The Aspergillus flavus hacA Gene in the Unfolded Protein Response Pathway Is a Candidate Target for Host-Induced Gene Silencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains, Media, and Culturing Conditions
2.2. Preparation of CRISP/Cas9 Constructs
2.3. Prediction of 3-Dimensional Structures
2.4. Construction of hacA RNAi Vectors
2.5. Generation of Protoplasts and Fungal Transformation
2.6. Analysis of hacA Sequence Defects of A. flavus Transformants and Colony PCR of hacA RNAi Transformants
2.7. Semi-Quantitative Thin Layer Chromatography (TLC) Analysis of Aflatoxin
3. Results
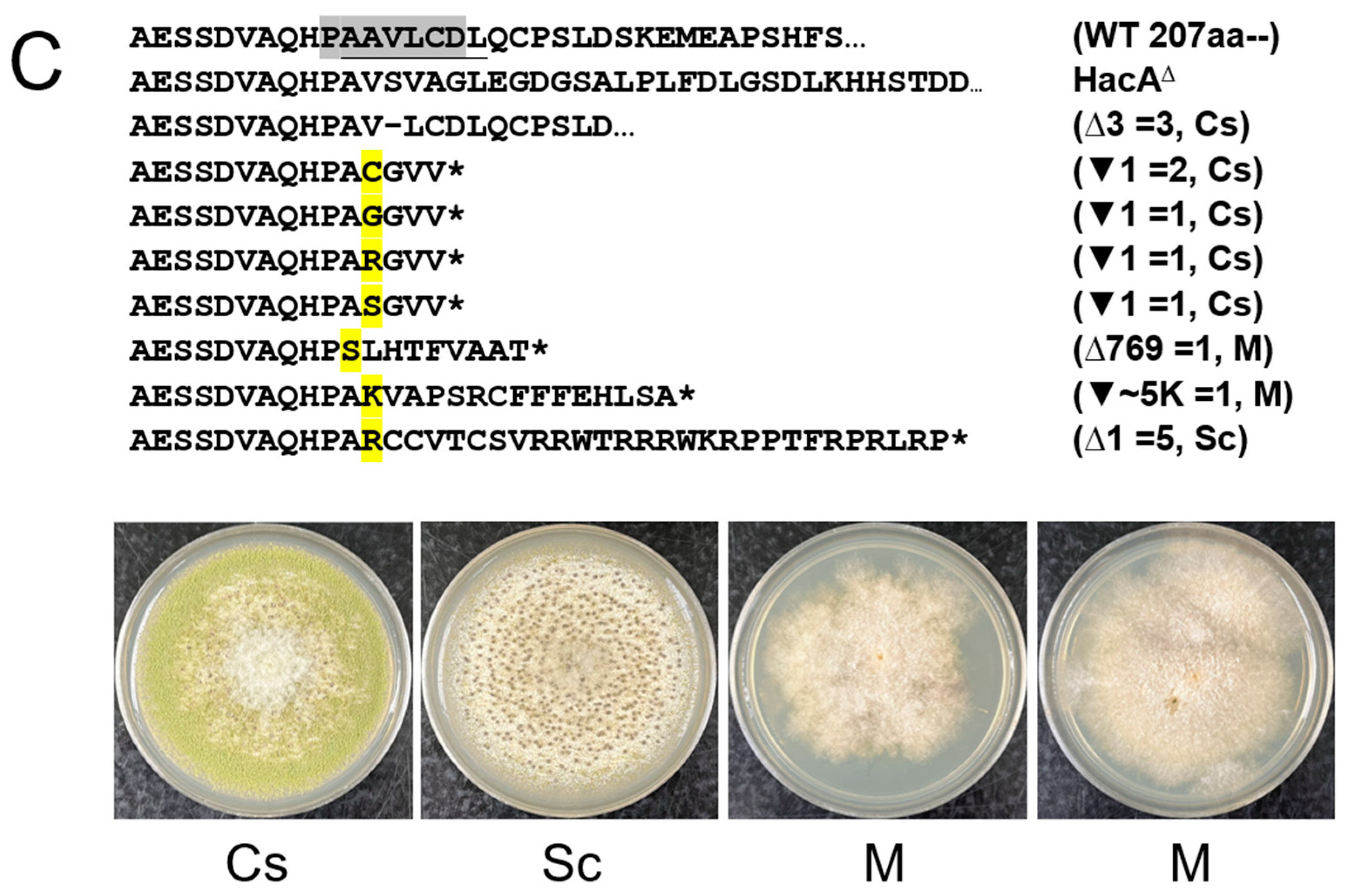
3.1. The Non-Conventional Intron in A. flavus hacA Is 20 Nucleotides Long and Its Removal Yields a Truncated HacA Lacking α-Helical Structures at the Carboxyl Terminal Region
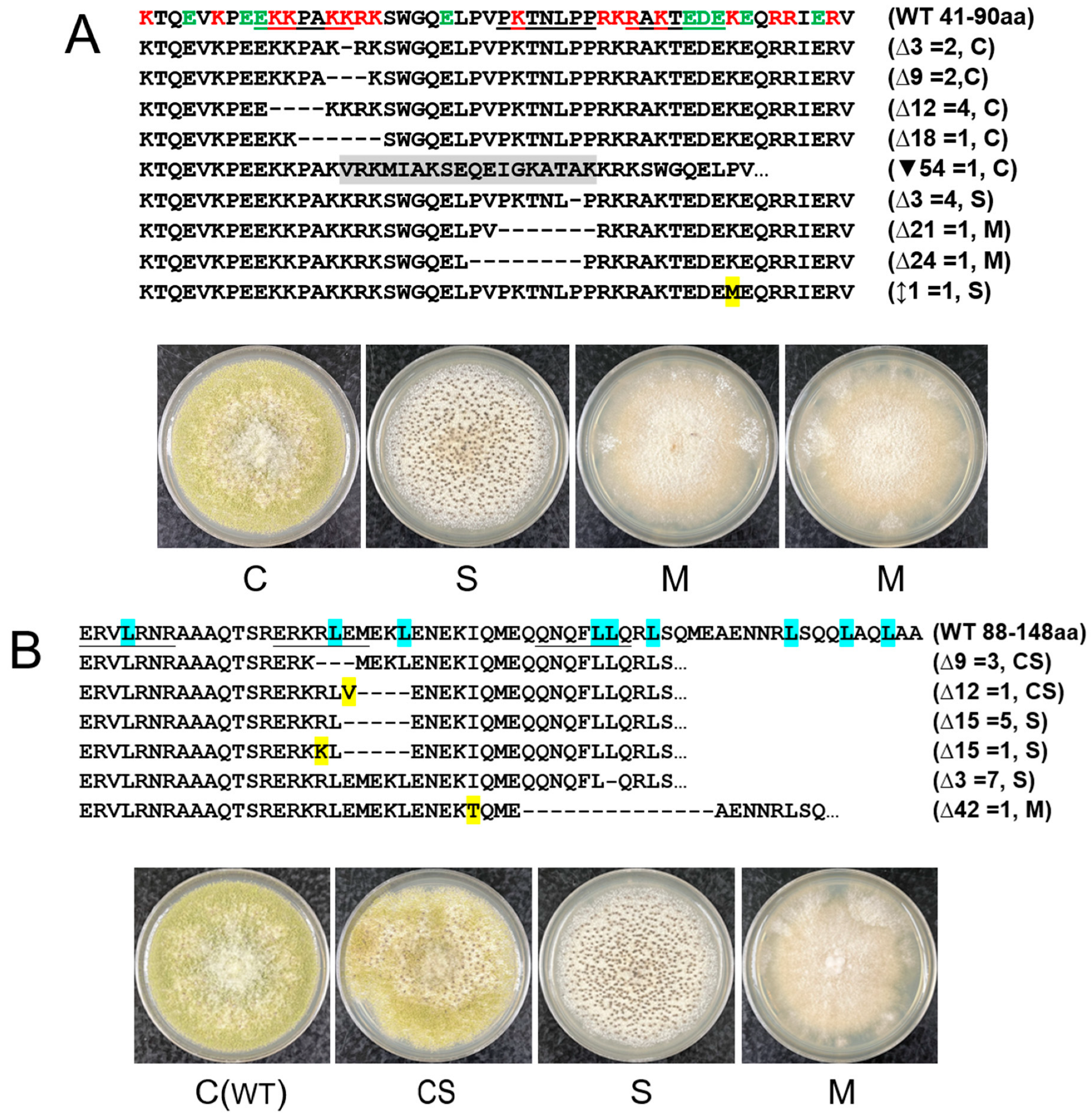
3.2. CRISPR/Cas9 Mutagenesis of the hacA Functional Domain-Coding Regions and the Non-Conventional Intron Generates Mutants Exhibiting Various Phenotypes
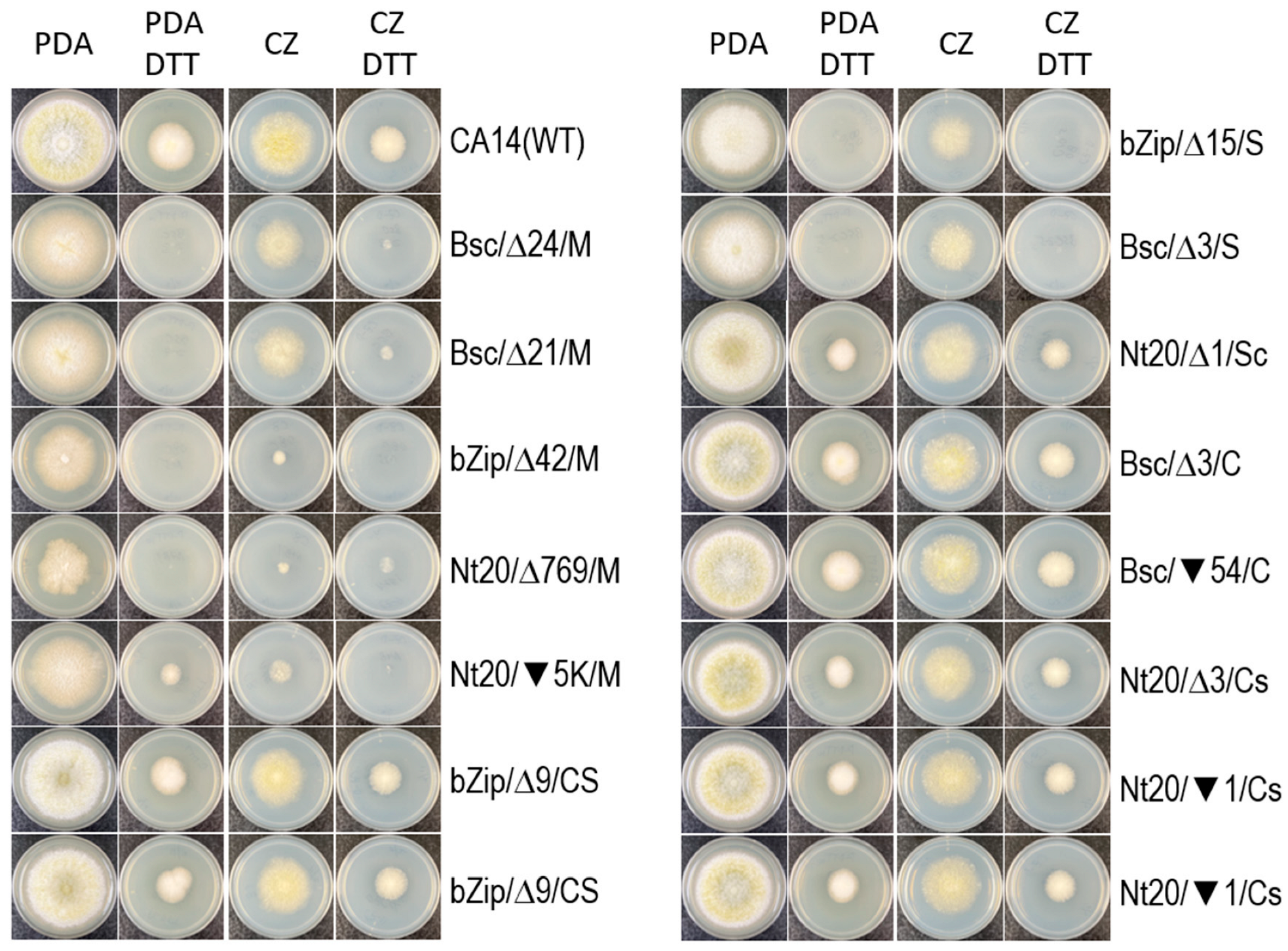
3.3. The hacA Mutants Display Different Sensitivities to DTT
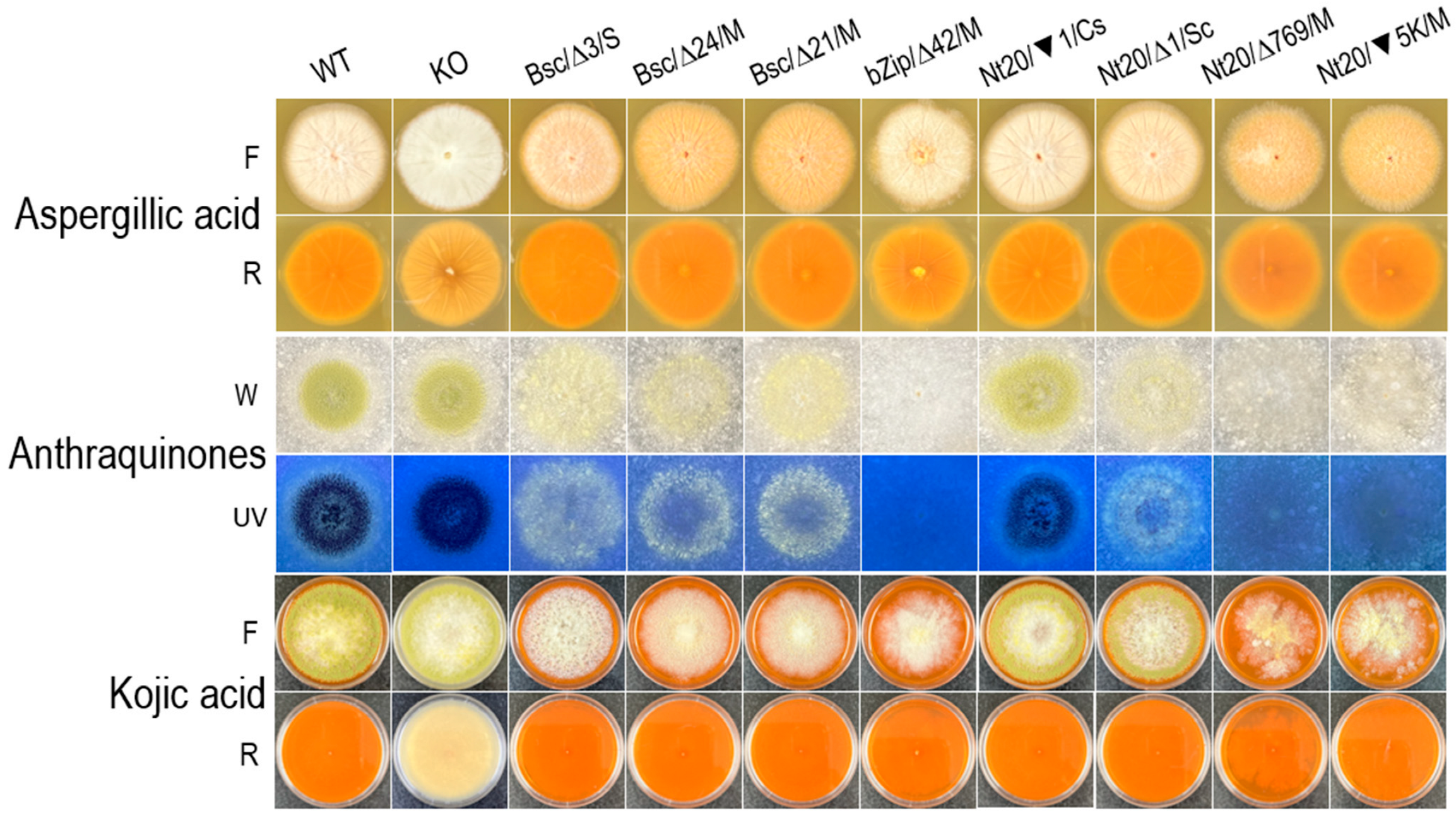
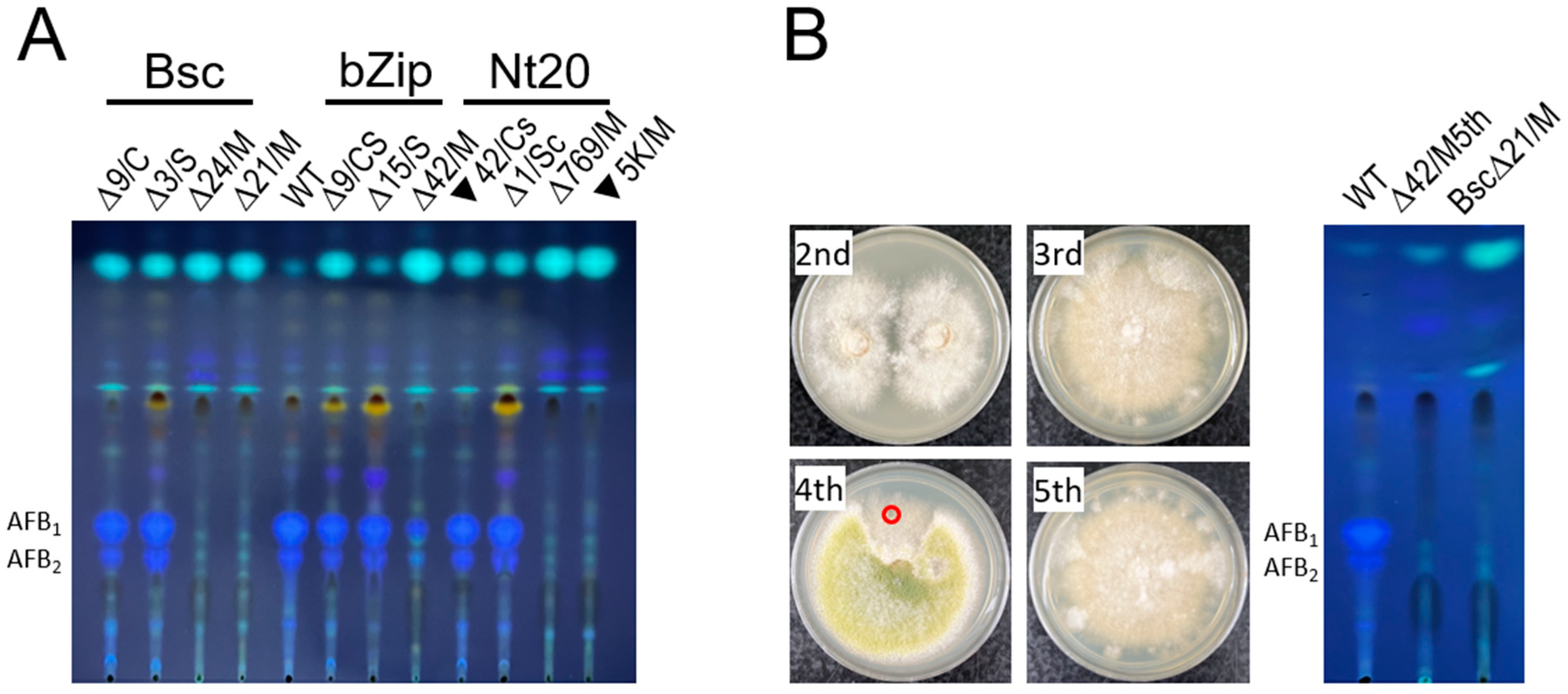
3.4. The hacA Mutants All Produced Aspergillic Acid and Kojic Acid but Not Anthraquinones or Aflatoxin
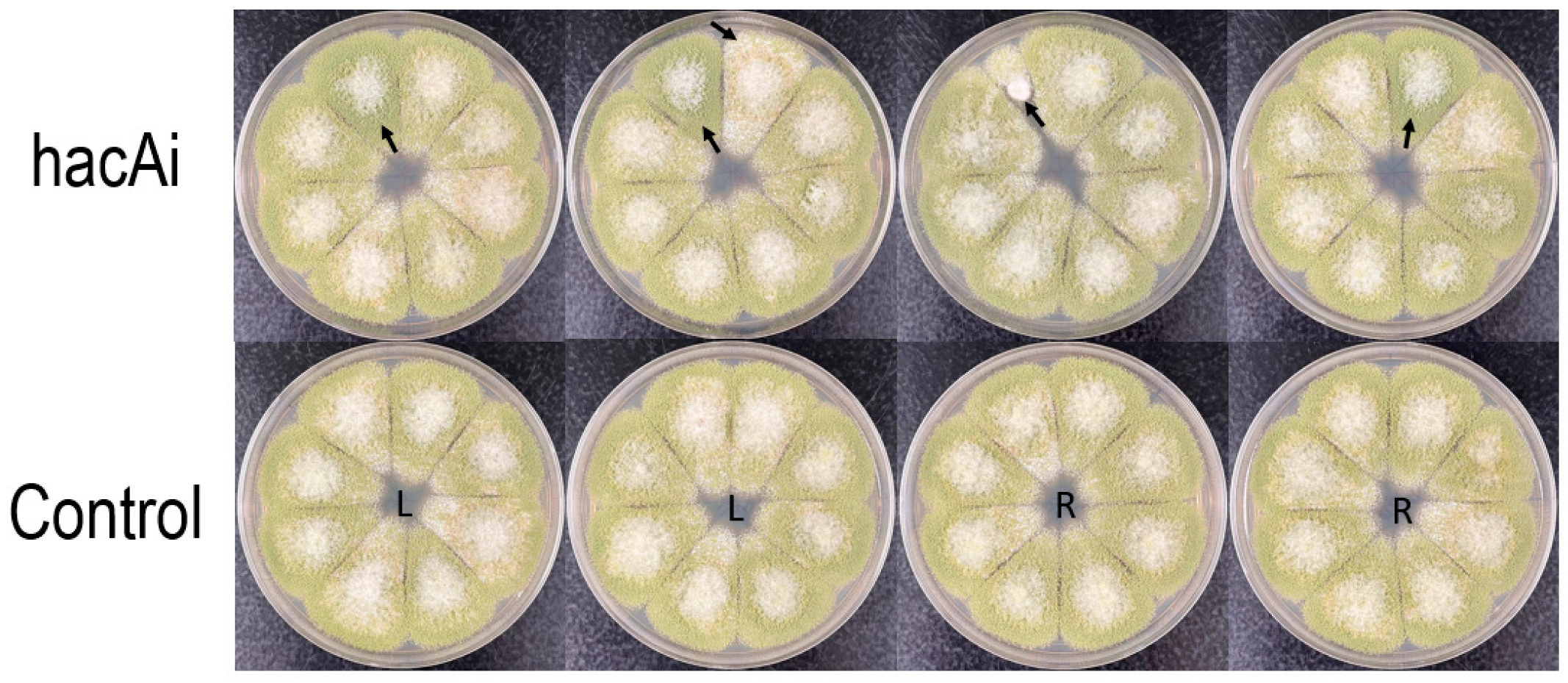
3.5. The hacA RNAi Affects Regeneration and Development of A. flavus Protoplasts
4. Discussion
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Read, A.; Schroder, M. The unfolded protein response: An overview. Biology 2021, 10, 384. [Google Scholar] [CrossRef]
- Hetz, C.; Zhang, K.; Kaufman, R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020, 21, 421–438. [Google Scholar] [CrossRef]
- Bashir, S.; Banday, M.; Qadri, O.; Bashir, A.; Hilal, N.; Nida, I.F.; Rader, S.; Fazili, K.M. The molecular mechanism and functional diversity of UPR signaling sensor IRE1. Life Sci. 2021, 265, 118740. [Google Scholar] [CrossRef]
- Hernandez-Elvira, M.; Torres-Quiroz, F.; Escamilla-Ayala, A.; Dominguez-Martin, E.; Escalante, R.; Kawasaki, L.; Ongay-Larios, L.; Coria, R. The unfolded protein response pathway in the yeast Kluyveromyces lactis. A comparative view among yeast species. Cells 2018, 7, 106. [Google Scholar] [CrossRef]
- Mulder, H.J.; Nikolaev, I.; Madrid, S.M. HACA, the transcriptional activator of the unfolded protein response (UPR) in Aspergillus niger, binds to partly palindromic UPR elements of the consensus sequence 5′-CAN(G/A)NTGT/GCCT-3′. Fungal Genet. Biol. 2006, 43, 560–572. [Google Scholar] [CrossRef]
