How Artificial Intelligence Can Enhance the Diagnosis of Cardiac Amyloidosis: A Review of Recent Advances and Challenges
Abstract
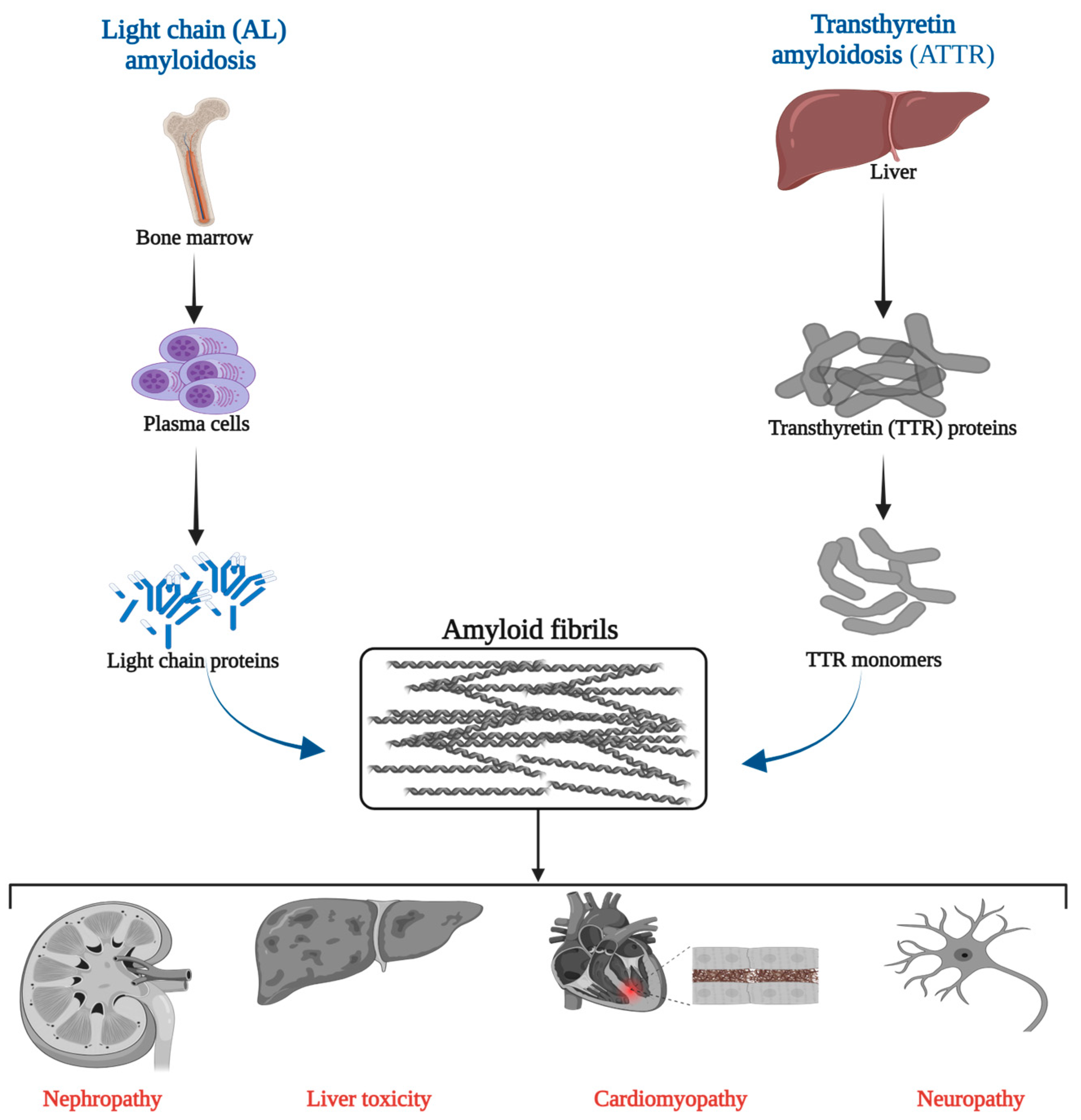
1. Introduction
2. AI Applications to Electrocardiogram (ECG) in Cardiac Amyloidosis
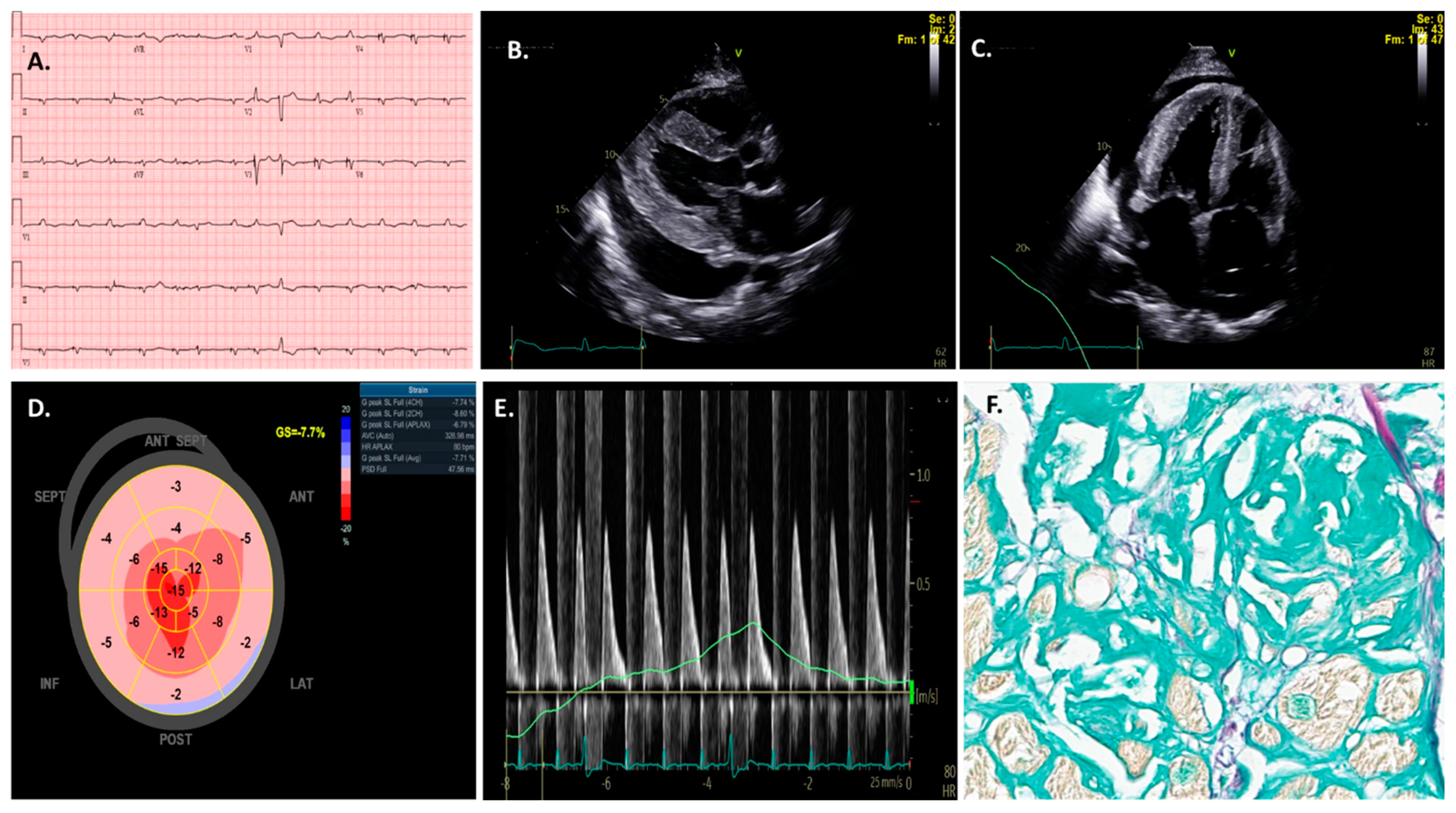
3. AI Applications to Echocardiography in Cardiac Amyloidosis
4. AI Applications to Cardiac Magnetic Resonance in Cardiac Amyloidosis
5. AI Applications to Scintigraphy in Cardiac Amyloidosis
6. AI Applications to Pathology in Cardiac Amyloidosis
7. Discussion and Clinical Implications
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bukhari, S. Cardiac amyloidosis: State-of-the-art review. J. Geriatr. Cardiol. 2023, 20, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Merlini, G.; Bellotti, V. Molecular mechanisms of amyloidosis. N. Engl. J. Med. 2003, 349, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Ruberg, F.L.; Grogan, M.; Hanna, M.; Kelly, J.W.; Maurer, M.S. Transthyretin Amyloid Cardiomyopathy: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2019, 73, 2872–2891. [Google Scholar] [CrossRef] [PubMed]
- Buxbaum, J.N.; Dispenzieri, A.; Eisenberg, D.S.; Fandrich, M.; Merlini, G.; Saraiva, M.J.M.; Sekijima, Y.; Westermark, P. Amyloid nomenclature 2022: Update, novel proteins, and recommendations by the International Society of Amyloidosis (ISA) Nomenclature Committee. Amyloid 2022, 29, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.M.; Rosenthal, J.L.; Grodin, J.L.; Maurer, M.S.; Grogan, M.; Cheng, R.K. ATTR Amyloidosis: Current and Emerging Management Strategies: JACC: CardioOncology State-of-the-Art Review. JACC CardioOncol. 2021, 3, 488–505. [Google Scholar] [CrossRef] [PubMed]
- Lousada, I.; Comenzo, R.L.; Landau, H.; Guthrie, S.; Merlini, G. Light Chain Amyloidosis: Patient Experience Survey from the Amyloidosis Research Consortium. Adv. Ther. 2015, 32, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Kastritis, E.; Palladini, G.; Minnema, M.C.; Wechalekar, A.D.; Jaccard, A.; Lee, H.C.; Sanchorawala, V.; Gibbs, S.; Mollee, P.; Venner, C.P.; et al. Daratumumab-Based Treatment for Immunoglobulin Light-Chain Amyloidosis. N. Engl. J. Med. 2021, 385, 46–58. [Google Scholar] [CrossRef]
- Garcia-Pavia, P.; Rapezzi, C.; Adler, Y.; Arad, M.; Basso, C.; Brucato, A.; Burazor, I.; Caforio, A.L.P.; Damy, T.; Eriksson, U.; et al. Diagnosis and treatment of cardiac amyloidosis: A position statement of the ESC Working Group on Myocardial and Pericardial Diseases. Eur. Heart J. 2021, 42, 1554–1568. [Google Scholar] [CrossRef]
- Writing, C.; Kittleson, M.M.; Ruberg, F.L.; Ambardekar, A.V.; Brannagan, T.H.; Cheng, R.K.; Clarke, J.O.; Dember, L.M.; Frantz, J.G.; Hershberger, R.E.; et al. 2023 ACC Expert Consensus Decision Pathway on Comprehensive Multidisciplinary Care for the Patient with Cardiac Amyloidosis: A Report of the American College of Cardiology Solution Set Oversight Committee. J. Am. Coll. Cardiol. 2023, 81, 1076–1126. [Google Scholar] [CrossRef]
- Martinez-Naharro, A.; Hawkins, P.N.; Fontana, M. Cardiac amyloidosis. Clin. Med. 2018, 18 (Suppl. 2), s30–s35. [Google Scholar] [CrossRef]
- Grogan, M.; Lopez-Jimenez, F.; Cohen-Shelly, M.; Dispenzieri, A.; Attia, Z.I.; Abou Ezzedine, O.F.; Lin, G.; Kapa, S.; Borgeson, D.D.; Friedman, P.A.; et al. Artificial Intelligence-Enhanced Electrocardiogram for the Early Detection of Cardiac Amyloidosis. Mayo Clin. Proc. 2021, 96, 2768–2778. [Google Scholar] [CrossRef] [PubMed]
