New Polyketide Congeners with Antibacterial Activities from an Endophytic Fungus Stemphylium globuliferum 17035 (China General Microbiological Culture Collection Center No. 40666)
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Experimental Procedures
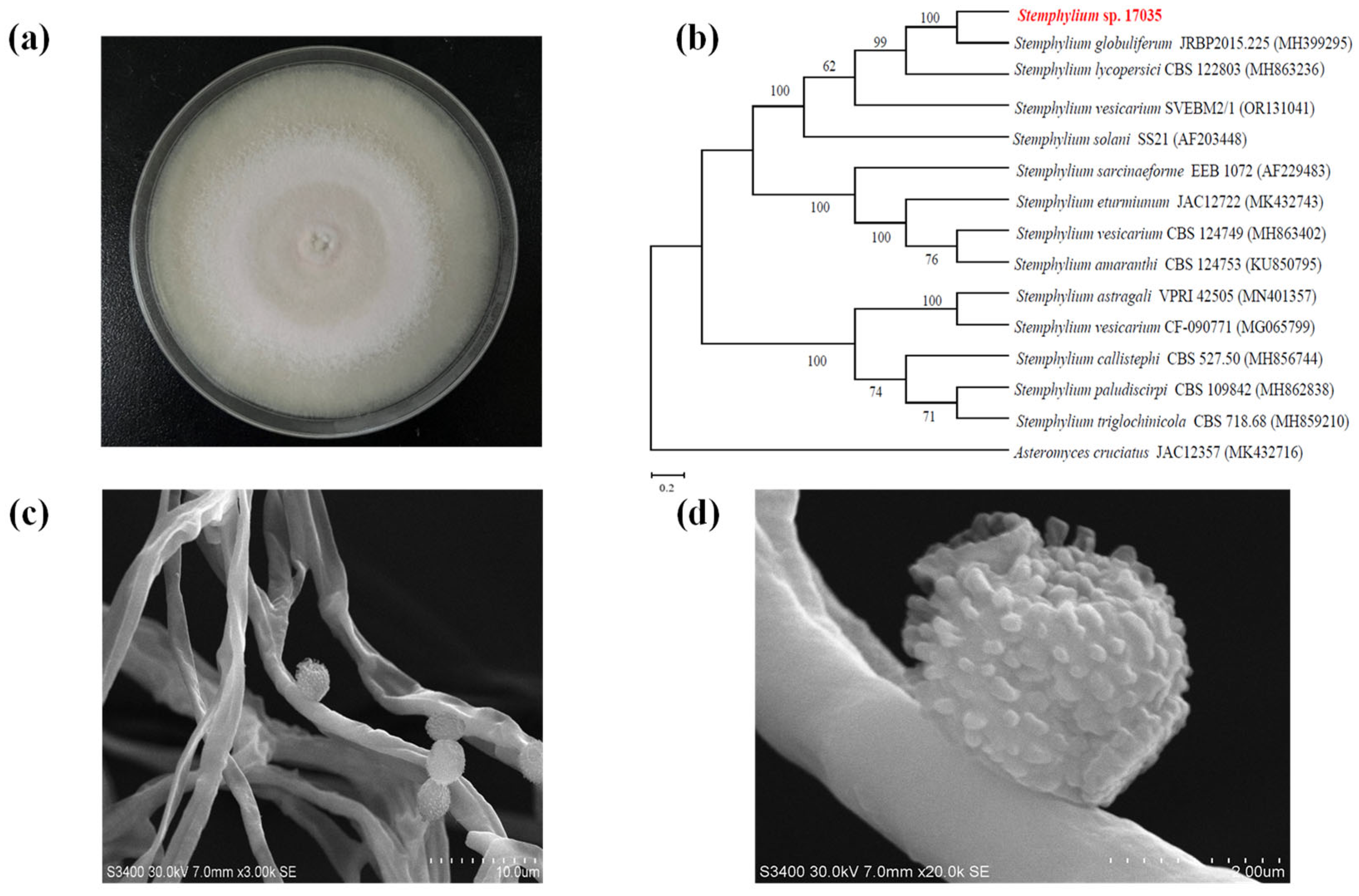
2.2. Characterization and Identification of Fungal Strain S. globuliferum 17035 (SG17035)
2.3. Fungal Material
2.4. Fermentation, Extraction, and Isolation
2.4.1. Fermentation
2.4.2. Compounds Extraction and Isolation
2.5. Spectral Data
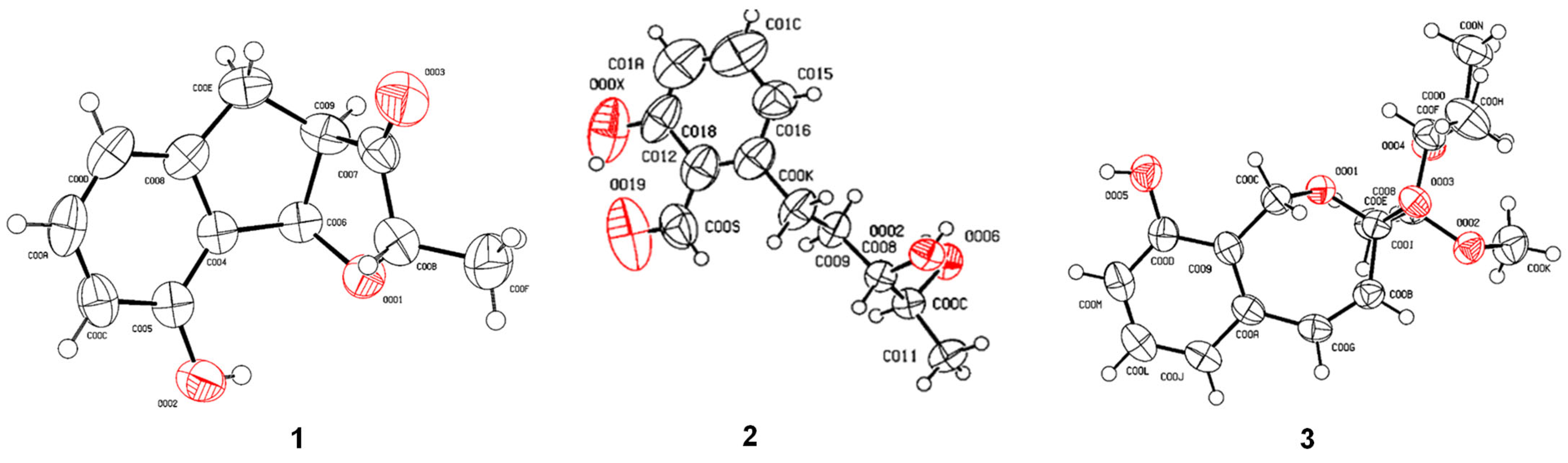
2.6. X-Ray Crystallographic Analysis
2.7. Calculation Details of 13C NMR and DP4 Analysis
2.8. Bioactivity Tests
2.8.1. General Antimicrobial Assays
2.8.2. Cytotoxic Assays
3. Results
3.1. Fungal Characterization and Identification
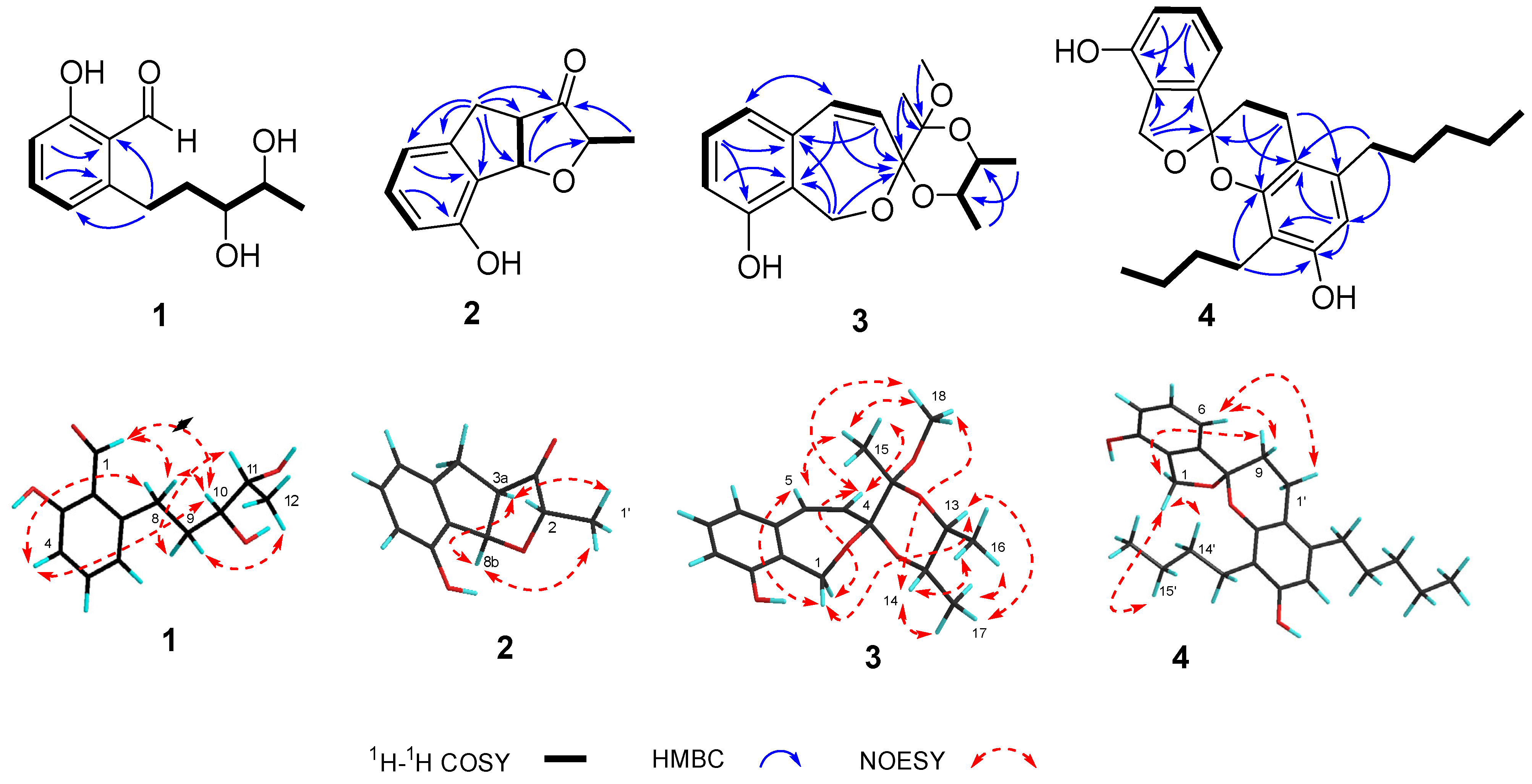
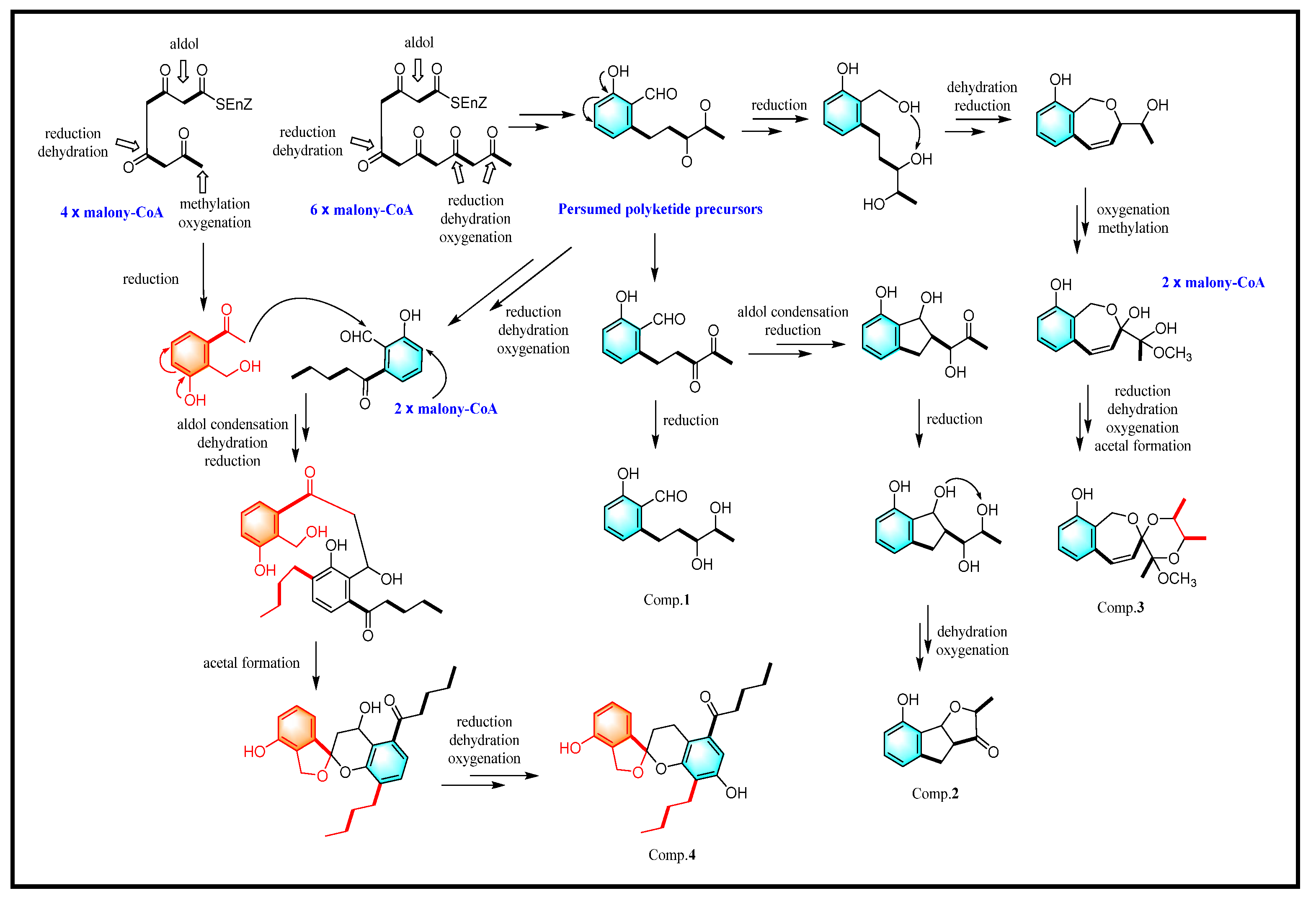
3.2. Structure Elucidation of Compounds 1–9
3.3. Results of Bioactivity Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nisa, H.; Kamili, A.N.; Nawchoo, I.A.; Shafi, S.; Shameem, N.; Bandh, S.A. Fungal endophytes as prolific source of phytochemicals and other bioactive natural products: A review. Microb. Pathog. 2015, 82, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Bull World Health Organ 2001, 79, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Mulder, K.C.; Mulinari, F.; Franco, O.L.; Soares, M.S.; Magalhães, B.S.; Parachin, N.S. Lovastatin production: From molecular basis to industrial process optimization. Biotechnol. Adv. 2015, 33, 648–665. [Google Scholar] [CrossRef] [PubMed]