- Mori, K.; Ogawa, N.; Kawahara, T.; Yanagi, H.; Yura, T. Palindrome with spacer of one nucleotide is characteristic of the cis-acting unfolded protein response element in Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 9912–9920. [Google Scholar] [CrossRef]
- Richie, D.L.; Hartl, L.; Aimanianda, V.; Winters, M.S.; Fuller, K.K.; Miley, M.D.; White, S.; McCarthy, J.W.; Latge, J.P.; Feldmesser, M.; et al. A role for the unfolded protein response (UPR) in virulence and antifungal susceptibility in Aspergillus fumigatus. PLoS Pathog. 2009, 5, e1000258. [Google Scholar] [CrossRef]
- Joubert, A.; Simoneau, P.; Campion, C.; Bataille-Simoneau, N.; Iacomi-Vasilescu, B.; Poupard, P.; Francois, J.M.; Georgeault, S.; Sellier, E.; Guillemette, T. Impact of the unfolded protein response on the pathogenicity of the necrotrophic fungus Alternaria brassicicola. Mol. Microbiol. 2011, 79, 1305–1324. [Google Scholar] [CrossRef]
- Heimel, K.; Freitag, J.; Hampel, M.; Ast, J.; Bolker, M.; Kamper, J. Crosstalk between the unfolded protein response and pathways that regulate pathogenic development in Ustilago maydis. Plant Cell 2013, 25, 4262–4277. [Google Scholar] [CrossRef]
- Schmitz, L.; Schwier, M.A.; Heimel, K. The unfolded protein response regulates pathogenic development of Ustilago maydis by rok1-dependent inhibition of mating-type signaling. mBio 2019, 10, e02756-19. [Google Scholar] [CrossRef]
- Saloheimo, M.; Valkonen, M.; Penttila, M. Activation mechanisms of the HAC1-mediated unfolded protein response in filamentous fungi. Mol. Microbiol. 2003, 47, 1149–1161. [Google Scholar] [CrossRef]
- Matabishi-Bibi, L.; Challal, D.; Barucco, M.; Libri, D.; Babour, A. Termination of the unfolded protein response is guided by ER stress-induced HAC1 mRNA nuclear retention. Nat. Commun. 2022, 13, 6331. [Google Scholar] [CrossRef]
- Charpentier, T.; Viault, G.; Le Ray, A.M.; Bataille-Simoneau, N.; Helesbeux, J.J.; Blon, N.; Bastide, F.; Marchi, M.; Aligon, S.; Bruguiere, A.; et al. Natural products targeting the fungal unfolded protein response as an alternative crop protection strategy. J. Agric. Food Chem. 2023, 71, 13706–13716. [Google Scholar] [CrossRef]
- Wiese, W.; Siwecka, N.; Wawrzynkiewicz, A.; Rozpedek-Kaminska, W.; Kucharska, E.; Majsterek, I. IRE1alpha Inhibitors as a promising therapeutic strategy in blood malignancies. Cancers 2022, 14, 2526. [Google Scholar] [CrossRef]
