Comparison of the Fecal Microbiota of Horses with Intestinal Disease and Their Healthy Counterparts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Horse Descriptions and Fecal Sampling
2.2. Microbial Community Analysis
2.3. Statistics
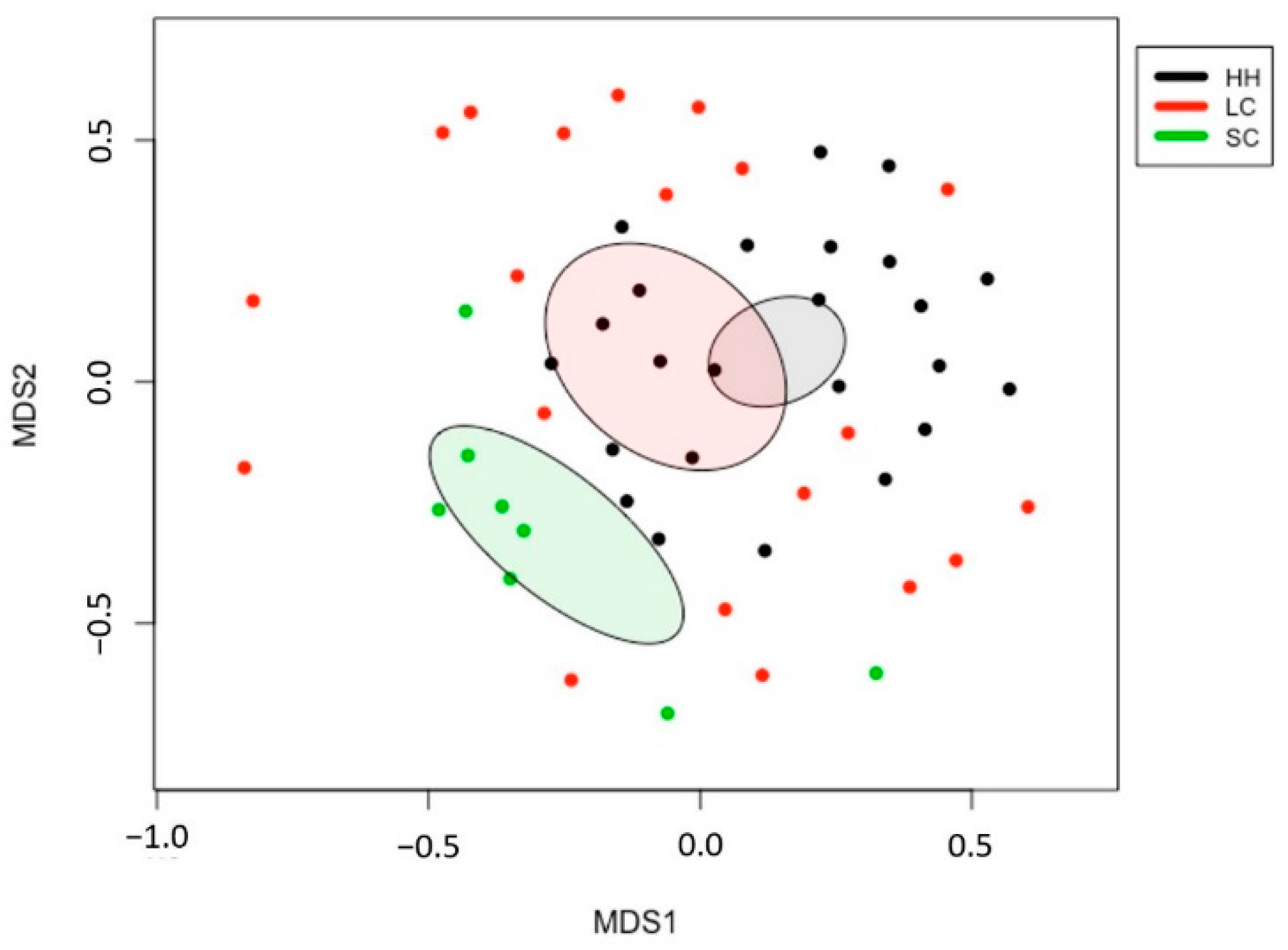
3. Results
3.1. α-Diversity Analysis
3.2. β-Diversity and Taxonomic Composition Analysis
3.3. Differentially Abundant Genera
3.4. Comparison of the Metabolic Activities between Horses with Intestinal Disease and Healthy Horses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Costa, M.C.; Weese, J.S. The equine intestinal microbiome. Anim. Health Res. Rev. 2012, 13, 121–128. [Google Scholar] [CrossRef]
- Julliand, V.; Grimm, P. Horse Species Symposium: The microbiome of the horse hindgut: History and current knowledge. J. Anim. Sci. 2016, 94, 2262–2274. [Google Scholar] [CrossRef] [PubMed]
- Julliand, V.; De Fombelle, A.; Drogoul, C.; Jacotot, E. Feeding and microbial disorders in horses: Part 3—Effects of three hay: Grain ratios on microbial profile and activities. J. Equine Vet. Sci. 2001, 21, 543–546. [Google Scholar] [CrossRef]
- Coverdale, J. Horse Species Symposium: Can the microbiome of the horse be altered to improve digestion? J. Anim. Sci. 2016, 94, 2275–2281. [Google Scholar] [CrossRef]
- Jensen, R.; Austbø, D.; Blache, D.; Knudsen, K.B.; Tauson, A.-H. The effect of feeding barley or hay alone or in combination with molassed sugar beet pulp on the metabolic responses in plasma and caecum of horses. Anim. Feed Sci. Technol. 2016, 214, 53–65. [Google Scholar] [CrossRef]
- Chapman, A.M. Acute diarrhea in hospitalized horses. Vet. Clin. N. Am. Equine Pract. 2009, 25, 363–380. [Google Scholar] [CrossRef] [PubMed]
- Vieira, S.M.; Hiltensperger, M.; Kumar, V.; Zegarra-Ruiz, D.; Dehner, C.; Khan, N.; Costa, F.R.C.; Tiniakou, E.; Greiling, T.; Ruff, W.; et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018, 359, 1156–1161. [Google Scholar] [CrossRef] [Green Version]
- Mazmanian, S.K. Capsular polysaccharides of symbiotic bacteria modulate immune responses during experimental colitis. J. Pediatric Gastroenterol. Nutr. 2008, 46, E11–E12. [Google Scholar] [CrossRef]
- Paun, A.; Danska, J.S. Immuno-ecology: How the microbiome regulates tolerance and autoimmunity. Curr. Opin. Immunol. 2015, 37, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.J.; Wu, E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 2012, 3, 4–14. [Google Scholar] [CrossRef] [Green Version]
- Milinovich, G.J.; Klieve, A.V.; Pollitt, C.C.; Trott, D.J. Microbial events in the hindgut during carbohydrate-induced equine laminitis. Vet. Clin. N. Am. Equine Pract. 2010, 26, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, T.; Imaeda, H.; Takahashi, K.; Kasumi, E.; Bamba, S.; Fujiyama, Y.; Andoh, A. Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn’s disease. J. Gastroenterol. Hepatol. 