Reference Whole Genome Sequence Analyses and Characterization of a Novel Carnobacterium maltaromaticum Distinct Sequence Type Isolated from a North American Gray Wolf (Canis lupus) Gastrointestinal Tract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Bacteria and Phenotypic Characterization
2.2. Bacterial Genomic DNA Isolation and Whole Genome Sequencing
2.3. Phylogenetic Analyses of the Wolf Bacterial Isolate
3. Results
3.1. Bacterial Isolation and Phenotypic Characterization
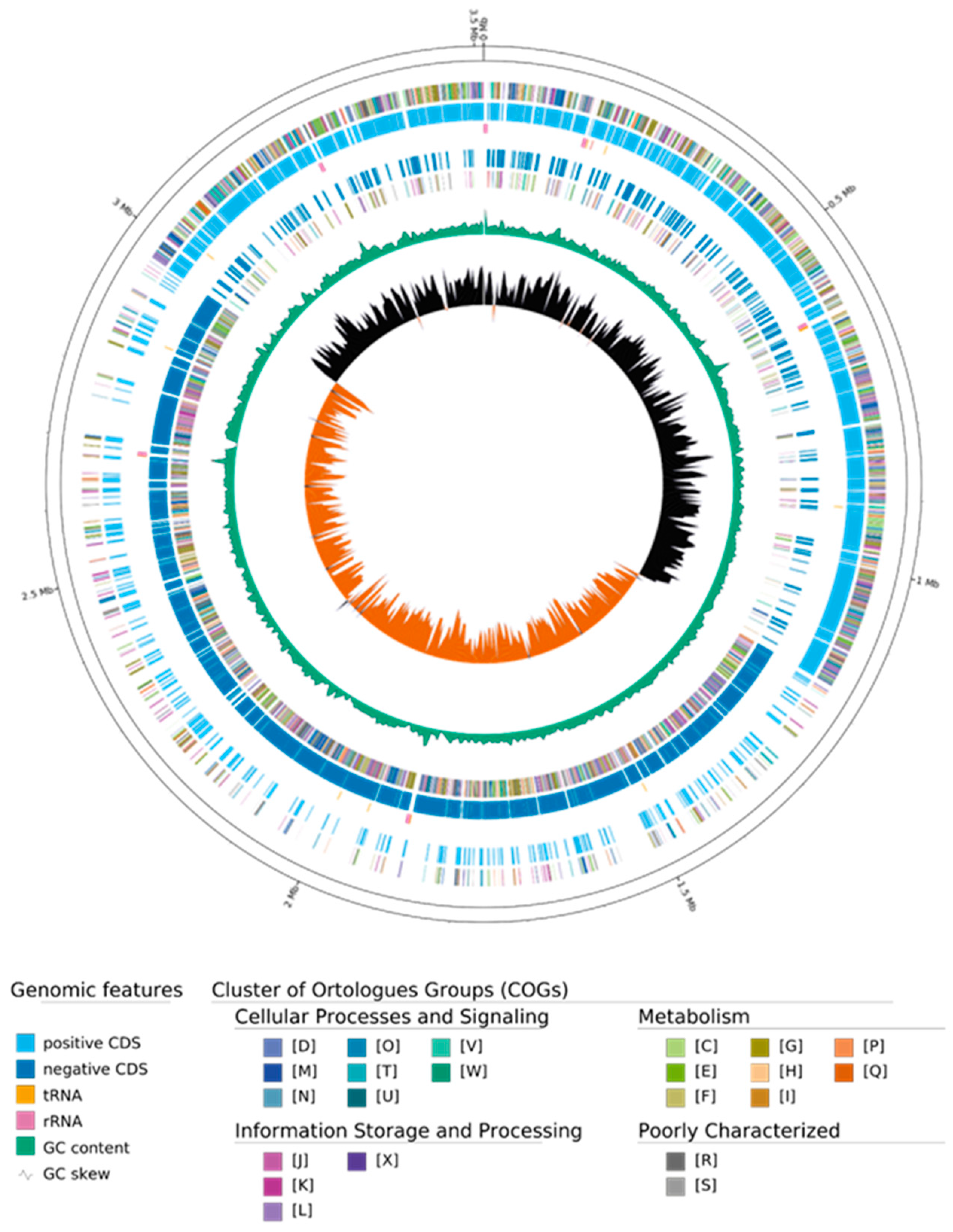
3.2. Whole Genome Sequence Characteristics of Bacterium ClWan1
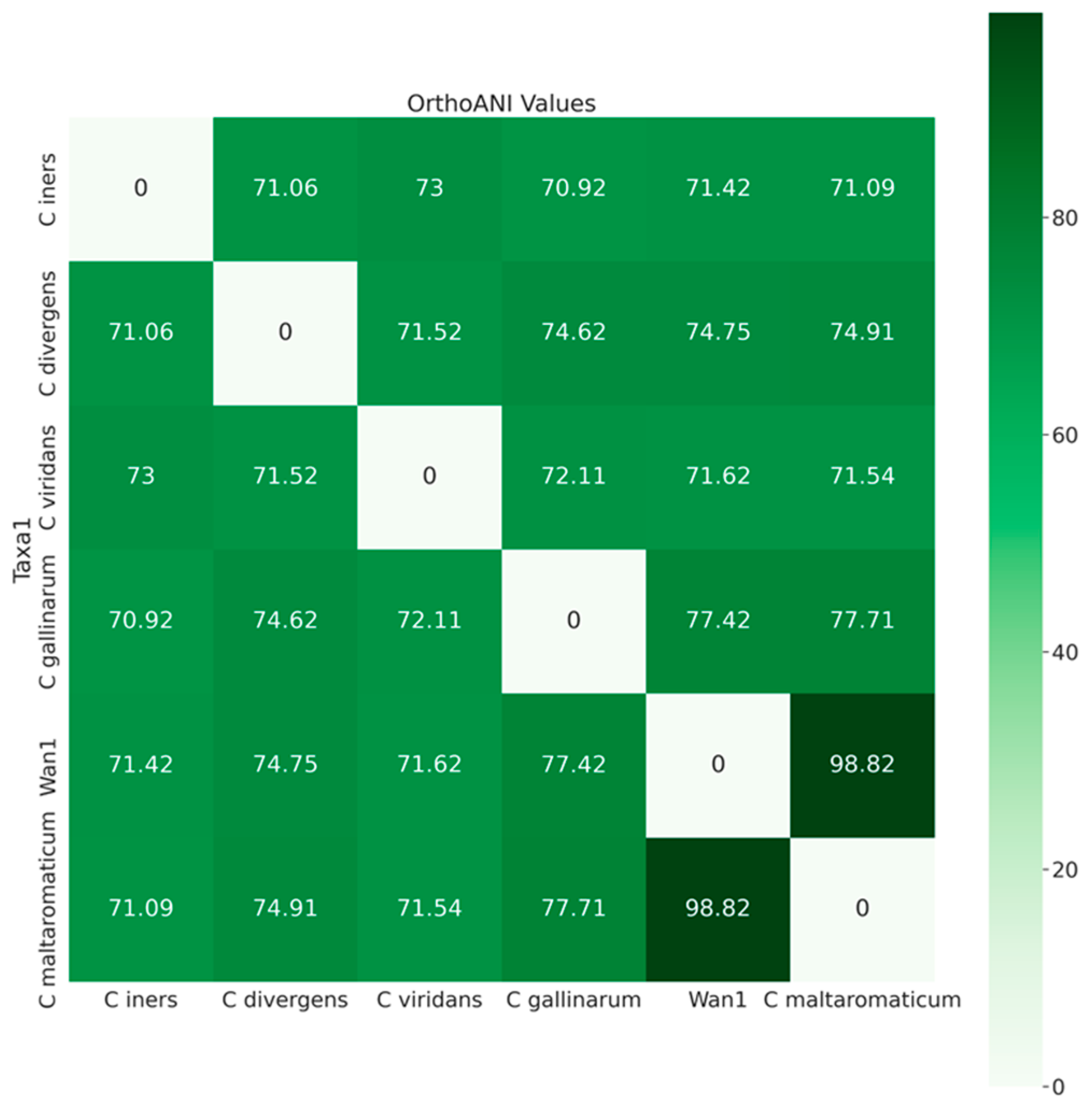
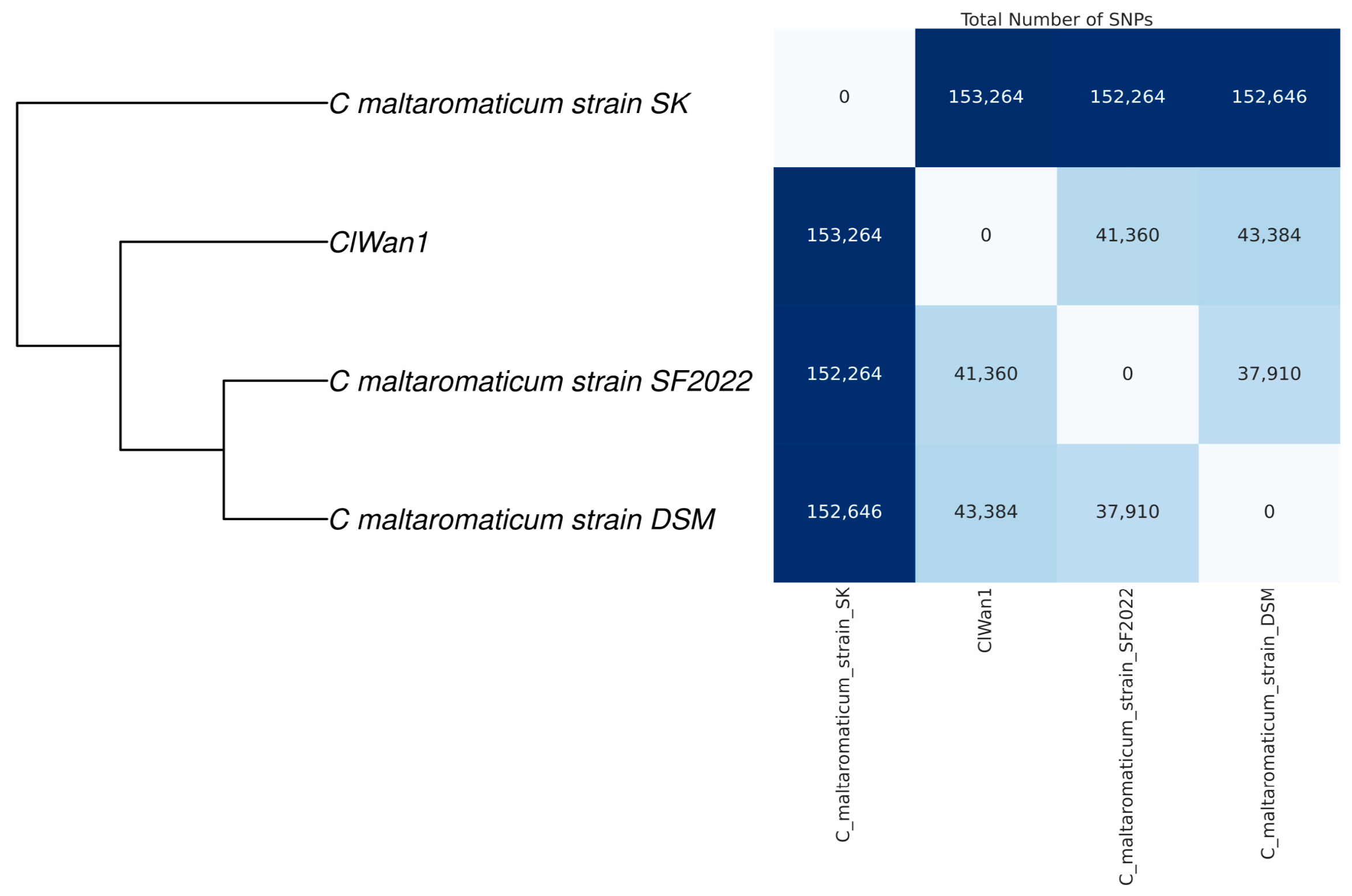
3.3. Phylogenetic Analyses of Gray Wolf Isolate ClWan1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A Taxonomic Note on the Genus Lactobacillus: Description of 23 Novel Genera, Emended Description of the Genus Lactobacillus Beijerinck 1901, and Union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Ayed, L.; M’hir, S.; Nuzzolese, D.; Di Cagno, R.; Filannino, P. Harnessing the Health and Techno-Functional Potential of Lactic Acid Bacteria: A Comprehensive Review. Foods 2024, 13, 1538. [Google Scholar] [CrossRef]
- Abdul Hakim, B.N.; Xuan, N.J.; Oslan, S.N.H. A Comprehensive Review of Bioactive Compounds from Lactic Acid Bacteria: Potential Functions as Functional Food in Dietetics and the Food Industry. Foods 2023, 12, 2850. [Google Scholar] [CrossRef] [PubMed]
- Leisner, J.J.; Laursen, B.G.; Prévost, H.; Drider, D.; Dalgaard, P. Carnobacterium: Positive and Negative Effects in the Environment and in Foods. FEMS Microbiol. Rev. 2007, 31, 592–613. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, A.G.; Danielski, G.M.; Corrêa, J.A.F.; Cavalari, C.M.A.; Souza, I.R.; Luciano, F.B.; Macedo, R.E.F. Carnobacterium as a bioprotective and potential probiotic culture to improve food quality, food safety, and human health—A scoping review. Crit. Rev. Food Sci. Nutr. 2023, 63, 6946–6959. [Google Scholar] [CrossRef]
- Begrem, S.; Ivaniuk, F.; Gigout-Chevalier, F.; Kolypczuk, L.; Bonnetot, S.; Leroi, F.; Grovel, O.; Delbarre-Ladrat, C.; Passerini, D. New Insight into Antimicrobial Compounds from Food and Marine-Sourced Carnobacterium Species through Phenotype and Genome Analyses. Microorganisms 2020, 8, 1093. [Google Scholar] [CrossRef]