- Nikawa, J.; Akiyoshi, M.; Hirata, S.; Fukuda, T. Saccharomyces cerevisiae IRE2/HAC1 is involved in IRE1-mediated KAR2 expression. Nucleic Acids Res. 1996, 24, 4222–4226. [Google Scholar] [CrossRef]
- Valkonen, M.; Penttila, M.; Saloheimo, M. Effects of inactivation and constitutive expression of the unfolded protein response pathway on protein production in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2003, 69, 2065–22072. [Google Scholar] [CrossRef]
- Oh, M.H.; Cheon, S.A.; Kang, H.A.; Kim, J.Y. Functional characterization of the unconventional splicing of Yarrowia lipolytica HAC1 mRNA induced by unfolded protein response. Yeast 2010, 27, 443–452. [Google Scholar] [CrossRef]
- Iracane, E.; Donovan, P.D.; Ola, M.; Butler, G.; Holland, L.M. Identification of an exceptionally long intron in the HAC1 gene of Candida parapsilosis. mSphere 2018, 3, e00532-18. [Google Scholar] [CrossRef]
- Bitencourt, T.A.; Lang, E.A.S.; Sanches, P.R.; Peres, N.T.A.; Oliveira, V.M.; Fachin, A.L.; Rossi, A.; Martinez-Rossi, N.M. HacA governs virulence traits and adaptive stress responses in Trichophyton rubrum. Front. Microbiol. 2020, 11, 193. [Google Scholar] [CrossRef]
- Montenegro-Montero, A.; Goity, A.; Larrondo, L.F. The bZIP transcription factor HAC-1 is involved in the unfolded protein response and is necessary for growth on cellulose in Neurospora crassa. PLoS ONE 2015, 10, e0131415. [Google Scholar] [CrossRef]
- Carvalho, N.D.; Arentshorst, M.; Jin Kwon, M.; Meyer, V.; Ram, A.F. Expanding the ku70 toolbox for filamentous fungi: Establishment of complementation vectors and recipient strains for advanced gene analyses. Appl. Microbiol. Biotechnol. 2010, 87, 1463–1473. [Google Scholar] [CrossRef]
- Zhou, B.; Xie, J.; Liu, X.; Wang, B.; Pan, L. Functional and transcriptomic analysis of the key unfolded protein response transcription factor HacA in Aspergillus oryzae. Gene 2016, 593, 143–153. [Google Scholar] [CrossRef]
- Yu, M.; Zhou, X.; Chen, D.; Jiao, Y.; Han, G.; Tao, F. HacA, a key transcription factor for the unfolded protein response, is required for fungal development, aflatoxin biosynthesis and pathogenicity of Aspergillus flavus. Int. J. Food Microbiol. 2024, 417, 110693. [Google Scholar] [CrossRef]
- Zhao, Q.; Pei, H.; Zhou, X.; Zhao, K.; Yu, M.; Han, G.; Fan, J.; Tao, F. Systematic characterization of bZIP transcription factors required for development and aflatoxin generation by high-throughput gene knockout in Aspergillus flavus. J. Fungi 2022, 8, 356. [Google Scholar] [CrossRef]