- Stierle, A.; Strobel, G.; Stierle, D. Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science 1993, 260, 214–216. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.C.; Jin, R.; Wen, T.C.; He, J.; Lei, B.X. Recent research advances on endophytic fungi producing taxol. Mycosystema 2011, 30, 168–179. [Google Scholar]
- Pimentel, M.R.; Molina, G.; Dionísio, A.P.; Maróstica Junior, M.R.; Pastore, G.M. The use of endophytes to obtain bioactive compounds and their application in biotransformation process. Biotechnol. Res. Int. 2011, 2011, 576286. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Singh, R.; Dubey, A. Endophytic actinomycetes as emerging source for therapeutic compounds. Indo. Global J. Pharm. Sci. 2015, 5, 106–116. [Google Scholar] [CrossRef]
- Zheng, L.; Lv, R.; Huang, J.; Jiang, D.; Hsiang, T. Isolation, Purification, and Biological Activity of a Phytotoxin Produced by Stemphylium solani. Plant. Dis. 2010, 94, 1231–1237. [Google Scholar] [CrossRef]
- Debbab, A.; Aly, A.H.; Edrada-Ebel, R.; Wray, V.; Pretsch, A.; Pescitelli, G.; Kurtan, T.; Proksch, P. New Anthracene Derivatives—Structure Elucidation and Antimicrobial Activity. Eur. J. Org. Chem. 2012, 2012, 1351–1359. [Google Scholar] [CrossRef]
- Aly, A.H.; Debbab, A.; Edrada-Ebel, R.A.; Müller, W.E.G.; Proksch, P. Protein kinase inhibitors and other cytotoxic metabolites from the fungal endophyte Stemphylium botryosum isolated from Chenopodium album. Mycosphere 2010, 1, 153–162. [Google Scholar]