- Vodanovic, M.; Subasic, M.; Milosevic, D.; Savic Pavicin, I. Artificial Intelligence in Medicine and Dentistry. Acta Stomatol. Croat. 2023, 57, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Grech, V.; Cuschieri, S.; Eldawlatly, A.A. Artificial intelligence in medicine and research—The good, the bad, and the ugly. Saudi J. Anaesth. 2023, 17, 401–406. [Google Scholar] [CrossRef]
- Itchhaporia, D. Artificial intelligence in cardiology. Trends Cardiovasc. Med. 2022, 32, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Krumholz, H.M. Screening ECGs in low-risk patients are associated with increased risk of downstream cardiac testing. BMJ Evid. Based Med. 2018, 23, 150–151. [Google Scholar] [CrossRef]
- Bhatia, R.S.; Bouck, Z.; Ivers, N.M.; Mecredy, G.; Singh, J.; Pendrith, C.; Ko, D.T.; Martin, D.; Wijeysundera, H.C.; Tu, J.V.; et al. Electrocardiograms in Low-Risk Patients Undergoing an Annual Health Examination. JAMA Intern. Med. 2017, 177, 1326–1333. [Google Scholar] [CrossRef] [PubMed]
- Hartnett, J.; Jaber, W.; Maurer, M.; Sperry, B.; Hanna, M.; Collier, P.; Patel, D.R.; Wazni, O.M.; Donnellan, E. Electrophysiological Manifestations of Cardiac Amyloidosis. JACC CardioOncol. 2021, 3, 506–515. [Google Scholar] [CrossRef] [PubMed]
- Quarta, C.C.; Solomon, S.D.; Uraizee, I.; Kruger, J.; Longhi, S.; Ferlito, M.; Gagliardi, C.; Milandri, A.; Rapezzi, C.; Falk, R.H. Left ventricular structure and function in transthyretin-related versus light-chain cardiac amyloidosis. Circulation 2014, 129, 1840–1849. [Google Scholar] [CrossRef] [PubMed]
- Isath, A.; Correa, A.; Siroky, G.P.; Perimbeti, S.; Mohammed, S.; Chahal, C.A.A.; Padmanabhan, D.; Mehta, D. Trends, burden, and impact of arrhythmia on cardiac amyloid patients: A 16-year nationwide study from 1999 to 2014. J. Arrhythm. 2020, 36, 727–734. [Google Scholar] [CrossRef]
- Assaf, A.; Mekhael, M.; Noujaim, C.; Chouman, N.; Younes, H.; Kreidieh, O.; Marrouche, N.; Donnellan, E. Conduction system disease in cardiac amyloidosis. Trends Cardiovasc. Med. 2023, in press. [CrossRef]
- Sharma, S.; Labib, S.B.; Shah, S.P. Electrocardiogram Criteria to Diagnose Cardiac Amyloidosis in Men with a Bundle Branch Block. Am. J. Cardiol. 2021, 146, 89–94. [Google Scholar] [CrossRef]
- Cyrille, N.B.; Goldsmith, J.; Alvarez, J.; Maurer, M.S. Prevalence and prognostic significance of low QRS voltage among the three main types of cardiac amyloidosis. Am. J. Cardiol. 2014, 114, 1089–1093. [Google Scholar] [CrossRef]
- Maurer, M.S.; Hanna, M.; Grogan, M.; Dispenzieri, A.; Witteles, R.; Drachman, B.; Judge, D.P.; Lenihan, D.J.; Gottlieb, S.S.; Shah, S.J.; et al. Genotype and Phenotype of Transthyretin Cardiac Amyloidosis: THAOS (Transthyretin Amyloid Outcome Survey). J. Am. Coll. Cardiol. 2016, 68, 161–172. [Google Scholar] [CrossRef]
- Siddiqi, O.K.; Ruberg, F.L. Cardiac amyloidosis: An update on pathophysiology, diagnosis, and treatment. Trends Cardiovasc. Med. 2018, 28, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Martini, N.; Sinigiani, G.; De Michieli, L.; Mussinelli, R.; Perazzolo Marra, M.; Iliceto, S.; Zorzi, A.; Perlini, S.; Corrado, D.; Cipriani, A. Electrocardiographic features and rhythm disorders in cardiac amyloidosis. Trends Cardiovasc. Med. 2023, in press. [CrossRef]
- Attia, Z.I.; Harmon, D.M.; Behr, E.R.; Friedman, P.A. Application of artificial intelligence to the electrocardiogram. Eur. Heart J. 2021, 42, 4717–4730. [Google Scholar] [CrossRef] [PubMed]