- Fan, F.; Ma, G.; Li, J.; Liu, Q.; Benz, J.P.; Tian, C.; Ma, Y. Genome-wide analysis of the endoplasmic reticulum stress response during lignocellulase production in Neurospora crassa. Biotechnol. Biofuels 2015, 8, 66. [Google Scholar] [CrossRef]
- IARC. Aflatoxins. In Monograph on the Evaluation of Carcinogenic Risks to Humans; Volume 82. Some Traditional Herbal Medicines, Some Mycotoxins; International Agency for Research on Cancer (IARC): Lyon, France, 2002. [Google Scholar]
- Dovenyi-Nagy, T.; Racz, C.; Molnar, K.; Bako, K.; Szlama, Z.; Jozwiak, A.; Farkas, Z.; Pocsi, I.; Dobos, A.C. Pre-harvest modelling and mitigation of aflatoxins in maize in a changing climatic environment-a review. Toxins 2020, 12, 768. [Google Scholar] [CrossRef]
- Koeppe, S.; Kawchuk, L.; Kalischuk, M. RNA interference past and future applications in plants. Int. J. Mol. Sci. 2023, 24, 9755. [Google Scholar] [CrossRef]
- Lam, J.K.; Chow, M.Y.; Zhang, Y.; Leung, S.W. siRNA versus miRNA as therapeutics for gene silencing. Mol. Ther. Nucleic Acids 2015, 4, e252. [Google Scholar] [CrossRef]
- Zeng, Y.; Yi, R.; Cullen, B.R. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. USA 2003, 100, 9779–9784. [Google Scholar] [CrossRef]
- Raruang, Y.; Omolehin, O.; Hu, D.; Wei, Q.; Han, Z.Q.; Rajasekaran, K.; Cary, J.W.; Wang, K.; Chen, Z.Y. Host induced gene silencing targeting Aspergillus flavus aflM reduced aflatoxin contamination in transgenic maize under field conditions. Front. Microbiol. 2020, 11, 754. [Google Scholar] [CrossRef]
- Thakare, D.; Zhang, J.; Wing, R.A.; Cotty, P.J.; Schmidt, M.A. Aflatoxin-free transgenic maize using host-induced gene silencing. Sci. Adv. 2017, 3, e1602382. [Google Scholar] [CrossRef]
- Masanga, J.O.; Matheka, J.M.; Omer, R.A.; Ommeh, S.C.; Monda, E.O.; Alakonya, A.E. Downregulation of transcription factor aflR in Aspergillus flavus confers reduction to aflatoxin accumulation in transgenic maize with alteration of host plant architecture. Plant Cell Rep. 2015, 34, 1379–1387. [Google Scholar] [CrossRef]
- Power, I.L.; Dang, P.M.; Sobolev, V.S.; Orner, V.A.; Powell, J.L.; Lamb, M.C.; Arias, R.S. Characterization of small RNA populations in non-transgenic and aflatoxin-reducing-transformed peanut. Plant Sci. 2017, 257, 106–125. [Google Scholar] [CrossRef]