- Hwang, J.Y.; Park, S.C.; Byun, W.S.; Oh, D.C.; Lee, S.K.; Oh, K.B.; Shin, J. Bioactive Bianthraquinones and Meroterpenoids from a Marine-Derived Stemphylium sp. Fungus. Mar. Drugs 2020, 18, 436. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.M.; Zheng, C.J.; Song, X.P.; Han, C.R.; Chen, W.H.; Chen, G.Y. Antibacterial α-pyrone derivatives from a mangrove-derived fungus Stemphylium sp. 33231 from the South China Sea. J. Antibiot. 2014, 67, 401–403. [Google Scholar] [CrossRef] [PubMed]
- Arnone, A.; Nasini, G.; Merlini, L.; Assante, G. Secondary mould metabolites. Part 16. Stemphyltoxins, new reduced perylenequinone metabolites from Stemphylium botryosum var. Lactucum. J. Chem. Soc. 1986, 1, 525–530. [Google Scholar] [CrossRef]
- Liu, F.; Cai, X.L.; Yang, H.; Xia, X.K.; Guo, Z.Y.; Yuan, J.; Li, M.F.; She, Z.G.; Lin, Y.C. The bioactive metabolites of the mangrove endophytic fungus Talaromyces sp. ZH-154 isolated from Kandelia candel (L.) Druce. Planta Med. 2010, 76, 185–189. [Google Scholar] [CrossRef]
- Zhou, X.M.; Zheng, C.J.; Chen, G.Y.; Song, X.P.; Han, C.R.; Tang, X.Z.; Liu, R.J.; Ren, L.L. Two new stemphol sulfates from the mangrove endophytic fungus Stemphylium sp. 33231. J. Antibiot. 2015, 68, 501–503. [Google Scholar] [CrossRef]
- Debbab, A.; Aly, A.H.; Edrada-Ebel, R.; Wray, V.; Müller, W.E.; Totzke, F.; Zirrgiebel, U.; Schächtele, C.; Kubbutat, M.H.; Lin, W.H.; et al. Bioactive metabolites from the endophytic fungus Stemphylium globuliferum isolated from Mentha pulegium. J. Nat. Prod. 2009, 72, 626–631. [Google Scholar] [CrossRef]