- Tison, G.H.; Zhang, J.; Delling, F.N.; Deo, R.C. Automated and Interpretable Patient ECG Profiles for Disease Detection, Tracking, and Discovery. Circ. Cardiovasc. Qual. Outcomes 2019, 12, e005289. [Google Scholar] [CrossRef]
- Goto, S.; Mahara, K.; Beussink-Nelson, L.; Ikura, H.; Katsumata, Y.; Endo, J.; Gaggin, H.K.; Shah, S.J.; Itabashi, Y.; MacRae, C.A.; et al. Artificial intelligence-enabled fully automated detection of cardiac amyloidosis using electrocardiograms and echocardiograms. Nat. Commun. 2021, 12, 2726. [Google Scholar] [CrossRef] [PubMed]
- Harmon, D.M.; Mangold, K.; Baez Suarez, A.; Scott, C.G.; Murphree, D.H.; Malik, A.; Attia, Z.I.; Lopez-Jimenez, F.; Friedman, P.A.; Dispenzieri, A.; et al. Postdevelopment Performance and Validation of the Artificial Intelligence-Enhanced Electrocardiogram for Detection of Cardiac Amyloidosis. JACC Adv. 2023, 2, 100612. [Google Scholar] [CrossRef]
- Cohen, O.C.; Ismael, A.; Pawarova, B.; Manwani, R.; Ravichandran, S.; Law, S.; Foard, D.; Petrie, A.; Ward, S.; Douglas, B.; et al. Longitudinal strain is an independent predictor of survival and response to therapy in patients with systemic AL amyloidosis. Eur. Heart J. 2022, 43, 333–341. [Google Scholar] [CrossRef]
- Cotella, J.I.; Slivnick, J.A.; Sanderson, E.; Singulane, C.; O’Driscoll, J.; Asch, F.M.; Addetia, K.; Woodward, G.; Lang, R.M. Artificial intelligence based left ventricular ejection fraction and global longitudinal strain in cardiac amyloidosis. Echocardiography 2023, 40, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Witteles, R.M.; Bokhari, S.; Damy, T.; Elliott, P.M.; Falk, R.H.; Fine, N.M.; Gospodinova, M.; Obici, L.; Rapezzi, C.; Garcia-Pavia, P. Screening for Transthyretin Amyloid Cardiomyopathy in Everyday Practice. JACC Heart Fail. 2019, 7, 709–716. [Google Scholar] [CrossRef]
- Kittleson, M.M.; Maurer, M.S.; Ambardekar, A.V.; Bullock-Palmer, R.P.; Chang, P.P.; Eisen, H.J.; Nair, A.P.; Nativi-Nicolau, J.; Ruberg, F.L.; American Heart Association Heart Failure and Transplantation Committee of the Council on Clinical Cardiology. Cardiac Amyloidosis: Evolving Diagnosis and Management: A Scientific Statement from the American Heart Association. Circulation 2020, 142, e7–e22. [Google Scholar] [CrossRef] [PubMed]
- Barros-Gomes, S.; Williams, B.; Nhola, L.F.; Grogan, M.; Maalouf, J.F.; Dispenzieri, A.; Pellikka, P.A.; Villarraga, H.R. Prognosis of Light Chain Amyloidosis with Preserved LVEF: Added Value of 2D Speckle-Tracking Echocardiography to the Current Prognostic Staging System. JACC Cardiovasc. Imaging 2017, 10, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Bellavia, D.; Pellikka, P.A.; Al-Zahrani, G.B.; Abraham, T.P.; Dispenzieri, A.; Miyazaki, C.; Lacy, M.; Scott, C.G.; Oh, J.K.; Miller, F.A., Jr. Independent predictors of survival in primary systemic (Al) amyloidosis, including cardiac biomarkers and left ventricular strain imaging: An observational cohort study. J. Am. Soc. Echocardiogr. 2010, 23, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Buss, S.J.; Emami, M.; Mereles, D.; Korosoglou, G.; Kristen, A.V.; Voss, A.; Schellberg, D.; Zugck, C.; Galuschky, C.; Giannitsis, E.; et al. Longitudinal left ventricular function for prediction of survival in systemic light-chain amyloidosis: Incremental value compared with clinical and biochemical markers. J. Am. Coll. Cardiol. 2012, 60, 1067–1076. [Google Scholar] [CrossRef] [PubMed]