- Prasad, K.; Yogendra, K.; Sanivarapu, H.; Rajasekaran, K.; Cary, J.W.; Sharma, K.K.; Bhatnagar-Mathur, P. Multiplexed host-induced gene silencing of Aspergillus flavus genes confers aflatoxin resistance in groundnut. Toxins 2023, 15, 319. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Pannunzio, N.R.; Watanabe, G.; Lieber, M.R. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10512–10523. [Google Scholar] [CrossRef]
- Jin, F.J.; Wang, B.T.; Wang, Z.D.; Jin, L.; Han, P. CRISPR/Cas9-based genome editing and its application in Aspergillus species. J. Fungi 2022, 8, 467. [Google Scholar] [CrossRef]
- Chang, P.-K. Creating large chromosomal segment deletions in Aspergillus flavus by a dual CRISPR/Cas9 system: Deletion of gene clusters for production of aflatoxin, cyclopiazonic acid, and ustiloxin B. Fungal Genet. Biol. 2024, 170, 103863. [Google Scholar] [CrossRef]
- Igarashi, T.; Katayama, T.; Maruyama, J.I. CRISPR/Cas9 genome editing for comparative genetic analysis related to soy sauce brewing in Aspergillus sojae industrial strains. Biosci. Biotechnol. Biochem. 2023, 87, 1236–1248. [Google Scholar] [CrossRef]
- Bothast, R.J.; Fennell, D.J. A medium for rapid identification and enumeration of Aspergillus flavus and related organisms. Mycologia 1974, 66, 365–369. [Google Scholar] [CrossRef]
- Macdonald, J.C. Biosynthesis of aspergillic acid. J. Biol. Chem. 1961, 236, 512–514. [Google Scholar] [CrossRef]
- Lin, M.; Dianese, J. A coconut agar medium for rapid detection of aflatoxin production by Aspergillus spp. Phytopathology 1976, 66, 1466–1469. [Google Scholar] [CrossRef]
- Terabayashi, Y.; Sano, M.; Yamane, N.; Marui, J.; Tamano, K.; Sagara, J.; Dohmoto, M.; Oda, K.; Ohshima, E.; Tachibana, K.; et al. Identification and characterization of genes responsible for biosynthesis of kojic acid, an industrially important compound from Aspergillus oryzae. Fungal Genet. Biol. 2010, 47, 953–961. [Google Scholar] [CrossRef]
- Chang, P.-K. A simple CRISPR/Cas9 system for efficiently targeting genes of Aspergillus section Flavi species, Aspergillus nidulans, Aspergillus fumigatus, Aspergillus terreus, and Aspergillus niger. Microbiol. Spectr. 2023, 11, e0464822. [Google Scholar] [CrossRef]
- Chang, P.-K.; Zhang, Q.; Scharfenstein, L.; Mack, B.; Yoshimi, A.; Miyazawa, K.; Abe, K. Aspergillus flavus GPI-anchored protein-encoding ecm33 has a role in growth, development, aflatoxin biosynthesis, and maize infection. Appl. Microbiol. Biotechnol. 2018, 102, 5209–5220. [Google Scholar] [CrossRef]
- Horng, J.S.; Chang, P.-K.; Pestka, J.J.; Linz, J.E. Development of a homologous transformation system for Aspergillus parasiticus with the gene encoding nitrate reductase. Mol. Gen. Genet. 1990, 224, 294–296. [Google Scholar] [CrossRef]
