers 2017, 5, e1373208. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen Recognition and Innate Immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, T.H. Pathogen Recognition and Inflammatory Signaling in Innate Immune Defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef] [PubMed]
- Hamad, S.K.; Kim, S.; El-Kadi, S.W.; Wong, E.A.; Dalloul, R.A. Comparative Expression of Host Defense Peptides in Turkey Poults. Poult. Sci. 2017, 96, 2083–2090. [Google Scholar] [CrossRef] [PubMed]
- Taha-Abdelaziz, K.; Perez-Casal, J.; Schott, C.; Hsiao, J.; Attah-Poku, S.; Slavić, D.; Caswell, J.L. Bactericidal Activity of Tracheal Antimicrobial Peptide against Respiratory Pathogens of Cattle. Vet. Immunol. Immunopathol. 2013, 152, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Hiemstra, P.S. Epithelial Antimicrobial Peptides and Proteins: Their Role in Host Defence and Inflammation. Paediatr. Respir. Rev. 2001, 2, 306–310. [Google Scholar] [CrossRef]
- Rengaraj, D.; Truong, A.D.; Lillehoj, H.S.; Han, J.Y.; Hong, Y.H. Expression and Regulation of Avian Beta-Defensin 8 Protein in Immune Tissues and Cell Lines of Chickens. Asian-Australas. J. Anim. Sci. 2018, 31, 1516–1524. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, M.; Shan, A.; Feng, X. Avian Host Defense Cathelicidins: Structure, Expression, Biological Functions, and Potential Therapeutic Applications. Poult. Sci. 2020, 99, 6434–6445. [Google Scholar] [CrossRef] [PubMed]
- Meade, K.G.; Higgs, R.; Lloyd, A.T.; Giles, S.; O’Farrelly, C. Differential Antimicrobial Peptide Gene Expression Patterns during Early Chicken Embryological Development. Dev. Comp. Immunol. 2009, 33, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Sunkara, L. Avian Antimicrobial Host Defense Peptides: From Biology to Therapeutic Applications. Pharmaceuticals 2014, 7, 220–247. [Google Scholar] [CrossRef]
- Kany, S.; Vollrath, J.T.; Relja, B. Cytokines in Inflammatory Disease. Int. J. Mol. Sci. 2019, 20, 6008. [Google Scholar] [CrossRef] [PubMed]
- Swaggerty, C.L.; Pevzner, I.Y.; Kaiser, P.; Kogut, M.H. Profiling Pro-Inflammatory Cytokine and Chemokine MRNA Expression Levels as a Novel Method for Selection of Increased Innate Immune Responsiveness. Vet. Immunol. Immunopathol. 2008, 126, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, M.; Bavananthasivam, J.; Shojadoost, B.; Astill, J.; Taha-Abdelaziz, K.; Alqazlan, N.; Boodhoo, N.; Shoja Doost, J.; Sharif, S. In Ovo and Oral Administration of Probiotic Lactobacilli Modulate Cell- and Antibody-Mediated Immune Responses in Newly Hatched Chicks. Front. Immunol. 2021, 12, 664387. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Brisbin, J.T.; Gong, J.; Parvizi, P.; Sharif, S. Effects of Lactobacilli on Cytokine Expression by Chicken Spleen and Cecal Tonsil Cells. Clin. Vaccine Immunol. 2010, 17, 1337–1343. [Google Scholar] [CrossRef]