2013, 28, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Giamarellos-Bourboulis, E.; Tang, J.; Pyleris, E.; Pistiki, A.; Barbatzas, C.; Brown, J.; Lee, C.C.; Harkins, T.T.; Kim, G.; Weitsman, S. Molecular assessment of differences in the duodenal microbiome in subjects with irritable bowel syndrome. Scand. J. Gastroenterol. 2015, 50, 1076–1087. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Sinha, R.; Pei, Z.; Dominianni, C.; Wu, J.; Shi, J.; Goedert, J.J.; Hayes, R.B.; Yang, L. Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 1907–1911. [Google Scholar] [CrossRef] [Green Version]
- Zheng, P.; Li, Z.; Zhou, Z. Gut microbiome in type 1 diabetes: A comprehensive review. Diabetes Metab. Res. Rev. 2018, 34, e3043. [Google Scholar] [CrossRef]
- Aydin, Ö.; Nieuwdorp, M.; Gerdes, V. The gut microbiome as a target for the treatment of type 2 diabetes. Curr. Diabetes Rep. 2018, 18, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Asano, Y.; Hiramoto, T.; Nishino, R.; Aiba, Y.; Kimura, T.; Yoshihara, K.; Koga, Y.; Sudo, N. Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G1288–G1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudo, N.; Chida, Y.; Aiba, Y.; Sonoda, J.; Oyama, N.; Yu, X.N.; Kubo, C.; Koga, Y. Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice. J. Physiol. 2004, 558, 263–275. [Google Scholar] [CrossRef]
- Stewart, H.L.; Pitta, D.; Indugu, N.; Vecchiarelli, B.; Hennessy, M.L.; Engiles, J.B.; Southwood, L.L. Changes in the faecal bacterial microbiota during hospitalisation of horses with colic and the effect of different causes of colic. Equine Vet. J. 2020, 1–13. [Google Scholar] [CrossRef]
- Al Jassim, R.A.; Andrews, F.M. The bacterial community of the horse gastrointestinal tract and its relation to fermentative acidosis, laminitis, colic, and stomach ulcers. Vet. Clin. N. Am. Equine Pract. 2009, 25, 199–215. [Google Scholar] [CrossRef]
- Weese, J.S.; Holcombe, S.J.; Embertson, R.M.; Kurtz, K.A.; Roessner, H.A.; Jalali, M.; Wismer, S.E. Changes in the faecal microbiota of mares precede the development of post partum colic. Equine Vet. J. 2015, 47, 641–649. [Google Scholar] [CrossRef]
- Costa, M.C.; Arroyo, L.G.; Allen-Vercoe, E.; Stampfli, H.R.; Kim, P.T.; Sturgeon, A.; Weese, J.S. Comparison of the fecal microbiota of healthy horses and horses with colitis by high throughput sequencing of the V3-V5 region of the 16S rRNA gene. PLoS ONE 2012, 7, e41484. [Google Scholar] [CrossRef] [Green Version]
- Salem, S.E.; Maddox, T.W.; Antczak, P.; Ketley, J.M.; Williams, N.J.; Archer, D.C. Acute changes in the colonic microbiota are associated with large intestinal forms of surgical colic. BMC Vet. Res. 2019, 15, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Dougal, K.; de la Fuente, G.; Harris, P.A.; Girdwood, S.E.; Pinloche, E.; Newbold, C.J. Identification of a core bacterial community within the large intestine of the horse. PLoS ONE 2013, 8, e77660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biddle, A.S.; Black, S.J.; Blanchard, J.L. An in vitro model of the horse gut microbiome