- Woudenberg, J.; Hanse, B.; Van Leeuwen, G.; Groenewald, J.Z.; Crous, P.W. Stemphylium revisited. Stud. Mycol. 2017, 87, 77–103. [Google Scholar] [CrossRef]
- Qian, N.; Feng, C.; Cheng, Y.; Zhang, G.; Bhadauria, V.; Lu, X.; Zhao, W. Two Species of Stemphylium, S. astragali and S. henanense sp. nov., Causing Leaf and Stem Spot Disease on Astragalus sinicus in China. Phytopathology 2023, 113, 945–952. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, J.; Gao, H.; Zhang, T. Two new species of Cadophora and Stemphylium isolated from soil. J. Fungal Res. 2018, 16, 201–204. [Google Scholar]
- Anwar, R.; Pratama, G.B.; Supratman, U.; Harneti, D.; Azhari, A.; Fajriah, S.; Azmi, M.N.; Shiono, Y. Steroids from Toona sureni-derived Endophytic Fungi Stemphylium sp. MAFF 241962 and Their Heme Polymerization Inhibition Activity. Nat. Life Sci. Commun. 2023, 22, e2023055. [Google Scholar] [CrossRef]
- Jiang, L.; Zhu, G.; Han, J.; Hou, C.; Zhang, X.; Wang, Z.; Yuan, W.; Lv, K.; Cong, Z.; Wang, X. Genome-guided investigation of anti-inflammatory sesterterpenoids with 5-15 trans-fused ring system from phytopathogenic fungi. Appl. Microbiol. Biotechnol. 2021, 105, 5407–5417. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Zhu, G.; Yuan, W.; Han, Z.; Dai, H.; Basiony, M.; Zhang, L.; Liu, X.; Hsiang, T.; Zhang, J. Two novel aliphatic unsaturated alcohols isolated from a pathogenic fungus Fusarium proliferatum. Synth. Syst. Biotechnol. 