- Taha-Abdelaziz, K.; Alkie, T.N.; Hodgins, D.C.; Yitbarek, A.; Shojadoost, B.; Sharif, S. Gene Expression Profiling of Chicken Cecal Tonsils and Ileum Following Oral Exposure to Soluble and PLGA-Encapsulated CpG ODN, and Lysate of Campylobacter Jejuni. Vet. Microbiol. 2017, 212, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Taha-Abdelaziz, K.; Astill, J.; Shojadoost, B.; Borrelli, S.; A Monteiro, M.; Sharif, S. Campylobacter-Derived Ligands Induce Cytokine and Chemokine Expression in Chicken Macrophages and Cecal Tonsil Mononuclear Cells. Vet. Microbiol. 2020, 246, 108732. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Scheenstra, M.R.; van Harten, R.M.; Haagsman, H.P.; Veldhuizen, E.J.A. The Immunomodulatory Effect of Cathelicidin-B1 on Chicken Macrophages. Vet. Res. 2020, 51, 122. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, L.; Li, S.; Zhang, G.; Ouyang, L.; Robinson, K.; Tang, Y.; Zhu, Q.; Li, D.; Hu, Y.; et al. 1,25-Dihydroxyvitamin-D3 Induces Avian β-Defensin Gene Expression in Chickens. PLoS ONE 2016, 11, e0154546. [Google Scholar] [CrossRef]
- St Paul, M.; Barjesteh, N.; Paolucci, S.; Pei, Y.; Sharif, S. Toll-like Receptor Ligands Induce the Expression of Interferon-Gamma and Interleukin-17 in Chicken CD4+ T Cells. BMC Res. Notes 2012, 5, 616. [Google Scholar] [CrossRef] [PubMed]
- Alkie, T.N.; Yitbarek, A.; Hodgins, D.C.; Kulkarni, R.R.; Taha-Abdelaziz, K.; Sharif, S. Development of Innate Immunity in Chicken Embryos and Newly Hatched Chicks: A Disease Control Perspective. Avian Pathol. 2019, 48, 288–310. [Google Scholar] [CrossRef]
- Eren, U.; Kum, S.; Nazligul, A.; Gules, O.; Aka, E.; Zorlu, S.; Yildiz, M. The Several Elements of Intestinal Innate Immune System at the Beginning of the Life of Broiler Chicks. Microsc. Res. Tech. 2016, 79, 604–614. [Google Scholar] [CrossRef]
- Friedman, A.; Bar-Shira, E.; Sklan, D. Ontogeny of Gut Associated Immune Competence in the Chick. Worlds Poult. Sci. J. 2003, 59, 209–219. [Google Scholar] [CrossRef]
- Oláh, I.; Nagy, N.; Vervelde, L. Avian Immunology; Academic Press: Cambridge, MA, USA, 2014. [Google Scholar]
- Kajiwara, E.; Shigeta, A.; Horiuchi, H.; Matsuda, H.; Furusawa, S. Development of Peyer’s Patch and Cecal Tonsil in Gut-Associated Lymphoid Tissues in the Chicken Embryo. J. Vet. Med. Sci. 2003, 65, 607–614. [Google Scholar] [CrossRef]