enables identification of lactate-utilizing bacteria that differentially respond to starch induction. PLoS ONE 2013, 8, e77599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, C.; Taminiau, B.; Brévers, B.; Avesani, V.; Van Broeck, J.; Leroux, A.; Gallot, M.; Bruwier, A.; Amory, H.; Delmée, M. Faecal microbiota characterisation of horses using 16 rdna barcoded pyrosequencing, and carriage rate of clostridium difficile at hospital admission. BMC Microbiol. 2015, 15, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Milinovich, G.J.; Trott, D.J.; Burrell, P.C.; Croser, E.L.; Al Jassim, R.A.; Morton, J.M.; Van Eps, A.W.; Pollitt, C.C. Fluorescence in situ hybridization analysis of hindgut bacteria associated with the development of equine laminitis. Environ. Microbiol. 2007, 9, 2090–2100. [Google Scholar] [CrossRef] [PubMed]
- Steelman, S.M.; Chowdhary, B.P.; Dowd, S.; Suchodolski, J.; Janečka, J.E. Pyrosequencing of 16S rRNA genes in fecal samples reveals high diversity of hindgut microflora in horses and potential links to chronic laminitis. BMC Vet. Res. 2012, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Daly, K.; Proudman, C.J.; Duncan, S.H.; Flint, H.J.; Dyer, J.; Shirazi-Beechey, S.P. Alterations in microbiota and fermentation products in equine large intestine in response to dietary variation and intestinal disease. Br. J. Nutr. 2012, 107, 989–995. [Google Scholar] [CrossRef] [Green Version]
- Uzal, F.A.; Diab, S.S. Gastritis, enteritis, and colitis in horses. Vet. Clin. N. Am. Equine Pract. 2015, 31, 337–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, H.L.; Pitta, D.; Indugu, N.; Vecchiarelli, B.; Engiles, J.B.; Southwood, L.L. Characterization of the fecal microbiota of healthy horses. Am. J. Vet. Res. 2018, 79, 811–819. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Midford, P.E.; Ong, W.K.; Paley, S.; Subhraveti, P.; Karp, P.D. The MetaCyc database of metabolic pathways and enzymes—A 2019 update. Nucleic Acids Res. 2020, 48, D445–D453. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, M.M.; Harris, H.M.; Ross, R.P.; O’Toole, P.W. Core fecal microbiota of domesticated herbivorous ruminant, hindgut fermenters, and monogastric animals. Microbiologyopen 2017, 6, e00509. [Google Scholar] [CrossRef]
- Salem, S.E.; Hough, R.; Probert, C.; Maddox, T.W.; Antczak, P.; Ketley, J.M.; Williams, N.J.; Stoneham, S.J.; Archer, D.C. A longitudinal study of the faecal microbiome and metabolome of periparturient mares. PeerJ 2019, 7, e6687. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, B.; Bai, D.; Huang, J.; Shiraigo, W.; Yang, L.; Zhao, Q.; Ren, X.; Wu, J.; Bao, W. Comparison of fecal microbiota of Mongolian and Thoroughbred Horses by high-throughput sequencing of the V4 Region of the 16S rRNA gene. Asian Aust. J. Anim. Sci. 2016, 29, 1345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, M.; Silva, G.; Ramos, R.; Staempfli, H.; Arroyo, L.; Kim, P.; Weese, J.S. Characterization and comparison of the bacterial microbiota in different gastrointestinal tract compartments in horses. Vet. J. 2015, 205, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Mach, N.; Foury, A.; Kittelmann, S.; Reigner, F.; Moroldo, M.; Ballester, M.; Esquerré, D.; Rivière, J.; Sallé, G.; Gérard, P. The effects of weaning methods on gut microbiota composition and horse physiology. Front. Physiol. 