- Gontijo, M.T.P.; Ramia, N.E.; Dijamentiuk, A.; Elfassy, A.; Taha, S.; Mangavel, C.; Revol-Junelles, A.M.; Borges, F. Mining Biosynthetic Gene Clusters in Carnobacterium maltaromaticum by Interference Competition Network and Genome Analysis. Microorganisms 2022, 10, 1794. [Google Scholar] [CrossRef]
- Cunningham, M.; Azcarate-Peril, M.A.; Barnard, A.; Benoit, V.; Grimaldi, R.; Guyonnet, D.; Holscher, H.D.; Hunter, K.; Manurung, S.; Obis, D.; et al. Shaping the Future of Probiotics and Prebiotics. Trends Microbiol. 2021, 29, 667–685. [Google Scholar] [CrossRef]
- Latif, A.; Shehzad, A.; Niazi, S.; Zahid, A.; Ashraf, W.; Iqbal, M.W.; Rehman, A.; Riaz, T.; Aadil, R.M.; Khan, I.M.; et al. Probiotics: Mechanism of Action, Health Benefits and their Application in Food Industries. Front. Microbiol. 2023, 1, 1216674. [Google Scholar] [CrossRef]
- Gul, S.; Durante-Mangoni, E. Unraveling the Puzzle: Health Benefits of Probiotics-A Comprehensive Review. J. Clin. Med. 2024, 13, 1436. [Google Scholar] [CrossRef]
- Markowiak, P.; Śliżewska, K. The Role of Probiotics, Prebiotics, and Synbiotics in Animal Nutrition. Gut Pathog. 2018, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Pilla, R.; Suchodolski, J.S. The Gut Microbiome of Dogs and Cats, and the Influence of Diet. Vet. Clin. N. Am. Small Anim. Pract. 2021, 51, 605–621. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, S.S. Evidence-based Use of Biotics in the Management of Gastrointestinal Disorders in Dogs and Cats. Vet. Rec. 2024, 195, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.M.; Swanson, K.S. The Influence of ‘Biotics’ on the Gut Microbiome of Dogs and Cats. Vet. Rec. 2024, 195, 2–12. [Google Scholar] [CrossRef]
- Garcias-Bonet, N.; Roik, A.; Tierney, B.; García, F.C.; Villela, H.D.M.; Dungan, A.M.; Quigley, K.M.; Sweet, M.; Berg, G.; Gram, L.; et al. Horizon Scanning the Application of Probiotics for Wildlife. Trends Microbiol. 2024, 32, 252–269. [Google Scholar] [CrossRef]
- Merenstein, D.; Pot, B.; Leyer, G.; Ouwehand, A.C.; Preidis, G.A.; Elkins, C.A.; Hill, C.; Lewis, Z.T.; Shane, A.L.; Zmora, N.; et al. Emerging Issues in Probiotic Safety: 2023 Perspectives. Gut Microbes 2023, 15, 2185034. [Google Scholar] [CrossRef]
