AoPrdx2 Regulates Oxidative Stress, Reactive Oxygen Species, Trap Formation, and Secondary Metabolism in Arthrobotrys oligospora
Abstract
:1. Introduction
2. Materials and Methods
2.1. Organisms and Media
2.2. Sequence Analysis of AoPrdx2
2.3. Deletion of Aoprdx2
2.4. Comparison of Mycelial Growth and Sporulation
2.5. Analysis of Stress Response
2.6. Analysis of Trap Formation and Nematode Predation Efficiency
2.7. Analysis of ROS Level and Endocytosis
2.8. Analysis of the Metabolites
2.9. Statistical Analysis
3. Results
3.1. AoPrdx2 Sequence and Phylogenetic Analysis
3.2. Verification of Knockout Strains
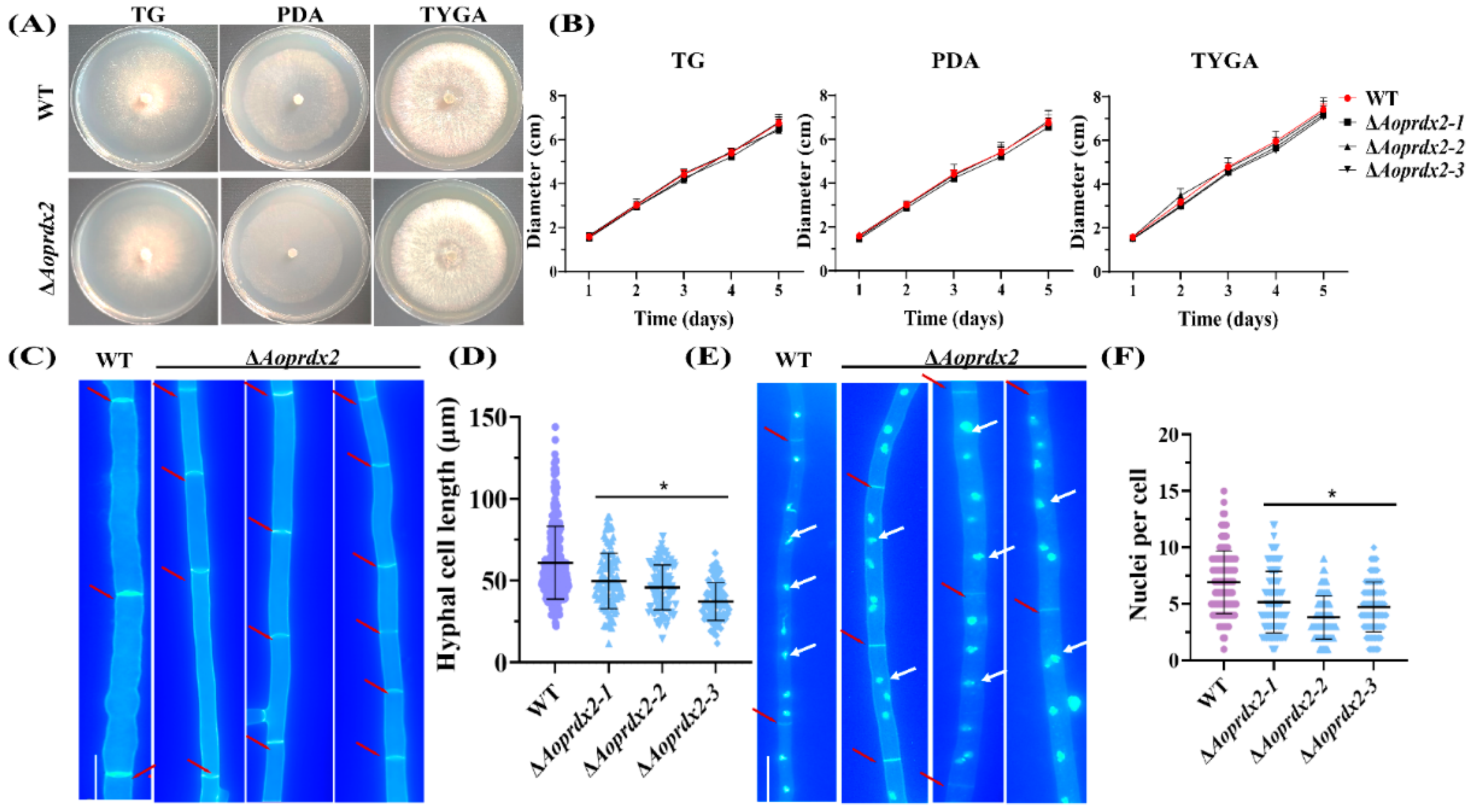
3.3. AoPrdx2 Impairs the Development of Mycelial Septa and Nuclei
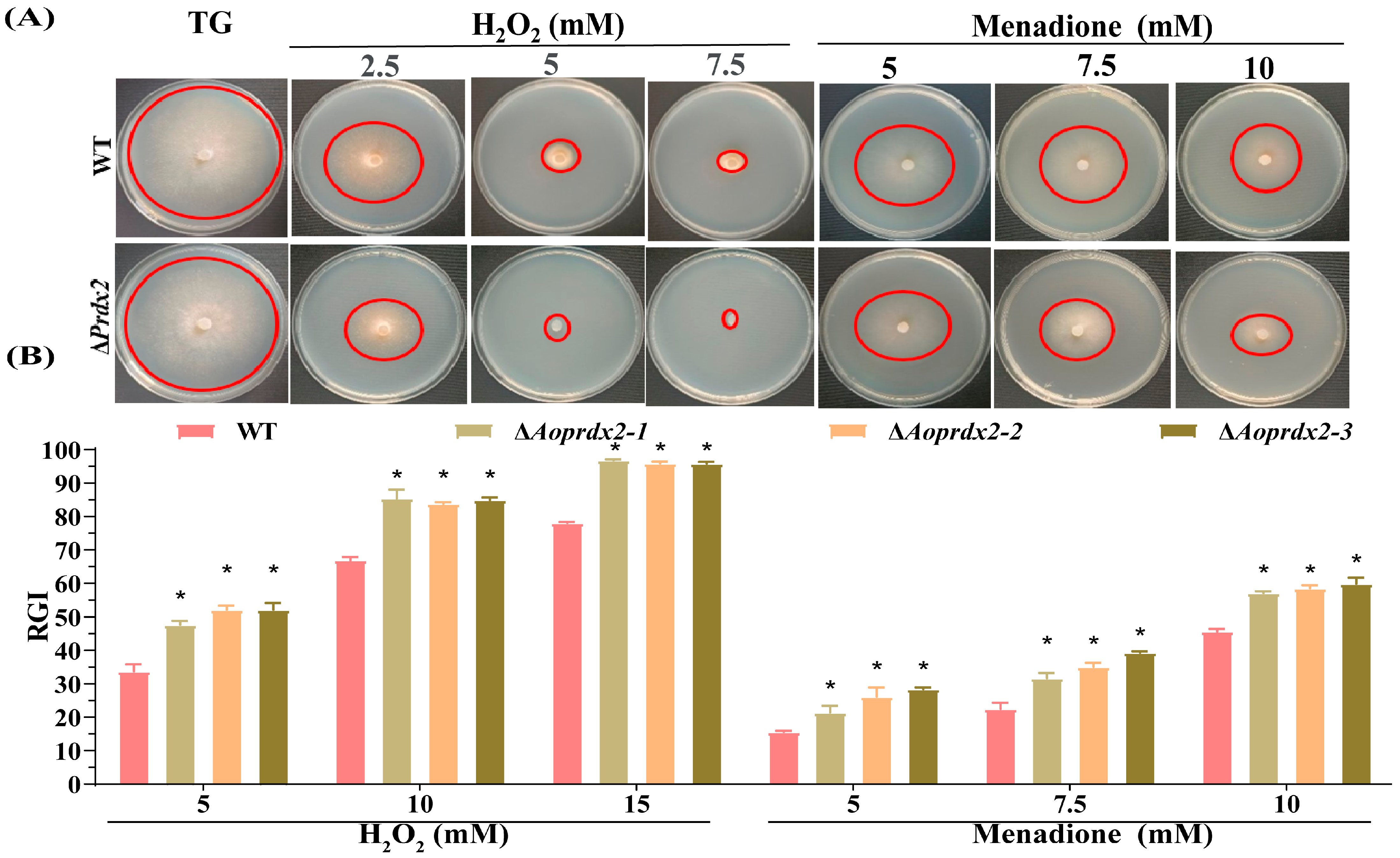
3.4. AoPrdx2 Regulates Tolerance to External Stresses
3.5. AoPrdx2 Regulates Sporulation
3.6. AoPrdx2 Regulates Trap Formation and Nematode Predation Efficiency
3.7. AoPrdx2 Regulates ROS Accumulation and Endocytosis
3.8. AoPrdx2 Impairs Secondary Metabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Araújo, J.V.; Braga, F.R.; Mendoza-de-Gives, P.; Paz-Silva, A.; Vilela, V.L.R. Recent advances in the control of helminths of domestic animals by helminthophagous fungi. Parasitologia 2021, 1, 168–176. [Google Scholar] [CrossRef]
- Jones, J.T.; Haegeman, A.; Danchin, E.G.; Gaur, H.S.; Helder, J.; Jones, M.G.; Kikuchi, T.; Manzanilla-López, R.; Palomares-Rius, J.E.; Wesemael, W.M.; et al. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Rosas-García, N.M. Biopesticide production from Bacillus thuringiensis: An environmentally friendly alternative. Recent Pat. Biotechnol. 2009, 3, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Lahm, G.P.; Desaeger, J.; Smith, B.K.; Pahutski, T.F.; Rivera, M.A.; Meloro, T.; Kucharczyk, R.; Lett, R.M.; Daly, A.; Smith, B.T.; et al. The discovery of fluazaindolizine: A new product for the control of plant parasitic nematodes. Bioorg. Med. Chem. Lett. 2017, 27, 1572–1575. [Google Scholar] [CrossRef]