- Cueto-Garcia, L.; Reeder, G.S.; Kyle, R.A.; Wood, D.L.; Seward, J.B.; Naessens, J.; Offord, K.P.; Greipp, P.R.; Edwards, W.D.; Tajik, A.J. Echocardiographic findings in systemic amyloidosis: Spectrum of cardiac involvement and relation to survival. J. Am. Coll. Cardiol. 1985, 6, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.L.; Hatle, L.K.; Taliercio, C.P.; Oh, J.K.; Kyle, R.A.; Gertz, M.A.; Bailey, K.R.; Seward, J.B.; Tajik, A.J. Prognostic significance of Doppler measures of diastolic function in cardiac amyloidosis. A Doppler echocardiography study. Circulation 1991, 83, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Koyama, J.; Falk, R.H. Prognostic significance of strain Doppler imaging in light-chain amyloidosis. JACC Cardiovasc. Imaging 2010, 3, 333–342. [Google Scholar] [CrossRef]
- Pagourelias, E.D.; Mirea, O.; Duchenne, J.; Van Cleemput, J.; Delforge, M.; Bogaert, J.; Kuznetsova, T.; Voigt, J.U. Echo Parameters for Differential Diagnosis in Cardiac Amyloidosis: A Head-to-Head Comparison of Deformation and Nondeformation Parameters. Circ. Cardiovasc. Imaging 2017, 10, e005588. [Google Scholar] [CrossRef]
- Maurer, M.S.; Elliott, P.; Comenzo, R.; Semigran, M.; Rapezzi, C. Addressing Common Questions Encountered in the Diagnosis and Management of Cardiac Amyloidosis. Circulation 2017, 135, 1357–1377. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Naharro, A.; Treibel, T.A.; Abdel-Gadir, A.; Bulluck, H.; Zumbo, G.; Knight, D.S.; Kotecha, T.; Francis, R.; Hutt, D.F.; Rezk, T.; et al. Magnetic Resonance in Transthyretin Cardiac Amyloidosis. J. Am. Coll. Cardiol. 2017, 70, 466–477. [Google Scholar] [CrossRef]
- Marwick, T.H. Ejection Fraction Pros and Cons: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2018, 72, 2360–2379. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.; Yoshitomi, H.; Ito, S.; Adachi, T.; Takahashi, N.; Tanabe, K. Single-beat determination of global longitudinal speckle strain in patients with atrial fibrillation. J. Echocardiogr. 2012, 10, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Tromp, J.; Seekings, P.J.; Hung, C.L.; Iversen, M.B.; Frost, M.J.; Ouwerkerk, W.; Jiang, Z.; Eisenhaber, F.; Goh, R.S.M.; Zhao, H.; et al. Automated interpretation of systolic and diastolic function on the echocardiogram: A multicohort study. Lancet Digit. Health 2022, 4, e46–e54. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Jin, P.; Joseph Raj, A.N.; Yuan, Y.; Zhuang, S. Automatic Segmentation of Left Ventricle in Echocardiography Based on YOLOv3 Model to Achieve Constraint and Positioning. Comput. Math. Methods Med. 2021, 2021, 3772129. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.J.; Jeong, J.; Arsanjani, R.; Kim, K.; Tsai, Y.L.; Yu, W.C.; Farina, J.M.; Mahmoud, A.K.; Ayoub, C.; Grogan, M.; et al. Echocardiography-Based Deep Learning Model to Differentiate Constrictive Pericarditis and Restrictive Cardiomyopathy. JACC Cardiovasc. Imaging 2023, 17, 349–360. [Google Scholar] [CrossRef]
- Li, J.; Chao, C.J.; Jeong, J.J.; Farina, J.M.; Seri, A.R.; Barry, T.; Newman, H.; Campany, M.; Abdou, M.; O’Shea, M.; et al. Developing an Echocardiography-Based, Automatic Deep Learning Framework for the Differentiation of Increased Left Ventricular Wall Thickness Etiologies. J. Imaging 2023, 9, 48. [Google Scholar] [CrossRef]
- Carvalho, F.P.; Erthal, F.; Azevedo, C.F. The Role of Cardiac MR Imaging in the Assessment of Patients with Cardiac Amyloidosis. Magn. Reson. Imaging Clin. N. Am. 2019, 27, 453–463. [Google Scholar] [CrossRef]