- Spanggaard, B.; Huber, I.; Nielsen, J.; Sick, E.B.; Pipper, C.B.; Martinussen, T.; Slierendrecht, W.J.; Gram, L. The Probiotic Potential against Vibriosis of the Indigenous Microflora of Rainbow Trout. Environ. Microbiol. 2001, 3, 755–765. [Google Scholar] [CrossRef]
- Kim, D.H.; Austin, B. Innate Immune Responses in Rainbow Trout (Oncorhynchus mykiss, Walbaum) Induced by Probiotics. Fish Shellfish Immunol. 2006, 21, 513–524. [Google Scholar] [CrossRef]
- Kim, D.H.; Austin, B. Characterization of Probiotic Carnobacteria Isolated from Rainbow Trout (Oncorhynchus mykiss) intestine. Lett. Appl. Microbiol. 2008, 47, 141–147. [Google Scholar] [CrossRef]
- Pilchová, T.; Pilet, M.F.; Cappelier, J.M.; Pazlarová, J.; Tresse, O. Protective Effect of Carnobacterium spp. against Listeria monocytogenes during Host Cell Invasion Using In vitro HT29 Model. Front. Cell Infect. Microbiol. 2016, 6, 88. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.K. Probiotics and Immunity: A Fish Perspective. Fish Shellfish Immunol. 2010, 29, 2–14. [Google Scholar] [CrossRef]
- Martínez Cruz, P.; Ibáñez, A.L.; Monroy Hermosillo, O.A.; Ramírez Saad, H.C. Use of probiotics in aquaculture. ISRN Microbiol. 2012, 2012, 916845. [Google Scholar] [CrossRef]
- Smialek, M.; Burchardt, S.; Koncicki, A. The influence of Probiotic Supplementation in Broiler Chickens on Population and Carcass Contamination with Campylobacter spp.—Field study. Res. Vet. Sci. 2018, 118, 312–316. [Google Scholar] [CrossRef]
- Bogucka, J.; Ribeiro, D.M.; Bogusławska-Tryk, M.; Dankowiakowska, A.; da Costa, R.P.R.; Bednarczyk, M. Microstructure of the Small Intestine in Broiler Chickens Fed a Diet with Probiotic or Synbiotic Supplementation. J. Anim. Physiol. Anim. Nutr. 2019, 103, 1785–1791. [Google Scholar] [CrossRef]
- McCabe, J.; Bryant, J.L.; Klews, C.C.; Johnson, M.; Atchley, A.N.; Cousins, T.W.; Dominguez, A.; Gabriel, M.; Middleton, K.; Bowles, N.A.; et al. Phenotypic and Draft Genome Sequence Analyses of a Paenibacillus sp. Isolated from the Gastrointestinal Tract of a North American Gray Wolf (Canis lupus). Appl. Microbiol. 2023, 3, 1120–1129. [Google Scholar] [CrossRef]
- Keshri, J.; Smith, K.M.; Svendsen, M.K.; Keillor, H.R.; Moss, M.L.; Jordan, H.J.; Larkin, A.M.; Garrish, J.K.; Line, J.E.; Ball, P.N.; et al. Phenotypic Characterization and Draft Genome Sequence Analyses of Two Novel Endospore-Forming Sporosarcina spp. Isolated from Canada Goose (Branta canadensis) Feces. Microorganisms 2024, 12, 70. [Google Scholar] [CrossRef]