- Wu, Z.; Hu, T.; Rothwell, L.; Vervelde, L.; Kaiser, P.; Boulton, K.; Nolan, M.J.; Tomley, F.M.; Blake, D.P.; Hume, D.A. Analysis of the Function of IL-10 in Chickens Using Specific Neutralising Antibodies and a Sensitive Capture ELISA. Dev. Comp. Immunol. 2016, 63, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The Antimicrobial Peptides and Their Potential Clinical Applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar] [PubMed]
- Prescott, J.F.; Parreira, V.R.; Gohari, I.M.; Lepp, D.; Gong, J. The Pathogenesis of Necrotic Enteritis in Chickens: What We Know and What We Need to Know: A Review. Avian Pathol. 2016, 45, 288–294. [Google Scholar] [CrossRef]
- Reddy, K.V.R.; Yedery, R.D.; Aranha, C. Antimicrobial Peptides: Premises and Promises. Int. J. Antimicrob. Agents 2004, 24, 536–547. [Google Scholar] [CrossRef]
- Jang, J.-H.; Shin, H.W.; Lee, J.M.; Lee, H.-W.; Kim, E.-C.; Park, S.H. An Overview of Pathogen Recognition Receptors for Innate Immunity in Dental Pulp. Mediat. Inflamm. 2015, 2015, 794143. [Google Scholar] [CrossRef]
- Rehman, M.S.; Rehman, S.U.; Yousaf, W.; Hassan, F.; Ahmad, W.; Liu, Q.; Pan, H. The Potential of Toll-Like Receptors to Modulate Avian Immune System: Exploring the Effects of Genetic Variants and Phytonutrients. Front. Genet. 2021, 12, 671235. [Google Scholar] [CrossRef]
| Gene | Primer Sequence (5′–3′) | Annealing Temperature (°C) | Reference |
|---|---|---|---|
| β-actin | F: CAACACAGTGCTGTCTGGTGGTA R: ATCGTACTCCTGCTTGCTGATCC | 60 | [23] |
| IFN-γ | F: ACACTGACAAGTCAAAGCCGCACA R: AGTCGTTCATCGGGAGCTTGGC | 60 | [23] |
| IL-6 | F: CGTGTGCGAGAACAGCATGGAGA R: TCAGGCATITCTCCTCGTCGAAGC | 60 | [24] |
| IL-8 | F: CCAAGCACACCTCTCTTCCA R: GCAAGGTAGGACGCTGGTAA | 64 | [24] |
| IL-10 | F: TTTGGCTGCCAGTCTGTGTC R: CTCATCCATCTTCTCGAACGTC | 64 | [25] |
| IL-13 | F: ACTTGTCCAAGCTGAAGCTGTC R: TCTTGCAGTCGGTCATGTTGTC | 60 | [25] |
| TGF-β | F: CGGCCGACGATGAGTGGCTC R: CGGGGCCCATCTCACAGGGA | 60 | [25] |
| CATH-1 | F: GCTGACCCTGTCCGCGTCA R: GAGGTTGTATCCTGCAATCAC | 60 | [26] |
| CATH-2 | F: CAAGGAGAATGGGGTCATCAG R: CGTGGCCCCATTTATTCATTCA | 60 | [26] |
| CATH-3 | F: CCATGGCTGACCCTGTCC R: TGATGGCTTTGTAGAGGTTGATG | 60 | [26] |
| AvBD-1 | F: GGATGCACGCTGTTCTTGGT R: TCCGCATGGTTTACGTCTGTC | 60 | [27] |
| AvBD-2 | F: CTGCTTCGGGTTCCGTTCCT R: TGCTGCTGAGGCTTTGCTGTA | 60 | [27] |
| AvBD-3 | F: AGGATTCTGTCGTGTTGGGAGC R: TTCCAGGAGCGAGAAGCCAC | 60 | [27] |
| AvBD-4 | F: GGCTATGCCGTCCCAAGTATT R: CCAAATCCAACAATGCAAGAAG | 60 | [27] |
| AvBD-6 | F: TGGCAGTGGACTAAAATCTTGC R: TTTCACAGGTGCTGATAGGGA | 60 | [27] |