- Cobb, N.A. Transference of nematodes (mononchs) from place to place for economic purposes. Science 1920, 51, 640–641. [Google Scholar] [CrossRef]
- Lin, H.C.; de Ulzurrun, G.V.; Chen, S.A.; Yang, C.T.; Tay, R.J.; Iizuka, T.; Huang, T.Y.; Kuo, C.Y.; Gonçalves, A.P.; Lin, S.Y.; et al. Key processes required for the different stages of fungal carnivory by a nematode-trapping fungus. PLoS Biol. 2023, 21, e3002400. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Zhao, Y.; Zhou, J.; Feng, H.; Jiang, D.; Zhang, K.Q.; Yang, J. Trapping devices of nematode-trapping fungi: Formation, evolution, and genomic perspectives. Biol. Rev. Camb. Philos. Soc. 2017, 92, 357–368. [Google Scholar] [CrossRef]
- Yang, J.; Wang, L.; Ji, X.; Feng, Y.; Li, X.; Zou, C.; Xu, J.; Ren, Y.; Mi, Q.; Wu, J.; et al. Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog. 2011, 7, e1002179. [Google Scholar] [CrossRef]
- Niu, X.M.; Zhang, K.Q. Arthrobotrys oligospora: A model organism for understanding the interaction between fungi and nematodes. Mycology 2011, 2, 59–78. [Google Scholar] [CrossRef]
- Liu, Q.; Li, D.; Jiang, K.; Zhang, K.Q.; Yang, J. AoPEX1 and AoPEX6 are required for mycelial growth, conidiation, stress response, fatty acid utilization, and trap formation in Arthrobotrys oligospora. Microbiol. Spectr. 2022, 10, e0027522. [Google Scholar] [CrossRef]
- Yang, L.; Li, X.; Bai, N.; Yang, X.; Zhang, K.Q.; Yang, J. Transcriptomic analysis reveals that Rho GTPases regulate trap development and lifestyle transition of the nematode-trapping fungus Arthrobotrys oligospora. Microbiol. Spectr. 2022, 10, e0175921. [Google Scholar] [CrossRef]
- Jiang, K.X.; Liu, Q.Q.; Bai, N.; Zhu, M.C.; Zhang, K.Q.; Yang, J.K. AoSsk1, a response regulator required for mycelial growth and development, stress responses, trap formation, and the secondary metabolism in Arthrobotrys oligospora. J. Fungi 2022, 8, 260. [Google Scholar] [CrossRef] [PubMed]