- Bryant, J.L.; McCabe, J.; Klews, C.C.; Johnson, M.; Atchley, A.N.; Cousins, T.W.; Barnard-Davidson, M.; Smith, K.M.; Ackermann, M.R.; Netherland, M., Jr.; et al. Phenotypic and Complete Reference Whole Genome Sequence Analyses of Two Paenibacillus spp. Isolates from a Gray Wolf (Canis lupus) Gastrointestinal Tract. Vet. Sci. 2025, 12, 51. [Google Scholar] [CrossRef]
- Berry, P.L.; Taylor, E.; Phillips, I. The Use of an Anaerobic Incubator for the Isolation of Anaerobes from Clinical Samples. J. Clin. Pathol. 1982, 35, 1158–1162. [Google Scholar] [CrossRef]
- Chapin, K.; Murray, P. Manual of Clinical Microbiology, 7th ed.; Murray, P.R., Baron, E.J., Pfaller, M.A., Tenover, F.C., Yolken, R.H., Eds.; American Society for Microbiology: Washington, DC, USA, 1999. [Google Scholar]
- Mahon, C.R.; Lehman, D.C.; Manuselis, G. Textbook of Diagnostic Microbiology, 5th ed.; W. B Saunders Co.: Philadelphia, PA, USA, 2014. [Google Scholar]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of Long, Error-prone Reads Using Repeat Graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Li, H. Toward a Better Understanding of Artifacts in Variant Calling from High-coverage Samples. Bioinformatics 2014, 30, 2843–2851. [Google Scholar] [CrossRef]
- Wick, R.R.; Holt, K.E. Polypolish: Short-read Polishing of Long-read Bacterial Genome Assemblies. PLoS Comput. Biol. 2022, 18, e1009802. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Cumsille, A.; Durán, R.E.; Rodríguez-Delherbe, A.; Saona-Urmeneta, V.; Cámara, B.; Seeger, M.; Araya, M.; Jara, N.; Buil-Aranda, C. GenoVi, an Open-source Automated Circular Genome Visualizer for Bacteria and Archaea. PLoS Comput. Biol. 2023, 19, e1010998. [Google Scholar] [CrossRef] [PubMed]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A User-Friendly Web Server to Thoroughly Mine RiPPs and Bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Medema, M.H.; Weber, T. The antiSMASH Database Version 4: Additional Genomes and BGCs, New Sequence-based Searches and More. Nucleic Acids Res. 2024, 52, D586–D589. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Han, S.; Saha, S.; Oler, E.; Peters, H.; Grant, J.R.; Stothard, P.; Gautam, V. PHASTEST: Faster than PHASTER, Better than PHAST. Nucleic Acids Res. 2023, 51, W443–W450. [Google Scholar] [CrossRef]