- Fontana, M.; Pica, S.; Reant, P.; Abdel-Gadir, A.; Treibel, T.A.; Banypersad, S.M.; Maestrini, V.; Barcella, W.; Rosmini, S.; Bulluck, H.; et al. Prognostic Value of Late Gadolinium Enhancement Cardiovascular Magnetic Resonance in Cardiac Amyloidosis. Circulation 2015, 132, 1570–1579. [Google Scholar] [CrossRef]
- Zhao, L.; Tian, Z.; Fang, Q. Diagnostic accuracy of cardiovascular magnetic resonance for patients with suspected cardiac amyloidosis: A systematic review and meta-analysis. BMC Cardiovasc. Disord. 2016, 16, 129. [Google Scholar] [CrossRef] [PubMed]
- Banypersad, S.M.; Sado, D.M.; Flett, A.S.; Gibbs, S.D.; Pinney, J.H.; Maestrini, V.; Cox, A.T.; Fontana, M.; Whelan, C.J.; Wechalekar, A.D.; et al. Quantification of myocardial extracellular volume fraction in systemic AL amyloidosis: An equilibrium contrast cardiovascular magnetic resonance study. Circ. Cardiovasc. Imaging 2013, 6, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Messroghli, D.R.; Moon, J.C.; Ferreira, V.M.; Grosse-Wortmann, L.; He, T.; Kellman, P.; Mascherbauer, J.; Nezafat, R.; Salerno, M.; Schelbert, E.B.; et al. Clinical recommendations for cardiovascular magnetic resonance mapping of T1, T2, T2* and extracellular volume: A consensus statement by the Society for Cardiovascular Magnetic Resonance (SCMR) endorsed by the European Association for Cardiovascular Imaging (EACVI). J. Cardiovasc. Magn. Reson. 2017, 19, 75. [Google Scholar] [CrossRef] [PubMed]
- Karamitsos, T.D.; Piechnik, S.K.; Banypersad, S.M.; Fontana, M.; Ntusi, N.B.; Ferreira, V.M.; Whelan, C.J.; Myerson, S.G.; Robson, M.D.; Hawkins, P.N.; et al. Noncontrast T1 mapping for the diagnosis of cardiac amyloidosis. JACC Cardiovasc. Imaging 2013, 6, 488–497. [Google Scholar] [CrossRef]
- Fontana, M.; Banypersad, S.M.; Treibel, T.A.; Maestrini, V.; Sado, D.M.; White, S.K.; Pica, S.; Castelletti, S.; Piechnik, S.K.; Robson, M.D.; et al. Native T1 mapping in transthyretin amyloidosis. JACC Cardiovasc. Imaging 2014, 7, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Baggiano, A.; Boldrini, M.; Martinez-Naharro, A.; Kotecha, T.; Petrie, A.; Rezk, T.; Gritti, M.; Quarta, C.; Knight, D.S.; Wechalekar, A.D.; et al. Noncontrast Magnetic Resonance for the Diagnosis of Cardiac Amyloidosis. JACC Cardiovasc. Imaging 2020, 13, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Kotecha, T.; Martinez-Naharro, A.; Treibel, T.A.; Francis, R.; Nordin, S.; Abdel-Gadir, A.; Knight, D.S.; Zumbo, G.; Rosmini, S.; Maestrini, V.; et al. Myocardial Edema and Prognosis in Amyloidosis. J. Am. Coll. Cardiol. 2018, 71, 2919–2931. [Google Scholar] [CrossRef] [PubMed]
- Agibetov, A.; Kammerlander, A.; Duca, F.; Nitsche, C.; Koschutnik, M.; Dona, C.; Dachs, T.M.; Rettl, R.; Stria, A.; Schrutka, L.; et al. Convolutional Neural Networks for Fully Automated Diagnosis of Cardiac Amyloidosis by Cardiac Magnetic Resonance Imaging. J. Pers. Med. 2021, 11, 1268. [Google Scholar] [CrossRef]
- Eckstein, J.; Moghadasi, N.; Korperich, H.; Weise Valdes, E.; Sciacca, V.; Paluszkiewicz, L.; Burchert, W.; Piran, M. A Machine Learning Challenge: Detection of Cardiac Amyloidosis Based on Bi-Atrial and Right Ventricular Strain and Cardiac Function. Diagnostics 2022, 12, 2696. [Google Scholar] [CrossRef]
- Xu, C.; Xu, L.; Ohorodnyk, P.; Roth, M.; Chen, B.; Li, S. Contrast agent-free synthesis and segmentation of ischemic heart disease images using progressive sequential causal GANs. Med. Image Anal. 2020, 62, 101668. [Google Scholar] [CrossRef]