- Chang, P.-K.; Bennett, J.W.; Cotty, P.J. Association of aflatoxin biosynthesis and sclerotial development in Aspergillus parasiticus. Mycopathologia 2002, 153, 41–48. [Google Scholar] [CrossRef]
- Mulder, H.J.; Saloheimo, M.; Penttila, M.; Madrid, S.M. The transcription factor HACA mediates the unfolded protein response in Aspergillus niger, and up-regulates its own transcription. Mol. Genet. Genom. 2004, 271, 130–140. [Google Scholar] [CrossRef]
- Starke, J.; Harting, R.; Maurus, I.; Leonard, M.; Bremenkamp, R.; Heimel, K.; Kronstad, J.W.; Braus, G.H. Unfolded protein response and scaffold independent pheromone map kinase signaling control Verticillium dahliae growth, development, and plant pathogenesis. J. Fungi 2021, 7, 305. [Google Scholar] [CrossRef]
- Rocha, M.C.; Godoy, K.F.; de Castro, P.A.; Hori, J.I.; Bom, V.L.; Brown, N.A.; Cunha, A.F.; Goldman, G.H.; Malavazi, I. The Aspergillus fumigatus pkcA G579R mutant is defective in the activation of the cell wall integrity pathway but is dispensable for virulence in a neutropenic mouse infection model. PLoS ONE 2015, 10, e0135195. [Google Scholar] [CrossRef]
- Ruegsegger, U.; Leber, J.H.; Walter, P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 2001, 107, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Mulder, H.J.; Nikolaev, I. HacA-dependent transcriptional switch releases hacA mRNA from a translational block upon endoplasmic reticulum stress. Eukaryot. Cell 2009, 8, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.-K.; Scharfenstein, L.L.; Li, P.; Ehrlich, K.C. Aspergillus flavus VelB acts distinctly from VeA in conidiation and may coordinate with FluG to modulate sclerotial production. Fungal Genet. Biol. 2013, 58–59, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Calvo, A.M.; Wilson, R.A.; Bok, J.W.; Keller, N.P. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 2002, 66, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Bayram, O.; Braus, G.H. Coordination of secondary metabolism and development in fungi: The velvet family of regulatory proteins. FEMS Microbiol. Rev. 2012, 36, 1–24. [Google Scholar] [CrossRef]
- Carvalho, N.D.; Jorgensen, T.R.; Arentshorst, M.; Nitsche, B.M.; van den Hondel, C.A.; Archer, D.B.; Ram, A.F. Genome-wide expression analysis upon constitutive activation of the HacA bZIP transcription factor in Aspergillus niger reveals a coordinated cellular response to counteract ER stress. BMC Genom. 2012, 13, 350. [Google Scholar] [CrossRef]