- Chalita, M.; Kim, Y.O.; Park, S.; Oh, H.S.; Cho, J.H.; Moon, J.; Baek, N.; Moon, C.; Lee, K.; Yang, J.; et al. EzBioCloud: A Genome-driven Database and Platform for Microbiome Identification and Discovery. Int. J. Syst. Evol. Microbiol. 2024, 74, 006421. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A General Classification Scheme for Bacterial Virulence Factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access Bacterial Population Genomics: BIGSdb Software, the PubMLST.org Website and Their Applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Lukjancenko, O.; Saputra, D.; Rasmussen, S.; Hasman, H.; Sicheritz-Pontén, T.; Aarestrup, F.M.; Ussery, D.W.; Lund, O. Benchmarking of Methods for Genomic Taxonomy. J. Clin. Microbiol. 2014, 52, 1529–1539. [Google Scholar] [CrossRef]
- Kim, J.; Na, S.I.; Kim, D.; Chun, J. UBCG2: Up-to-Date Bacterial Core Genes and Pipeline for Phylogenomic Analysis. J. Microbiol. 2021, 59, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A Fast, Scalable and User-friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A Large-scale Evaluation of Algorithms to Calculate Average Nucleotide Identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. ape 5.0: An Environment for Modern Phylogenetics and Evolutionary Analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest Suite for Rapid Core-genome Alignment and Visualization of Thousands of Intraspecific Microbial Genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A Database Tandem for Fast and Reliable Genome-based Classification and Nomenclature of Prokaryotes. Nucleic Acids Res. 2022, 7, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Iskandar, C.F.; Borges, F.; Taminiau, B.; Daube, G.; Zagorec, M.; Remenant, B.; Leisner, J.J.; Hansen, M.A.; Sørensen, S.J.; Mangavel, C.; et al. Comparative Genomic Analysis Reveals Ecological Differentiation in the Genus Carnobacterium. Front Microbiol. 2017, 8, 357. [Google Scholar] [CrossRef]
- Borges, F. Carnobacterium maltaromaticum. Available online: https://pubmlst.org/organisms/carnobacterium-maltaromaticum (accessed on 19 December 2024).