- Martinez-Naharro, A.; Baksi, A.J.; Hawkins, P.N.; Fontana, M. Diagnostic imaging of cardiac amyloidosis. Nat. Rev. Cardiol. 2020, 17, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Timoteo, A.T.; Rosa, S.A.; Bras, P.G.; Ferreira, M.J.V.; Bettencourt, N. Multimodality imaging in cardiac amyloidosis: State-of-the-art review. J. Clin. Ultrasound 2022, 50, 1084–1096. [Google Scholar] [CrossRef] [PubMed]
- Pilebro, B.; Suhr, O.B.; Naslund, U.; Westermark, P.; Lindqvist, P.; Sundstrom, T. (99m)Tc-DPD uptake reflects amyloid fibril composition in hereditary transthyretin amyloidosis. Ups. J. Med. Sci. 2016, 121, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Stats, M.A.; Stone, J.R. Varying levels of small microcalcifications and macrophages in ATTR and AL cardiac amyloidosis: Implications for utilizing nuclear medicine studies to subtype amyloidosis. Cardiovasc. Pathol. 2016, 25, 413–417. [Google Scholar] [CrossRef]
- Weiler-Sagie, M.; Ben-Haim, S. Variability in bone-seeking tracers and imaging protocols for the diagnosis of cardiac amyloidosis: The more the merrier? J. Nucl. Cardiol. 2022, 29, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Ahluwalia, N.; Roshankar, G.; Draycott, L.; Jimenez-Zepeda, V.; Fine, N.; Chan, D.; Han, D.; Miller, R.J.H. Diagnostic accuracy of bone scintigraphy imaging for transthyretin cardiac amyloidosis: Systematic review and meta-analysis. J. Nucl. Cardiol. 2023, 30, 2464–2476. [Google Scholar] [CrossRef] [PubMed]
- Cappelli, F.; Gallini, C.; Di Mario, C.; Costanzo, E.N.; Vaggelli, L.; Tutino, F.; Ciaccio, A.; Bartolini, S.; Angelotti, P.; Frusconi, S.; et al. Accuracy of 99mTc-Hydroxymethylene diphosphonate scintigraphy for diagnosis of transthyretin cardiac amyloidosis. J. Nucl. Cardiol. 2019, 26, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Dorbala, S.; Ando, Y.; Bokhari, S.; Dispenzieri, A.; Falk, R.H.; Ferrari, V.A.; Fontana, M.; Gheysens, O.; Gillmore, J.D.; Glaudemans, A.; et al. ASNC/AHA/ASE/EANM/HFSA/ISA/SCMR/SNMMI Expert Consensus Recommendations for Multimodality Imaging in Cardiac Amyloidosis: Part 1 of 2-Evidence Base and Standardized Methods of Imaging. Circ. Cardiovasc. Imaging 2021, 14, e000029. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Falk, R.; Di Carli, M.F.; Kijewski, M.; Rapezzi, C.; Dorbala, S. State-of-the-art radionuclide imaging in cardiac transthyretin amyloidosis. J. Nucl. Cardiol. 2019, 26, 158–173. [Google Scholar] [CrossRef]
- Hutt, D.F.; Quigley, A.M.; Page, J.; Hall, M.L.; Burniston, M.; Gopaul, D.; Lane, T.; Whelan, C.J.; Lachmann, H.J.; Gillmore, J.D.; et al. Utility and limitations of 3,3-diphosphono-1,2-propanodicarboxylic acid scintigraphy in systemic amyloidosis. Eur. Heart J. Cardiovasc. Imaging 2014, 15, 1289–1298. [Google Scholar] [CrossRef]
- Halme, H.L.; Ihalainen, T.; Suomalainen, O.; Loimaala, A.; Matzke, S.; Uusitalo, V.; Sipila, O.; Hippelainen, E. Convolutional neural networks for detection of transthyretin amyloidosis in 2D scintigraphy images. EJNMMI Res. 2022, 12, 27. [Google Scholar] [CrossRef] [PubMed]
- Delbarre, M.A.; Girardon, F.; Roquette, L.; Blanc-Durand, P.; Hubaut, M.A.; Hachulla, E.; Semah, F.; Huglo, D.; Garcelon, N.; Marchal, E.; et al. Deep Learning on Bone Scintigraphy to Detect Abnormal Cardiac Uptake at Risk of Cardiac Amyloidosis. JACC Cardiovasc. Imaging 2023, 16, 1085–1095. [Google Scholar] [CrossRef] [PubMed]
- Vrana, J.A.; Gamez, J.D.; Madden, B.J.; Theis, J.D.; Bergen, H.R., 3rd; Dogan, A. Classification of amyloidosis by laser microdissection and mass spectrometry-based proteomic analysis in clinical biopsy specimens. Blood 2009, 114, 4957–4959. [Google Scholar] [CrossRef] [PubMed]