2021, 6, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, Z.; Song, Z.; Karthik, L.; Hou, C.; Zhu, G.; Jiang, L.; Han, J.; Ma, R.; Li, L. Brocaeloid D, a novel compound isolated from a wheat pathogenic fungus, Microdochium majus 99049. Synth. Syst. Biotechnol. 2019, 4, 173–179. [Google Scholar] [CrossRef]
- Liu, Y.; Wray, V.; Abdel-Aziz, M.S.; Wang, C.Y.; Lai, D.; Proksch, P. Trimeric anthracenes from the endophytic fungus Stemphylium globuliferum. J. Nat. Prod. 2014, 77, 1734–1738. [Google Scholar] [CrossRef]
- Lei, H.; Lin, X.; Han, L.; Ma, J.; Dong, K.; Wang, X.; Zhong, J.; Mu, Y.; Liu, Y.; Huang, X. Polyketide derivatives from a marine-sponge-associated fungus Pestalotiopsis heterocornis. Phytochemistry 2017, 142, 51–59. [Google Scholar] [CrossRef]
- Quévrain, E.; Domart Coulon, I.; Pernice, M.; Bourguet Kondracki, M.L. Novel natural parabens produced by a Microbulbifer bacterium in its calcareous sponge host Leuconia nivea. Environ. Microbiol. 2010, 11, 1527–1539. [Google Scholar] [CrossRef]
- Hussain, R.; Ali, B.; Imran, M.; Malik, A. Thymofolinoates A and B, new cinnamic acid derivatives from Euphorbia thymifolia. Nat. Prod. Commun. 2012, 7, 1351–1352. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Zhang, D.; Ding, N.; Chen, S.; Song, C.; Luo, Y.; Fu, X.; Bi, X.; Niu, H. New cytotoxic natural products from the marine sponge-derived fungus Pestalotiopsis sp. by epigenetic modification. RSC Adv. 2020, 10, 37982–37988. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Yao, G.S.; Ma, Z.L.; Zheng, Y.Y.; Lv, L.; Mao, J.Q.; Wang, C.Y. Bioactive Alkaloids from the Marine-Derived Fungus Metarhizium sp. P2100. J. Fungi 2022, 8, 1218. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gong, N.; Zhou, L.; Yang, Z.; Zhang, H.; Gu, Y.; Ma, J.; Ju, J. OSMAC-Based Discovery and Biosynthetic Gene Clusters Analysis of Secondary Metabolites from Marine-Derived Streptomyces globisporus SCSIO LCY30. Mar. Drugs 2023, 22, 21. [Google Scholar] [CrossRef] [PubMed]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Frisch, G.W.; Trucks, H.B.; Schlegel, G.E.; Scuseria, M.A.; Robb, J.R.; Cheeseman, G.; Scalmani, V.; Barone, B.; Mennucci, G.A.; Petersson, H.N.; et al. Gaussian 09, Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2009; Available online: https://gaussian.com/g09citation/ (accessed on 1 October 2024).
- Song, F.; Liu, X.; Guo, H.; Ren, B.; Chen, C.; Piggott, A.M.; Yu, K.; Gao, H.; Wang, Q.; Liu, M.; et al. Brevianamides with antitubercular potential from a marine-derived isolate of Aspergillus versicolor. Org. Lett. 2012, 14, 4770–4773. [Google Scholar] [CrossRef]
- Xu, M.Y.; Lee, S.Y.; Kang, S.S.; Kim, Y.S. Antitumor activity of jujuboside B and the underlying mechanism via induction of apoptosis and autophagy. J. Nat. Prod. 2014, 77, 370–376. [Google Scholar] [CrossRef]
- Wu, J.T. Effect of 3,3′-Biisofraxidin on Apoptosis of Human Gastric Cancer BGC-823 Cells. Trop. J. Pharm. Res. 2015, 14, 1803–1811. [Google Scholar] [CrossRef]
- Wang, Y. Taxonomic Studies of two Hyphomycete Genera. In Ulocladium Preuss and Stemphylium Wallr; Agricultural University: Tai’an, China.