- Gradisteanu Pircalabioru, G.; Popa, L.I.; Marutescu, L.; Gheorghe, I.; Popa, M.; Czobor Barbu, I.; Cristescu, R.; Chifiriuc, M.C. Bacteriocins in the Era of Antibiotic Resistance: Rising to the Challenge. Pharmaceutics 2021, 13, 196. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ramos, A.; Madi-Moussa, D.; Coucheney, F.; Drider, D. Current Knowledge of the Mode of Action and Immunity Mechanisms of LAB-Bacteriocins. Microorganisms 2021, 9, 2107. [Google Scholar] [CrossRef]
- González-Gragera, E.; García-López, J.D.; Teso-Pérez, C.; Jiménez-Hernández, I.; Peralta-Sánchez, J.M.; Valdivia, E.; Montalban-Lopez, M.; Martín-Platero, A.M.; Baños, A.; Martínez-Bueno, M. Genomic Characterization of Piscicolin CM22 Produced by Carnobacterium maltaromaticum CM22 Strain Isolated from Salmon (Salmo salar). Probiotics Antimicrob Proteins 2024. [Google Scholar] [CrossRef]
- Quadri, L.; Sailer, M.; Roy, K.L.; Vederas, J.C.; Stiles, M.E. Chemical and Genetic Characterization of Bacteriocins Produced by Carnobacterium piscicola LV17B. J. Biol. Chem. 1994, 269, 12204–12211. [Google Scholar] [CrossRef]
- Shaw, B.G.; Harding, C.D. Atypical Lactobacilli from Vacuum-packaged Meats: Comparison by DNA Hybridization, Cell Composition and Biochemical Tests with a Description of Lactobacillus carnis sp. nov. Syst. Appl. Microbiol. 1985, 6, 291297. [Google Scholar] [CrossRef]
- Mora, D.; Scarpellini, M.; Franzetti, L.; Colombo, S.; Galli, A. Reclassification of Lactobacillus maltaromicus (Miller et al. 1974) DSM 20342T and DSM 20344 and Carnobacterium piscicola (Collins et al. 1987) DSM 20730T and DSM 20722 as Carnobacterium maltaromaticum comb. nov. Int. J. Syst. Evol. Microbiol. 2003, 53, 675–678. [Google Scholar] [CrossRef]
- Miller, A., III; Morgan, M.E.; Libbey, L.M. Lactobacillus maltaromicus, a new species producing a malty aroma. Int. J. Syst. Bacteriol. 1974, 24, 346–354. [Google Scholar] [CrossRef]
- Wiedmann, M.; Bruce, J.L.; Knorr, R.; Bodis, M.; Cole, E.M.; McDowell, C.I.; McDonough, P.L.; Batt, C.A. Ribotype Diversity of Listeria monocytogenes Strains Associated with Outbreaks of Listeriosis in Ruminants. J. Clin. Microbiol. 1996, 34, 1086–1090. [Google Scholar] [CrossRef] [PubMed]
- Schlech, W.F. Epidemiology and Clinical Manifestations of Listeria monocytogenes Infection. Microbiol. Spectr. 2019, 7, 3. [Google Scholar] [CrossRef]
- Weyna, A.A.W.; Niedringhaus, K.D.; Kunkel, M.R.; Fenton, H.M.A.; Keel, M.K.; Webb, A.H.; Bahnson, C.; Radisic, R.; Munk, B.; Sánchez, S.; et al. Listeriosis with Viral Coinfections in 8 Gray Foxes, 8 Wild Turkeys, and 2 Young Cervids in the Southeastern United States. J. Vet. Diagn. Investig. 2022, 34, 654–661. [Google Scholar] [CrossRef] [PubMed]
- Stahler, D.R.; Smith, D.W.; Guernsey, D.S. Foraging and Feeding Ecology of the Gray Wolf (Canis lupus): Lessons from Yellowstone National Park, Wyoming, USA. J. Nutr. 2006, 136, 1923S–1926S. [Google Scholar] [CrossRef] [PubMed]
- Lyu, T.; Liu, G.; Zhang, H.; Wang, L.; Zhou, S.; Dou, H.; Pang, B.; Sha, W.; Zhang, H. Changes in Feeding Habits Promoted the Differentiation of the Composition and Function of Gut Microbiotas Between Domestic Dogs (Canis lupus familiaris) and Gray Wolves (Canis lupus). AMB Express. 2018, 8, 123. [Google Scholar] [CrossRef]
- Wetzels, S.U.; Strachan, C.R.; Conrady, B.; Wagner, M.; Burgener, I.A.; Virányi, Z.; Selberherr, E. Wolves, Dogs and Humans in Regular Contact Can Mutually Impact Each Other’s Skin Microbiota. Sci. Rep. 2021, 11, 17106. [Google Scholar] [CrossRef]
- Podar, N.A.; Carrell, A.A.; Cassidy, K.A.; Klingeman, D.M.; Yang, Z.; Stahler, E.A.; Smith, D.W.; Stahler, D.R.; Podar, M. From Wolves to Humans: Oral Microbiome Resistance to Transfer Across Mammalian Hosts. mBio 2024, 15, e0334223. [Google Scholar] [CrossRef]
- Li, Q.; Chan, H.; Liu, W.X.; Liu, C.A.; Zhou, Y.; Huang, D.; Wang, X.; Li, X.; Xie, C.; Liu, W.Y.; et al. Carnobacterium maltaromaticum Boosts Intestinal Vitamin D Production to Suppress Colorectal Cancer in Female Mice. Cancer Cell. 2023, 41, 1450–1465.e8. [Google Scholar] [CrossRef]