- Dasari, S.; Theis, J.D.; Vrana, J.A.; Rech, K.L.; Dao, L.N.; Howard, M.T.; Dispenzieri, A.; Gertz, M.A.; Hasadsri, L.; Highsmith, W.E.; et al. Amyloid Typing by Mass Spectrometry in Clinical Practice: A Comprehensive Review of 16,175 Samples. Mayo Clin. Proc. 2020, 95, 1852–1864. [Google Scholar] [CrossRef] [PubMed]
- Finsterer, J.; Iglseder, S.; Wanschitz, J.; Topakian, R.; Loscher, W.N.; Grisold, W. Hereditary transthyretin-related amyloidosis. Acta Neurol. Scand. 2019, 139, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Falk, R.H.; Alexander, K.M.; Liao, R.; Dorbala, S. AL (Light-Chain) Cardiac Amyloidosis: A Review of Diagnosis and Therapy. J. Am. Coll. Cardiol. 2016, 68, 1323–1341. [Google Scholar] [CrossRef]
- Solomon, A.; Murphy, C.L.; Westermark, P. Unreliability of immunohistochemistry for typing amyloid deposits. Arch. Pathol. Lab. Med. 2008, 132, 14, author reply 14–15. [Google Scholar] [CrossRef]
- Satoskar, A.A.; Burdge, K.; Cowden, D.J.; Nadasdy, G.M.; Hebert, L.A.; Nadasdy, T. Typing of amyloidosis in renal biopsies: Diagnostic pitfalls. Arch. Pathol. Lab. Med. 2007, 131, 917–922. [Google Scholar] [CrossRef]
- Palstrom, N.B.; Rojek, A.M.; Moller, H.E.H.; Hansen, C.T.; Matthiesen, R.; Rasmussen, L.M.; Abildgaard, N.; Beck, H.C. Classification of Amyloidosis by Model-Assisted Mass Spectrometry-Based Proteomics. Int. J. Mol. Sci. 2021, 23, 319. [Google Scholar] [CrossRef]
- Kim, J.H.; Zhang, C.; Sperati, C.J.; Bagnasco, S.M.; Barman, I. Non-Perturbative Identification and Subtyping of Amyloidosis in Human Kidney Tissue with Raman Spectroscopy and Machine Learning. Biosensors 2023, 13, 466. [Google Scholar] [CrossRef]
- Cazzaniga, G.; Bolognesi, M.M.; Stefania, M.D.; Mascadri, F.; Eccher, A.; Alberici, F.; Mescia, F.; Smith, A.; Fraggetta, F.; Rossi, M.; et al. Congo Red Staining in Digital Pathology: The Streamlined Pipeline for Amyloid Detection Through Congo Red Fluorescence Digital Analysis. Lab. Investig. 2023, 103, 100243. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Y.; Sharma, V.; Saini, H.; Tingen, J.N.; Flores, A.; Liu, D.; Safain, M.G.; Kryzanski, J.; McPhail, E.D.; Arkun, K.; et al. Machine Learning Quantification of Amyloid Deposits in Histological Images of Ligamentum Flavum. J. Pathol. Inform. 2022, 13, 100013. [Google Scholar] [CrossRef] [PubMed]
- Kessel, K.; Mattila, J.; Linder, N.; Kivela, T.; Lundin, J. Deep Learning Algorithms for Corneal Amyloid Deposition Quantitation in Familial Amyloidosis. Ocul. Oncol. Pathol. 2020, 6, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, M.; Piccoli, L.; Romeo, M.; Barzago, M.M.; Ravasio, S.; Foglierini, M.; Matkovic, M.; Sgrignani, J.; De Gasparo, R.; Prunotto, M.; et al. Machine learning analyses of antibody somatic mutations predict immunoglobulin light chain toxicity. Nat. Commun. 2021, 12, 3532. [Google Scholar] [CrossRef] [PubMed]
- David, M.P.; Concepcion, G.P.; Padlan, E.A. Using simple artificial intelligence methods for predicting amyloidogenesis in antibodies. BMC Bioinform. 2010, 11, 79. [Google Scholar] [CrossRef] [PubMed]
- Hanna, M.; Ruberg, F.L.; Maurer, M.S.; Dispenzieri, A.; Dorbala, S.; Falk, R.H.; Hoffman, J.; Jaber, W.; Soman, P.; Witteles, R.M.; et al. Cardiac Scintigraphy with Technetium-99m-Labeled Bone-Seeking Tracers for Suspected Amyloidosis: JACC Review Topic of the Week. J. Am. Coll. Cardiol. 2020, 75, 2851–2862. [Google Scholar] [CrossRef]