| TLR-2 | F: ATCCTGCTGGAGCCCATTCAGAG R: TTGCTCTTCATCAGGAGGCCACTC | 60 | [28] |
| TLR-3 | F: TCAGTACATTTGTAACACCCCGCC R: GGCGTCATAATCAAACACTCC | 60 | [28] |
| TLR-4 | F: TGCCATCCCAACCCAACCACAG R: ACACCCACTGAGCAGCACCAA | 60 | [28] |
| TLR-21 | F: CCTGCGCAAGTGTCCGCTCA R: GCCCCAGGTCCAGGAAGCAG | 60 | [28] |
| Gene | Embryonic Day | Duodenum | Jejunum | Ileum | Cecum |
|---|---|---|---|---|---|
| IFN-γ | ED14 | 4.11 × 10−6 | 1.98 × 10−6 | 1.51 × 10−5 | 4.08 × 10−6 |
| ED17 | 3.69 × 10−6 | 8.13 × 10−6 | 1.56 × 10−6 | 2.41 × 10−6 | |
| ED20 | 1.68 × 10−5 | 1.98 × 10−6 | 2.05 × 10−6 | 7.79 × 10−5 | |
| IL-13 | ED14 | 6.03 × 10−7 | 2.83 × 10−7 | 1.66 × 10−6 | 1.15 × 10−6 |
| ED17 | 2.55 × 10−6 | 1.34 × 10−6 | 1.38 × 10−6 | 1.30 × 10−6 | |
| ED20 | 2.70 × 10−6 | 1.81 × 10−6 | 1.92 × 10−6 | 1.36 × 10−5 | |
| IL-6 | ED14 | 1.22 × 10−7 | 1.75 × 10−6 | 3.55 × 10−7 | 1.43 × 10−7 |
| ED17 | 9.15 × 10−8 | 8.52 × 10−8 | 1.24 × 10−7 | 2.96 × 10−7 | |
| ED20 | 8.10 × 10−7 | 1.19 × 10−12 | 3.55 × 10−7 | 2.25 × 10−5 | |
| TGF-β | ED14 | 6.34 × 10−6 | 9.08 × 10−6 | 6.55 × 10−5 | 1.43 × 10−5 |
| ED17 | 8.11 × 10−6 | 7.93 × 10−5 | 5.16 × 10−6 | 9.14 × 10−6 | |
| ED20 | 1.87 × 10−5 | 4.63 × 10−5 | 4.62 × 10−5 | 1.30 × 10−4 | |
| IL-10 | ED14 | 1.64 × 10−8 | 6.40 × 10−8 | 5.73 × 10−8 | |
| ED17 | 2.54 × 10−8 | 4.88 × 10−8 | 7.37 × 10−09 | 2.74 × 10−8 | |
| ED20 | 1.11 × 10−6 | 2.44 × 10−7 | 1.96 × 10−6 | ||
| IL-8 | ED14 | 8.60 × 10−5 | 1.12 × 10−4 | 4.44 × 10−5 | 3.94 × 10−5 |
| ED17 | 8.23 × 10−5 | 8.64 × 10−6 | 1.49 × 10−5 | 2.40 × 10−5 | |
| ED20 | 1.81 × 10−5 | 2.22 × 10−5 | 5.05 × 10−6 | 6.36 × 10−5 | |
| AvBD-1 | ED14 | 7.46 × 10−3 | 1.78 × 10−3 | 3.32 × 10−3 | 5.78 × 10−3 |
| ED17 | 2.80 × 10−2 | 9.18 × 10−3 | 5.25 × 10−3 | 1.34 × 10−2 | |
| ED20 | 2.17 × 10−5 | 3.00 × 10−2 | 1.26 × 10−2 | 4.53 × 10−5 | |
| AvBD-2 | ED14 | 2.42 × 10−4 | 6.06 × 10−5 | 7.40 × 10−4 | 2.72 × 10−4 |
| ED17 | 1.87 × 10−3 | 1.87 × 10−4 | 3.89 × 10−4 | 1.04 × 10−3 | |
| ED20 | 2.17 × 10−6 | 2.00 × 10−6 | 1.20 × 10−7 | 5.00 × 10−8 | |
| AvBD-3 | ED14 | 5.87 × 10−6 | 3.70 × 10−6 | 5.35 × 10−5 | 1.87 × 10−5 |
| ED17 | 2.82 × 10−5 | 4.86 × 10−6 | 3.95 × 10−6 | 1.83 × 10−5 | |
| ED20 | 1.10 × 10−6 | 3.63 × 10−5 | 1.35 × 10−5 | 3.91 × 10−6 | |
| AvBD-4 | ED14 | 8.96 × 10−5 | 2.60 × 10−5 | 4.81 × 10−3 | 6.54 × 10−5 |
| ED17 | 5.05 × 10−4 | 1.05 × 10−4 | 8.74 × 10−5 | 1.78 × 10−4 | |
| ED20 | 1.87 × 10−5 | 6.82 × 10−4 | 2.56 × 10−4 | 2.77 × 10−4 | |
| AvBD-6 | ED14 | 3.26 × 10−5 | 8.49 × 10−6 | 1.37 × 10−1 | 3.77 × 10−5 |
| ED17 | 2.73 × 10−4 | 1.51 × 10−5 | 1.29 × 10−5 | 7.16 × 10−5 | |