- Wang, J.; Wang, T.; Sun, Y.; Feng, Y.; Kisseberth, W.C.; Henry, C.J.; Mok, I.; Lana, S.E.; Dobbin, K.; Northrup, N.; et al. Proliferative and Invasive Colorectal Tumors in Pet Dogs Provide Unique Insights into Human Colorectal Cancer. Cancers 2018, 10, 330. [Google Scholar] [CrossRef]
- Olaimat, A.N.; Al-Holy, M.A.; Shahbaz, H.M.; Al-Nabulsi, A.A.; Abu Ghoush, M.H.; Osaili, T.M.; Ayyash, M.M.; Holley, R.A. Emergence of Antibiotic Resistance in Listeria monocytogenes Isolated from Food Products: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1277–1292. [Google Scholar] [CrossRef]
- Baquero, F.; Lanza, V.F.; Duval, M.; Coque, T.M. Ecogenetics of Antibiotic Resistance in Listeria monocytogenes. Mol. Microbiol. 2020, 113, 570–579. [Google Scholar] [CrossRef]
- Díaz-Martínez, C.; Bolívar, A.; Mercanoglu Taban, B.; Kanca, N.; Pérez-Rodríguez, F. Exploring the Antibiotic Resistance of Listeria monocytogenes in Food Environments—A Review. Crit. Rev. Microbiol. 2024, 15, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.-J.; Ciufolini, M.A. Therapies from Thiopeptides. Molecules 2023, 28, 7579. [Google Scholar] [CrossRef] [PubMed]
- Claesen, J.; Spagnolo, J.B.; Ramos, S.F.; Kurita, K.L.; Byrd, A.L.; Aksenov, A.A.; Melnik, A.V.; Wong, W.R.; Wang, S.; Hernandez, R.D.; et al. A Cutibacterium acnes Antibiotic Modulates Human Skin Microbiota Composition in Hair Follicles. Sci Transl Med. 2020, 12, eaay5445. [Google Scholar] [CrossRef] [PubMed]
- Carnio, M.C.; Stachelhaus, T.; Francis, K.P.; Scherer, S. Pyridinyl Polythiazole Class Peptide Antibiotic Micrococcin P1, Secreted by Foodborne Staphylococcus equorum WS2733, is Biosynthesized Nonribosomally. Eur J Biochem. 2001, 268, 6390–6401. [Google Scholar] [CrossRef]
- Widodo, W.S.; Billerbeck, S. Natural and Engineered Cyclodipeptides: Biosynthesis, Chemical Diversity, and Engineering Strategies for Diversification and High-yield Bioproduction. Eng. Microbiol. 2022, 3, 100067. [Google Scholar] [CrossRef]
- Wyatt, M.A.; Wang, W.; Roux, C.M.; Beasley, F.C.; Heinrichs, D.E.; Dunman, P.M.; Magarvey, N.A. Staphylococcus aureus Nonribosomal Peptide Secondary Metabolites Regulate Virulence. Science 2010, 29, 294–296. [Google Scholar] [CrossRef]
- Zimmermann, M.; Fischbach, M.A. A Family of Pyrazinone Natural Products from a Conserved Nonribosomal Peptide Synthetase in Staphylococcus aureus. Chem. Biol. 2010, 17, 925–930. [Google Scholar] [CrossRef]
- Chen, J.; Kuipers, O.P. Isolation and Analysis of the Nisin Biosynthesis Complex NisBTC: Further Insights into Their Cooperative Action. mBio 2021, 12, e0258521. [Google Scholar] [CrossRef]
- Vargas-González, A.; Barajas, M.; Pérez-Sánchez, T. Isolation of Lactic Acid Bacteria (LAB) from Salmonids for Potential Use as Probiotics: In Vitro Assays and Toxicity Assessment of Salmo trutta Embryonated Eggs. Animals 2024, 14, 200. [Google Scholar] [CrossRef]
- Elzinga, D.C.; Kulwicki, R.; Iselin, S.; Spence, L.; Capaldi, A. Rapid Evolution of Prehistoric Dogs from Wolves by Natural and Sexual Selection Emerges from an Agent-based Model. Proc. Biol. Sci. 2025, 292, 20242646. [Google Scholar] [CrossRef] [PubMed]
- Bornbusch, S.L.; Power, M.L.; Schulkin, J.; Drea, C.M.; Maslanka, M.T.; Muletz-Wolz, C.R. Integrating Microbiome Science and Evolutionary Medicine into Animal Health and Conservation. Biol. Rev. Camb. Philos. Soc. 2024, 99, 458–477. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, B.; Liu, C.; Li, X.; Li, X.; Zhang, X.; Irwin, D.M.; Wu, Z.; Chen, Z.; Jin, Q.; et al. Differences in the gut microbiomes of dogs and wolves: Roles of antibiotics and starch. BMC Vet Res. 2021, 17, 112. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Sun, M.; Xu, D.; Gao, Z.; Shi, Y.; Wang, S.; Zhou, Y. Gut microbiome of captive wolves is more similar to domestic dogs than wild wolves indicated by metagenomics study. Front Microbiol. 2022, 13, 1027188. [Google Scholar] [CrossRef]

| ID# | # of Contigs | Longest Contig | Assembly Length | GC % | N50 |
|---|---|---|---|---|---|