- Kamath, M.M.; Lightfoot, J.D.; Adams, E.M.; Kiser, R.M.; Wells, B.L.; Fuller, K.K. The Aspergillus fumigatus UPR is variably activated across nutrient and host environments and is critical for the establishment of corneal infection. PLoS Pathog. 2023, 19, e1011435. [Google Scholar] [CrossRef]
- Gressel, J.; Polturak, G. Suppressing aflatoxin biosynthesis is not a breakthrough if not useful. Pest. Manag. Sci. 2018, 74, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.K.; Majumdar, R.; Rajasekaran, K.; Chen, Z.Y.; Wei, Q.; Sickler, C.M.; Lebar, M.D.; Cary, J.W.; Frame, B.R.; Wang, K. RNA interference-based silencing of the alpha-amylase (amy1) gene in Aspergillus flavus decreases fungal growth and aflatoxin production in maize kernels. Planta 2018, 247, 1465–1473. [Google Scholar] [CrossRef]
- Omolehin, O.; Raruang, Y.; Hu, D.; Han, Z.Q.; Wei, Q.; Wang, K.; Rajasekaran, K.; Cary, J.W.; Chen, Z.Y. Resistance to aflatoxin accumulation in maize mediated by host-induced silencing of the Aspergillus flavus alkaline protease (alk) gene. J. Fungi 2021, 7, 904. [Google Scholar] [CrossRef]
- Raruang, Y.; Omolehin, O.; Hu, D.; Wei, Q.; Promyou, S.; Parekattil, L.J.; Rajasekaran, K.; Cary, J.W.; Wang, K.; Chen, Z.Y. Targeting the Aspergillus flavus p2c gene through host-induced gene silencing reduces A. flavus infection and aflatoxin contamination in transgenic maize. Front. Plant Sci. 2023, 14, 1150086. [Google Scholar] [CrossRef] [PubMed]
- Rajasekaran, K.; Sayler, R.J.; Sickler, C.M.; Majumdar, R.; Jaynes, J.M.; Cary, J.W. Control of Aspergillus flavus growth and aflatoxin production in transgenic maize kernels expressing a tachyplesin-derived synthetic peptide, AGM182. Plant Sci. 2018, 270, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Mocellin, S.; Provenzano, M. RNA interference: Learning gene knock-down from cell physiology. J. Transl. Med. 2004, 2, 39. [Google Scholar] [CrossRef] [PubMed]
- Han, H. RNA interference to knock down gene expression. Methods Mol. Biol. 2018, 1706, 293–302. [Google Scholar] [CrossRef]
- Dorsett, Y.; Tuschl, T. siRNAs: Applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004, 3, 318–329. [Google Scholar] [CrossRef]

| Primer | Sequence (5′→3′) | |
|---|---|---|
| U6-F-P a | ATACTGCAGTTCTCTTTAGAATTCAACTGTGGGT | |
| U6-R-K a | TATGGTACCACATATTTAAAAAAAGTCTCCTGCC | |
| gdFd_gene | Fd_seqGTTTTAGAGCTAGAAATAGCAAGTTAA | |
| gdRC_gene | RC_seqACTTGTTCTTCTTTACAATGATTTATATACC | |