- Simmons, E.G. Perfect States of Stemphylium. Mycologia 1969, 61, 1–26. [Google Scholar] [CrossRef]
- Badrinarayanan, S.; Squire, C.J.; Sperry, J.; Brimble, M.A. Bioinspired Total Synthesis and Stereochemical Revision of the Fungal Metabolite Pestalospirane B. Org. Lett. 2017, 19, 3414–3417. [Google Scholar] [CrossRef]
- Meng, C.W.; Zhao, H.Y.; Zhu, H.; Peng, C.; Zhou, Q.M.; Xiong, L. Novel Indane Derivatives with Antioxidant Activity from the Roots of Anisodus tanguticus. Molecules 2023, 28, 1493. [Google Scholar] [CrossRef]
- Kesting, J.R.; Olsen, L.; Staerk, D.; Tejesvi, M.V.; Kini, K.R.; Prakash, H.S.; Jaroszewski, J.W. Production of unusual dispiro metabolites in Pestalotiopsis virgatula endophyte cultures: HPLC-SPE-NMR, electronic circular dichroism, and time-dependent density-functional computation study. J. Nat. Prod. 2011, 74, 2206–2215. [Google Scholar] [CrossRef]
- Namikoshi, M.; Kobayashi, H.; Yoshimoto, T.; Meguro, S.; Akano, K. Isolation and characterization of bioactive metabolites from marine-derived filamentous fungi collected from tropical and sub-tropical coral reefs. Chem. Pharm. Bull. 2000, 48, 1452–1457. [Google Scholar] [CrossRef] [PubMed]
- Namikoshi, M.; Kobayashi, H.; Yoshimoto, T.; Meguro, S. Paecilospirone, a Unique Spiro[chroman-2,1′(3′H)-isobenzofuran] Derivative Isolated from Tropical Marine Fungus Paecilomyces sp. Chem. Lett. 2000, 2000, 308–309. [Google Scholar] [CrossRef]
- Radić, N.; Strukelj, B. Endophytic fungi: The treasure chest of antibacterial substances. Phytomedicine 2012, 19, 1270–1284. [Google Scholar] [CrossRef] [PubMed]
- Marumo, S.; Hattori, H.; Katayama, M.J.A. Stemphol from Pleospora herbarum as a self-inhibitor. Agric. Biol. Chem. 1985, 49, 1521–1522. [Google Scholar] [CrossRef]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D.W. Extending the “One Strain Many Compounds” (OSMAC) Principle to Marine Microorganisms. Mar. Drugs 2018, 16, 244–273. [Google Scholar] [CrossRef]

| Pos. | 1 | Pos. | 2 | ||
|---|---|---|---|---|---|
| δH a Mult (J in Hz) | δC b, Type | δH a Mult (J in Hz) | δC b, Type | ||
| 1 | 10.41, s | 197.3, CH | 2 | 3.62, q (6.8) | 75.6, CH |
| 2 | 119.4, C | 3 | 220.9, C | ||
| 3 | 164.3, C | 3a | 3.20, m | 49.3, CH | |
| 4 | 6.83, d (8.1) | 116.6, CH | 4 | 3.17, m 3.08, m | 35.5, CH2 |
| 5 | 7.43, t (8.1) | 138.3, CH | 4a | 145.3, C | |
| 6 | 6.77, d (8.1) | 112.4, CH | 5 | 6.68, d (7.8) | 116.7, CH |
| 7 | 148.8, C | 6 | 7.12, t (7.8) | 132.1, CH | |
| 8 | 3.20, ddd (13.8, 10.4, 4.9) 2.99, ddd (13.8, 10.1, 6.7) | 29.0, CH2 | 7 | 6.65, d (7.8) | 114.7, CH |
| 9 | 1.80, m 1.72, dtd (14.2, 9.8, 4.8) | 36.9, CH2 | 8 | 156.5, C | |
| 10 | 3.38, ddd (9.6, 4.9, 3.2) | 75.6, CH | 8a | 127.5, C | |
| 11 | 3.62, qd (6.3, 4.8) | 71.3, CH | 8b | 6.07, m | 84.2, CH |
| 12 | 1.13, d (6.4) | 116.8, CH3 | 1′ | 1.28, d (6.8) | 16.3, CH3 |
| Pos. | 3 | Pos. | 4 | ||
|---|---|---|---|---|---|
| δH a Mult (J in Hz) | δC b, Type | δH a Mult (J in Hz) | δC b, Type | ||
| 1a | 4.57, d (13.7) | 56.5, CH2 | 1a | 5.09, d (12.9) | 69.8, CH2 |
| 1b | 5.15, d (13.7) | 1b | 5.22, d (12.9) | ||
| 3 | 100.9, C | 2 | 126.5, C | ||
| 4 | 6.06, d (12.7) | 132.2, CH | 3 | 149.9, C | |
| 5 | 6.61, d (12.7) | 131.5, CH | 4 | 6.78, d (7.8) | 115.6, CH |