| Wan1 | 1 | 3,512,202 | 3,512,202 | 34.48 | 3,512,202 |
| SF2022 | 2 | 3,410,292 | 3,410,292 | 34.50 | 3,400,000 |
| Multilocus Sequence Type Genes | ||||||||
|---|---|---|---|---|---|---|---|---|
| Bacterial Species | dapE | ddlA | glpQ | ilvE | leuS | pyc | pyrE | ST |
| Carnobacterium maltaromaticum | 20 * | 2 ^ | 19 | 2 ^ | 22 | 10 * | 16 | 63 |
| COG | Category Type | Number of Genes | % of Total |
|---|---|---|---|
| D | Cell cycle control, cell division, chromosome partitioning | 89 | 2.6 |
| M | Cell wall/membrane/envelope biogenesis | 261 | 7.5 |
| N | Cell motility | 44 | 1.3 |
| O | Post-translational modification, protein turnover, chaperones | 145 | 4.2 |
| T | Signal transduction mechanisms | 192 | 5.5 |
| U | Intracellular trafficking, secretion, and vesicular transport | 49 | 1.4 |
| V | Defense mechanisms | 125 | 3.6 |
| W | Extracellular structures | 25 | 0.7 |
| Y | Nuclear structure | 0 | 0.0 |
| Z | Cytoskeleton | 3 | 0.1 |
| A | RNA processing and modification | 0 | 0.0 |
| B | Chromatin structure and dynamics | 0 | 0.0 |
| J | Translation, ribosomal structure, and biogenesis | 263 | 7.5 |
| K | Transcription | 353 | 10.1 |
| L | Replication, recombination, and repair | 146 | 4.2 |
| X | Mobilome: prophages, transposons | 64 | 1.8 |
| C | Energy production and conversion | 137 | 3.9 |
| E | Amino acid transport and metabolism | 232 | 6.6 |
| F | Nucleotide transport and metabolism | 105 | 3.0 |
| G | Carbohydrate transport and metabolism | 310 | 8.9 |
| H | Coenzyme transport and metabolism | 123 | 3.5 |
| I | Lipid transport and metabolism | 124 | 3.6 |
| P | Inorganic ion transport and metabolism | 149 | 4.3 |
| Q | Secondary metabolites biosynthesis, transport, and catabolism | 42 | 1.2 |
| R | General function prediction only | 290 | 8.3 |
| S | Function unknown | 218 | 6.2 |
| Unclassified | Unclassified | 0 | 0.0 |
| Location | Sequence Name | Class |
|---|---|---|
| 602492…604366 | ABC-F type ribosomal protection protein | MACROLIDE |
| 313798…314697 | class A beta-lactamase | BETA-LACTAM |
| 508539…508952 | NAD(+)—rifampin ADP-ribosyltransferase | RIFAMYCIN |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klews, C.C.; Bryant, J.L.; McCabe, J.; Atchley, A.N.; Cousins, T.W.; Barnard-Davidson, M.; Ackermann, M.R.; Netherland, M., Jr.; Hasan, N.A.; Jordan, P.A.; et al. Reference Whole Genome Sequence Analyses and Characterization of a Novel Carnobacterium maltaromaticum Distinct Sequence Type Isolated from a North American Gray Wolf (Canis lupus) Gastrointestinal Tract. Vet. Sci. 2025, 12, 410. https://doi.org/10.3390/vetsci12050410
Klews CC, Bryant JL, McCabe J, Atchley AN, Cousins TW, Barnard-Davidson M, Ackermann MR, Netherland M Jr., Hasan NA, Jordan PA, et al. Reference Whole Genome Sequence Analyses and Characterization of a Novel Carnobacterium maltaromaticum Distinct Sequence Type Isolated from a North American Gray Wolf (Canis lupus) Gastrointestinal Tract. Veterinary Sciences. 2025; 12(5):410. https://doi.org/10.3390/vetsci12050410
Chicago/Turabian StyleKlews, C. Cristoph, Jessika L. Bryant, Jennifer McCabe, Ariel N. Atchley, Thomas W. Cousins, Maya Barnard-Davidson, Mark R. Ackermann, Michael Netherland, Jr., Nur A. Hasan, Peter A. Jordan, and et al. 2025. "Reference Whole Genome Sequence Analyses and Characterization of a Novel Carnobacterium maltaromaticum Distinct Sequence Type Isolated from a North American Gray Wolf (Canis lupus) Gastrointestinal Tract" Veterinary Sciences 12, no. 5: 410. https://doi.org/10.3390/vetsci12050410
APA StyleKlews, C. C., Bryant, J. L., McCabe, J., Atchley, A. N., Cousins, T. W., Barnard-Davidson, M., Ackermann, M. R., Netherland, M., Jr., Hasan, N. A., Jordan, P. A., Forsythe, E. S., Ball, P. N., & Seal, B. S. (2025). Reference Whole Genome Sequence Analyses and Characterization of a Novel Carnobacterium maltaromaticum Distinct Sequence Type Isolated from a North American Gray Wolf (Canis lupus) Gastrointestinal Tract. Veterinary Sciences, 12(5), 410. https://doi.org/10.3390/vetsci12050410