2017, 8, 535. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Mao, B.; Gu, J.; Wu, J.; Cui, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Blautia—A new functional genus with potential probiotic properties? Gut Microbes 2021, 13, 1–21. [Google Scholar]
- Nueno-Palop, C.; Narbad, A. Probiotic assessment of Enterococcus faecalis CP58 isolated from human gut. Int. J. Food Microbiol. 2011, 145, 390–394. [Google Scholar] [CrossRef]
- Zupancic, K.; Kriksic, V.; Kovacevic, I.; Kovacevic, D. Influence of oral probiotic Streptococcus salivarius K12 on ear and oral cavity health in humans: Systematic review. Probiotics Antimicrob. Proteins 2017, 9, 102–110. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Stewart, H.L.; Southwood, L.L.; Indugu, N.; Vecchiarelli, B.; Engiles, J.B.; Pitta, D. Differences in the equine faecal microbiota between horses presenting to a tertiary referral hospital for colic compared with an elective surgical procedure. Equine Vet. J. 2019, 51, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Hussein, H.; Vogedes, L.; Fernandez, G.; Frankeny, R. Effects of cereal grain supplementation on apparent digestibility of nutrients and concentrations of fermentation end-products in the feces and serum of horses consuming alfalfa cubes. J. Anim. Sci. 2004, 82, 1986–1996. [Google Scholar] [CrossRef]
- Geor, R.J. Current concepts on the pathophysiology of pasture-associated laminitis. Vet. Clin. N. Am. Equine Pract. 2010, 26, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Horn, M.A.; Matthies, C.; Küsel, K.; Schramm, A.; Drake, H.L. Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl. Environ. Microbiol. 2003, 69, 74–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venable, E.; Kerley, M.; Raub, R. Assessment of equine fecal microbial profiles during and after a colic episode using pyrosequencing. J. Equine Vet. Sci. 2013, 5, 347–348. [Google Scholar] [CrossRef]
- Kim, I.S.; Hwang, M.H.; Jang, N.J.; Hyun, S.H.; Lee, S.T. Effect of low pH on the activity of hydrogen utilizing methanogen in bio-hydrogen process. Int. J. Hydrog. Energy 2004, 29, 1133–1140. [Google Scholar] [CrossRef]
- Candela, M.; Perna, F.; Carnevali, P.; Vitali, B.; Ciati, R.; Gionchetti, P.; Rizzello, F.; Campieri, M.; Brigidi, P. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: Adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int. J. Food Microbiol. 2008, 125, 286–292. [Google Scholar] [CrossRef]
- Dougal, K.; Harris, P.A.; Edwards, A.; Pachebat, J.A.; Blackmore, T.M.; Worgan, H.J.; Newbold, C.J. A comparison of the microbiome and the metabolome of different regions of the equine hindgut. FEMS Microbiol. Ecol. 2012, 82, 642–652. [Google Scholar] [CrossRef] [Green Version]