| ED20 | 1.44 × 10−5 | 2.38 × 10−3 | 3.42 × 10−5 | 2.66 × 10−5 | |
| CATH-1 | ED14 | 1.04 × 101 | 7.52 × 10−5 | 2.23 × 10−2 | 2.96 × 10−4 |
| ED17 | 1.32 × 10−3 | 3.67 × 10−4 | 1.67 × 10−4 | 4.99 × 10−4 | |
| ED20 | 8.56 × 10−5 | 2.95 × 10−3 | 6.58 × 10−5 | 1.47 × 10−3 | |
| CATH-2 | ED14 | 3.97 × 10−1 | 5.92 × 10−6 | 7.37 × 10−6 | 2.10 × 10−5 |
| ED17 | 1.68 × 10−4 | 5.29 × 10−7 | 2.10 × 10−5 | 5.20 × 10−5 | |
| ED20 | 1.22 × 10−6 | 4.39 × 10−4 | 2.96 × 10−6 | 9.71 × 10−6 | |
| CATH-3 | ED14 | 6.24 × 10−4 | 1.89 × 103 | 4.42 × 10−3 | 1.04 × 10−3 |
| ED17 | 4.11 × 10−3 | 1.70 × 10−3 | 5.47 × 10−4 | 1.85 × 10−3 | |
| ED20 | 1.56 × 10−4 | 6.65 × 10−3 | 4.33 × 10−4 | 2.00 × 10−3 | |
| TLR-2 | ED14 | 3.59 × 10−4 | 4.63 × 10−4 | 5.45 × 10−4 | 6.64 × 10−4 |
| ED17 | 6.12 × 10−4 | 3.21 × 10−4 | 5.24 × 10−4 | 6.30 × 10−4 | |
| ED20 | 1.12 × 10−4 | 3.08 × 10−4 | 9.91 × 10−8 | 1.11 × 10−4 | |
| TLR-3 | ED14 | 7.43 × 10−5 | 4.18 × 10−4 | 4.01 × 10−4 | 1.86 × 10−3 |
| ED17 | 1.52 × 10−3 | 9.95 × 10−4 | 5.91 × 10−4 | 1.86 × 10−3 | |
| ED20 | 6.10 × 10−4 | 1.11 × 10−3 | 3.80 × 10−3 | 3.14 × 10−3 | |
| TLR-4 | ED14 | 1.23 × 10−4 | 2.47 × 10−4 | 3.17 × 10−4 | 1.77 × 10−4 |
| ED17 | 2.27 × 10−4 | 5.52 × 10−5 | 9.52 × 10−5 | 1.51 × 10−4 | |
| ED20 | 2.27 × 10−4 | 3.66 × 10−5 | 4.56 × 10−5 | 4.07 × 10−3 | |
| TLR-21 | ED14 | 6.49 × 10−6 | 1.50 × 10−5 | 1.60 × 10−5 | 1.87 × 10−5 |
| ED17 | 6.99 × 10−6 | 5.29 × 10−7 | 6.07 × 10−6 | 5.03 × 10−6 | |
| ED20 | 6.17 × 10−7 | 2.96 × 10−7 | 7.65 × 10−10 | 1.18 × 10−6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, S.; Alizadeh, M.; Pratt, S.; Stamatikos, A.; Abdelaziz, K. Differential Expression of Key Immune Markers in the Intestinal Tract of Developing Chick Embryos. Vet. Sci. 2025, 12, 186. https://doi.org/10.3390/vetsci12020186
Sharma S, Alizadeh M, Pratt S, Stamatikos A, Abdelaziz K. Differential Expression of Key Immune Markers in the Intestinal Tract of Developing Chick Embryos. Veterinary Sciences. 2025; 12(2):186. https://doi.org/10.3390/vetsci12020186
Chicago/Turabian StyleSharma, Shreeya, Mohammadali Alizadeh, Scott Pratt, Alexis Stamatikos, and Khaled Abdelaziz. 2025. "Differential Expression of Key Immune Markers in the Intestinal Tract of Developing Chick Embryos" Veterinary Sciences 12, no. 2: 186. https://doi.org/10.3390/vetsci12020186
APA StyleSharma, S., Alizadeh, M., Pratt, S., Stamatikos, A., & Abdelaziz, K. (2025). Differential Expression of Key Immune Markers in the Intestinal Tract of Developing Chick Embryos. Veterinary Sciences, 12(2), 186. https://doi.org/10.3390/vetsci12020186