- Bai, N.; Xie, M.; Liu, Q.; Zhu, Y.; Yang, X.; Zhang, K.Q.; Yang, J. AoMedA has a complex regulatory relationship with AoBrlA, AoAbaA, and AoWetA in conidiation, trap formation, and secondary metabolism in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Environ. Microbiol. 2023, 89, e0098323. [Google Scholar] [CrossRef] [PubMed]
- D’Autréaux, B.; Toledano, M.B. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007, 8, 813–824. [Google Scholar] [CrossRef]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef] [PubMed]
- Devasagayam, T.P.; Tilak, J.C.; Boloor, K.K.; Sane, K.S.; Ghaskadbi, S.S.; Lele, R.D. Free radicals and antioxidants in human health: Current status and future prospects. J. Assoc. Physicians India 2004, 52, 794–804. [Google Scholar] [PubMed]
- Novo, E.; Parola, M. Redox mechanisms in hepatic chronic wound healing and fibrogenesis. Fibrogenesis Tissue Repair 2008, 1, 5. [Google Scholar] [CrossRef] [PubMed]
- Chae, H.Z.; Chung, S.J.; Rhee, S.G. Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem. 1994, 269, 27670–27678. [Google Scholar] [CrossRef]
- Kim, K.; Kim, I.H.; Lee, K.Y.; Rhee, S.G.; Stadtman, E.R. The isolation and purification of a specific “protector” protein which inhibits enzyme inactivation by a thiol/ Fe (III)/O2 mixed-function oxidation system. J. Biol. Chem. 1988, 263, 4704–4711. [Google Scholar] [CrossRef]
- Wood, Z.A.; Schröder, E.; Robin Harris, J.; Poole, L.B. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 2003, 28, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Link, A.J.; Robison, K.; Church, G.M. Comparing the predicted and observed properties of proteins encoded in the genome of Escherichia coli K-12. Electrophoresis 1997, 18, 1259–1313. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008, 4, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Chae, H.Z.; Uhm, T.B.; Rhee, S.G. Dimerization of thiol-specific antioxidant and the essential role of cysteine 47. Proc. Natl. Acad. Sci. USA 1994, 91, 7022–7026. [Google Scholar] [CrossRef] [PubMed]