- Joblin, K.; Campbell, G.P.; Richardson, A.J.; Stewart, C. Fermentation of barley straw by anaerobic rumen bacteria and fungi in axenic culture and in co-culture with methanogens. Lett. Appl. Microbiol. 1989, 9, 195–197. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinberg, L.M.; Regan, J.M. mcrA-targeted real-time quantitative PCR method to examine methanogen communities. Appl. Environ. Microbiol. 2009, 75, 4435–4442. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Pathways (MetaCyc) | Enriched Pathways in SC | ALDEx Diff. | Metabolism |
|---|---|---|---|
| ALL-CHORISMATE-PWY | Superpathway of chorismate metabolism | 8.16 | Enterobactin biosynthesis |
| ENTBACSYN-PWY | Enterobactin biosynthesis | 8.02 | |
| PWY0-321 | Phenylacetate degradation I (aerobic) | 5.05 | TCA cycle |
| PWY-5178 | Toluene degradation IV (aerobic) (via catechol) | 5.01 | |
| PWY-6185 | 4-methylcatechol degradation (ortho cleavage) | 4.77 | |
| PWY0-1277 | 3-phenylpropanoate and 3-(3-hydroxyphenyl) propanoate degradation | 4.60 | |
| PWY-5417 | Catechol degradation III (ortho-cleavage pathway) | 4.53 | |
| PWY-6182 | Superpathway of salicylate degradation | 4.38 | |
| PWY-5431 | Aromatic compounds degradation via & beta;-ketoadipate | 4.21 | |
| TCA-GLYOX-BYPASS | Superpathway of glyoxylate bypass and TCA | 4.11 |
| Pathways (MetaCyc) | Depleted Pathways in SC | ALDEx Diff. | Metabolism |
|---|---|---|---|
| PWY-6141 | Archaetidylserine and archaetidylethanolamine biosynthesis | −2.52 | Methanogen lipid membrane biosynthesis |
| PWY-7286 | 7-(3-amino-3-carboxypropyl)-wyosine biosynthesis | −2.39 | |
| PWY-6167 | Flavin biosynthesis II (archaea) | −2.35 | |
| PWY-6350 | Archaetidylinositol biosynthesis | −2.32 | |
| PWY-6349 | CDP-archaeol biosynthesis | −2.31 | |
| P261-PWY | Coenzyme M biosynthesis I | −2.49 | Methanogenesis |
| METHANOGENESIS-PWY | Methanogenesis from H2 and CO2 | −2.40 | |
| PWY-6148 | Tetrahydromethanopterin biosynthesis | −2.29 | |
| PWY-5198 | Factor 420 biosynthesis | −2.27 | |
| TCA-GLYOX-BYPASS | Coenzyme B biosynthesis | −2.21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, T.; Cheong, H.; Yoon, J.; Kim, A.; Yun, Y.; Unno, T. Comparison of the Fecal Microbiota of Horses with Intestinal Disease and Their Healthy Counterparts. Vet. Sci. 2021, 8, 113. https://doi.org/10.3390/vetsci8060113
Park T, Cheong H, Yoon J, Kim A, Yun Y, Unno T. Comparison of the Fecal Microbiota of Horses with Intestinal Disease and Their Healthy Counterparts. Veterinary Sciences. 2021; 8(6):113. https://doi.org/10.3390/vetsci8060113
Chicago/Turabian StylePark, Taemook, Heetae Cheong, Jungho Yoon, Ahram Kim, Youngmin Yun, and Tatsuya Unno. 2021. "Comparison of the Fecal Microbiota of Horses with Intestinal Disease and Their Healthy Counterparts" Veterinary Sciences 8, no. 6: 113. https://doi.org/10.3390/vetsci8060113