| 6 | 6.89, d (7.7) | 123.4, CH | 5 | 7.24, t (7.8) | 129.7, CH |
| 7 | 7.12, t (7.7) | 128.1, CH | 6 | 6.93, d (7.8) | 114.6, CH |
| 8 | 6.70, d (7.7) | 114.8, CH | 7 | 143.1, C | |
| 9 | 152.0, C | 8 | 108.2, C | ||
| 9-OH | 4.98, br | 9a | 2.21, ddd (13.2, 6.1, 2.3) | 30.3, CH2 | |
| 10 | 127.2, C | 9b | 2.34 td (13.2, 6.1) | ||
| 11 | 137.3, C | 1′a | 2.85 ddd (16.0, 13.0, 2.3) | 18.9, CH2 | |
| 12 | 99.6, C | 1′b | 2.99 ddd (16.0, 13.0, 6.0) | ||
| 13 | 3.91, m | 68.1, CH | 2′ | 112.2, C | |
| 14 | 3.68, m | 69.6, CH | 3′ | 151.7, C | |
| 15 | 1.09, s | 17.7, CH3 | 4′ | 114.4, C | |
| 16 | 1.19, d (6.5) | 17.2, CH3 | 5′ | 152.3, C | |
| 17 | 1.15, d (6.5) | 17.1, CH3 | 6′ | 6.32, s | 108.6, CH |
| 18 | 3.26, s | 48.1, CH3 | 7′ | 139.3, C | |
| 8′ | 2.51, m | 32.7, CH2 | |||
| 9′ | 1.58, m | 29.9, CH2 | |||
| 10′ | 1.37, m | 32.1, CH2 | |||
| 11′ | 1.37, m | 22.8, CH2 | |||
| 12′ | 0.92, t (7.3) | 14.2, CH3 | |||
| 13′ | 2.45, t (7.4) | 22.6, CH2 | |||
| 14′ | 1.37, m | 31.6, CH2 | |||
| 15′ | 1.22, m | 22.5, CH2 | |||
| 16′ | 0.76, t (7.3) | 14.0, CH3 | |||
| Compound | Pathogenic Bacteria (MIC, μM) | ||||
|---|---|---|---|---|---|
| C. albicans | S. mutans | P. gingivalis | S. aureus | MRSA | |
| 1 | >100 | >100 | >100 | >100 | >100 |
| 2 | >100 | >100 | 50 | >100 | >100 |
| 3 | >100 | >100 | 50 | >100 | >100 |
| 4 | >100 | 50 | 6.25 | 3.125 | 12.5 |
| 5 | >100 | >100 | >100 | >100 | - |
| 6 | 100 | 100 | 25 | 16 | - |
| 7 | >100 | >100 | >100 | >100 | - |
| 8 | >100 | >100 | >100 | >100 | - |
| 9 | >100 | >100 | >100 | >100 | - |
| Amphotericin B a | 1 | - | - | - | - |
| Chlorhexidine b | - | 2 | 1 | 1 | - |
| Vancomycin c | - | - | - | - | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Zhu, G.; Wang, J.; Yu, J.; Ye, K.; Xing, C.; Ren, B.; Zhu, B.; Chen, S.; Lai, L.; et al. New Polyketide Congeners with Antibacterial Activities from an Endophytic Fungus Stemphylium globuliferum 17035 (China General Microbiological Culture Collection Center No. 40666). J. Fungi 2024, 10, 737. https://doi.org/10.3390/jof10110737
Li Y, Zhu G, Wang J, Yu J, Ye K, Xing C, Ren B, Zhu B, Chen S, Lai L, et al. New Polyketide Congeners with Antibacterial Activities from an Endophytic Fungus Stemphylium globuliferum 17035 (China General Microbiological Culture Collection Center No. 40666). Journal of Fungi. 2024; 10(11):737. https://doi.org/10.3390/jof10110737
Chicago/Turabian StyleLi, Yingying, Guoliang Zhu, Jing Wang, Junjie Yu, Ke Ye, Cuiping Xing, Biao Ren, Bin Zhu, Simin Chen, Lijun Lai, and et al. 2024. "New Polyketide Congeners with Antibacterial Activities from an Endophytic Fungus Stemphylium globuliferum 17035 (China General Microbiological Culture Collection Center No. 40666)" Journal of Fungi 10, no. 11: 737. https://doi.org/10.3390/jof10110737
APA StyleLi, Y., Zhu, G., Wang, J., Yu, J., Ye, K., Xing, C., Ren, B., Zhu, B., Chen, S., Lai, L., Li, Y., Hsiang, T., Zhang, L., Liu, X., & Zhang, J. (2024). New Polyketide Congeners with Antibacterial Activities from an Endophytic Fungus Stemphylium globuliferum 17035 (China General Microbiological Culture Collection Center No. 40666). Journal of Fungi, 10(11), 737. https://doi.org/10.3390/jof10110737