| Target | Protospacer sequence b | |
| Fd_seq | RC_seq | |
| Bsc1 | AGAAGAAGCCTGCAAAGAAG | CTTCTTTGCAGGCTTCTTCT |
| Bsc2 | CCAAGACCAACTTGCCTCCG | CGGAGGCAAGTTGGTCTTGG |
| Bsc3 | TGCCAAAACAGAAGATGAGA | TCTCATCTTCTGTTTTGGCA |
| bZip1 | ACGCGTCCTTCGAAATCGTG | CACGATTTCGAAGGACGCGT |
| bZip2 | GGAGCGCAAAAGGCTGGAAA | TTTCCAGCCTTTTGCGCTCC |
| bZip3 | AGAATCAATTCCTCCTTCAG | CTGAAGGAGGAATTGATTCT |
| Nt20 | AGGTCACACAACACCGCTGC | GCAGCGGTGTTGTGTGACCT |
| CRISPR | Domain | AA Position a | Transformant # | Sequenced/Indel Type c |
|---|---|---|---|---|
| Controls | 816 b, 850, 900 | - | ||
| HacA | Bsc1 | 49–55 | 48 | 10/10 in-frame |
| Bsc2 | 66–72 | 6 | 6/6 in-frame | |
| Bsc3 | 75–81 | 1 | 1/single base substitution | |
| bZip1 | 88–94 | 0 | - | |
| bZip2 | 102–108 | 55 | 11/10 in-frame, 1 wild-type | |
| bZip3 | 121–127 | 132 | 8/8 in-frame | |
| Nt20 | 217–223 | 59 | 17/16 in-frame and out-of-frame, 1 wild-type |
| Linearized DNA a | Protoplast Amount | Total Transformant Number b |
|---|---|---|
| P70-U6 | 2.0 × 106 | >1000 |
| R_bZip | >1000 | |
| RNAi 1 | 712 | |
| RNAi 2 | 575 | |
| R_bZip | 1.0 × 106 | 611 |
| RNAi 1 | 351 | |
| RNAi 2 | 190 |
| Linearized DNA a | Protoplast Amount | Transformant Number | Survival Rate (%) |
|---|---|---|---|
| L_bZip | ~3.0 × 104 | 590/740 b | 75 |
| R_bZip | 519/700 | ||
| hacAi 1 | 334/463 | ||
| hacAi 2 | 449/612 | ||
| R_bZip | ~1.0 × 104 c | 104/134 | 71 |
| hacAi 1 | 59/75 | ||
| hacAi 2 | 85/116 | ||
| L_bZip | ~1.0 × 104 d | 61/81 | 93 |
| hacAi 1 | 61/85 | ||
| hacAi 2 | 51/66 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, P.-K. The Aspergillus flavus hacA Gene in the Unfolded Protein Response Pathway Is a Candidate Target for Host-Induced Gene Silencing. J. Fungi 2024, 10, 719. https://doi.org/10.3390/jof10100719
Chang P-K. The Aspergillus flavus hacA Gene in the Unfolded Protein Response Pathway Is a Candidate Target for Host-Induced Gene Silencing. Journal of Fungi. 2024; 10(10):719. https://doi.org/10.3390/jof10100719
Chicago/Turabian StyleChang, Perng-Kuang. 2024. "The Aspergillus flavus hacA Gene in the Unfolded Protein Response Pathway Is a Candidate Target for Host-Induced Gene Silencing" Journal of Fungi 10, no. 10: 719. https://doi.org/10.3390/jof10100719
APA StyleChang, P.-K. (2024). The Aspergillus flavus hacA Gene in the Unfolded Protein Response Pathway Is a Candidate Target for Host-Induced Gene Silencing. Journal of Fungi, 10(10), 719. https://doi.org/10.3390/jof10100719