- Poterucha, T.J.; Elias, P.; Ruberg, F.L.; DeLuca, A.; Kinkhabwala, M.; Johnson, L.L.; Griffin, J.M.; Pandey, S.; Einstein, A.J.; Maurer, M.S. False Positive 99mTc-Pyrophosphate Scanning Leading to Inappropriate Tafamidis Prescriptions. JACC Cardiovasc. Imaging 2021, 14, 2042–2044. [Google Scholar] [CrossRef] [PubMed]
- Manolis, A.S.; Manolis, A.A.; Manolis, T.A.; Melita, H. Cardiac amyloidosis: An underdiagnosed/underappreciated disease. Eur. J. Intern. Med. 2019, 67, 1–13. [Google Scholar] [CrossRef]
- Farina, J.M.; Pereyra, M.; Mahmoud, A.K.; Scalia, I.G.; Abbas, M.T.; Chao, C.J.; Barry, T.; Ayoub, C.; Banerjee, I.; Arsanjani, R. Artificial Intelligence-Based Prediction of Cardiovascular Diseases from Chest Radiography. J. Imaging 2023, 9, 236. [Google Scholar] [CrossRef]
- Kagiyama, N.; Shrestha, S.; Farjo, P.D.; Sengupta, P.P. Artificial Intelligence: Practical Primer for Clinical Research in Cardiovascular Disease. J. Am. Heart Assoc. 2019, 8, e012788. [Google Scholar] [CrossRef]
- Goto, S.; Solanki, D.; John, J.E.; Yagi, R.; Homilius, M.; Ichihara, G.; Katsumata, Y.; Gaggin, H.K.; Itabashi, Y.; MacRae, C.A.; et al. Multinational Federated Learning Approach to Train ECG and Echocardiogram Models for Hypertrophic Cardiomyopathy Detection. Circulation 2022, 146, 755–769. [Google Scholar] [CrossRef] [PubMed]
- Allegra, A.; Mirabile, G.; Tonacci, A.; Genovese, S.; Pioggia, G.; Gangemi, S. Machine Learning Approaches in Diagnosis, Prognosis and Treatment Selection of Cardiac Amyloidosis. Int. J. Mol. Sci. 2023, 24, 5680. [Google Scholar] [CrossRef] [PubMed]

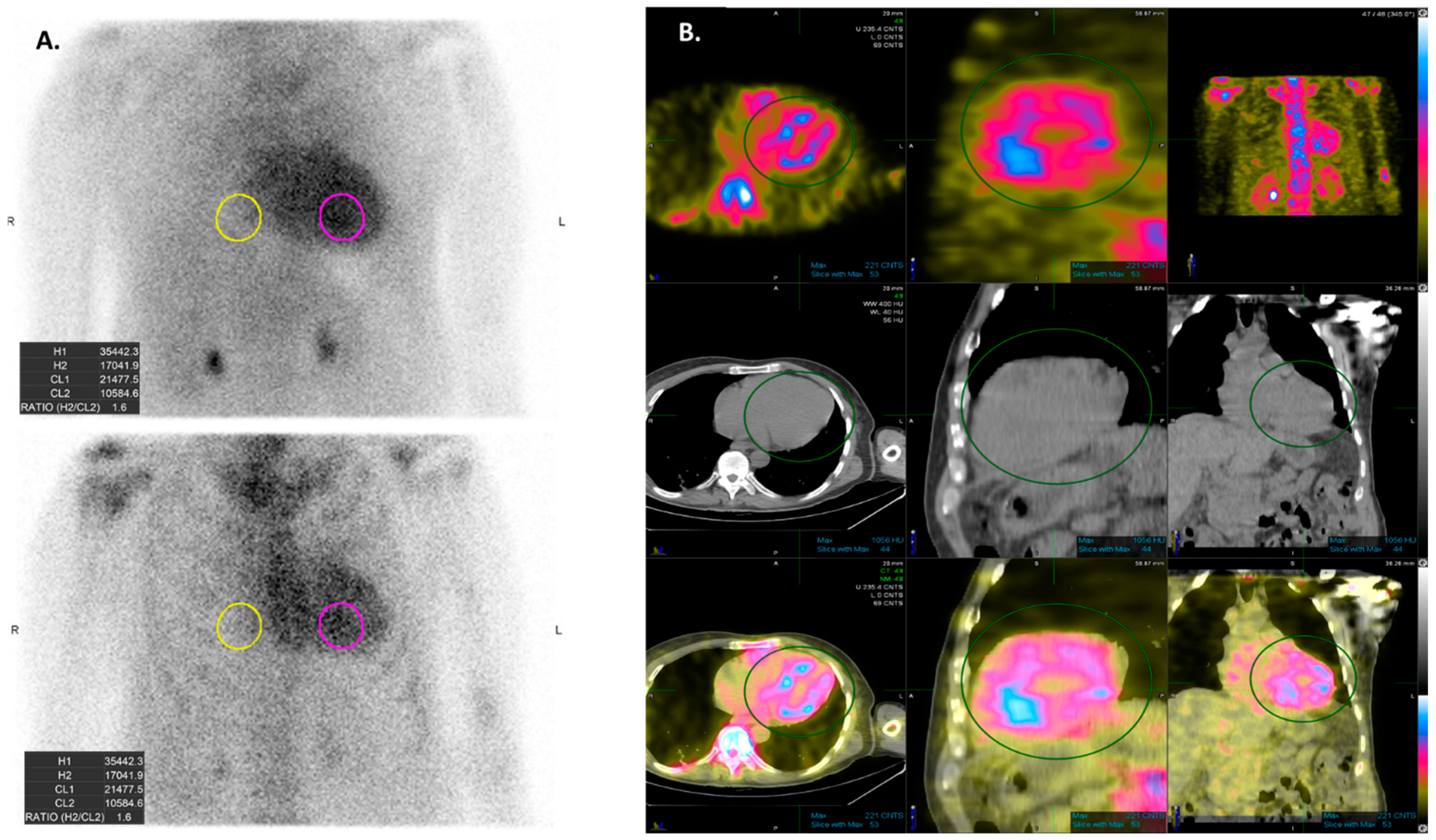
| Grade 0 | No cardiac uptake. |
| Grade 1 | Mild cardiac uptake, less than that in ribs. |
| Garde 2 | Moderate cardiac uptake is similar to that in ribs, but uptake in ribs remains clearly visible. |
| Garde 3 | Intense cardiac uptake greater than that in ribs with weak or no signal evident in ribs. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamel, M.A.; Abbas, M.T.; Kanaan, C.N.; Awad, K.A.; Baba Ali, N.; Scalia, I.G.; Farina, J.M.; Pereyra, M.; Mahmoud, A.K.; Steidley, D.E.; et al. How Artificial Intelligence Can Enhance the Diagnosis of Cardiac Amyloidosis: A Review of Recent Advances and Challenges. J. Cardiovasc. Dev. Dis. 2024, 11, 118. https://doi.org/10.3390/jcdd11040118
Kamel MA, Abbas MT, Kanaan CN, Awad KA, Baba Ali N, Scalia IG, Farina JM, Pereyra M, Mahmoud AK, Steidley DE, et al. How Artificial Intelligence Can Enhance the Diagnosis of Cardiac Amyloidosis: A Review of Recent Advances and Challenges. Journal of Cardiovascular Development and Disease. 2024; 11(4):118. https://doi.org/10.3390/jcdd11040118
Chicago/Turabian StyleKamel, Moaz A., Mohammed Tiseer Abbas, Christopher N. Kanaan, Kamal A. Awad, Nima Baba Ali, Isabel G. Scalia, Juan M. Farina, Milagros Pereyra, Ahmed K. Mahmoud, D. Eric Steidley, and et al. 2024. "How Artificial Intelligence Can Enhance the Diagnosis of Cardiac Amyloidosis: A Review of Recent Advances and Challenges" Journal of Cardiovascular Development and Disease 11, no. 4: 118. https://doi.org/10.3390/jcdd11040118
APA StyleKamel, M. A., Abbas, M. T., Kanaan, C. N., Awad, K. A., Baba Ali, N., Scalia, I. G., Farina, J. M., Pereyra, M., Mahmoud, A. K., Steidley, D. E., Rosenthal, J. L., Ayoub, C., & Arsanjani, R. (2024). How Artificial Intelligence Can Enhance the Diagnosis of Cardiac Amyloidosis: A Review of Recent Advances and Challenges. Journal of Cardiovascular Development and Disease, 11(4), 118. https://doi.org/10.3390/jcdd11040118