- Knoops, B.; Loumaye, E.; Van Der Eecken, V. Evolution of the peroxiredoxins. Subcell. Biochem. 2007, 44, 27–40. [Google Scholar] [PubMed]
- Hall, A.; Nelson, K.; Poole, L.B.; Karplus, P.A. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox Signal. 2011, 15, 795–815. [Google Scholar] [CrossRef]
- Aranda-Caño, L.; Valderrama, R.; Pedrajas, J.R.; Begara-Morales, J.C.; Chaki, M.; Padilla, M.N.; Melguizo, M.; López-Jaramillo, F.J.; Barroso, J.B. Nitro-oleic acid-mediated nitroalkylation modulates the antioxidant function of cytosolic peroxiredoxin Tsa1 during heat stress in Saccharomyces cerevisiae. Antioxidants 2022, 11, 972. [Google Scholar] [CrossRef]
- Roger, F.; Picazo, C.; Reiter, W.; Libiad, M.; Asami, C.; Hanzén, S.; Gao, C.X.; Lagniel, G.; Welkenhuysen, N.; Labarre, J.; et al. Peroxiredoxin promotes longevity and H2O2-resistance in yeast through redox-modulation of protein kinase A. eLife 2020, 9, e60346. [Google Scholar] [CrossRef]
- Molin, M.; Yang, J.; Hanzén, S.; Toledano, M.B.; Labarre, J.; Nyström, T. Life span extension and H2O2 resistance elicited by caloric restriction require the reroxiredoxin Tsa1 in Saccharomyces cerevisiae. Mol. Cell 2011, 43, 823–833. [Google Scholar] [CrossRef]
- Paulo, E.; García-Santamarina, S.; Calvo, I.A.; Carmona, M.; Boronat, S.; Domènech, A.; Ayté, J.; Hidalgo, E. A genetic approach to study H2O2 scavenging in fission yeast-distinct roles of peroxiredoxin and catalase. Mol. Microbiol. 2014, 92, 246–257. [Google Scholar] [CrossRef]
- Morgan, B.A.; Veal, E.A. Functions of typical 2-cys peroxiredoxins in yeast. Subcell. Biochem. 2007, 44, 253–265. [Google Scholar]
- Mendoza-Martínez, A.E.; Sánchez, O.; Aguirre, J. The role of peroxiredoxins PrxA and PrxB in the antioxidant response, carbon utilization and development in Aspergillus nidulans. Fungal Biol. 2023, 127, 1198–1208. [Google Scholar] [CrossRef] [PubMed]
- Longo, L.V.G.; Breyer, C.A.; Novaes, G.M.; Gegembauer, G.; Leitão, N.P., Jr.; Octaviano, C.E.; Toyama, M.H.; de Oliveira, M.A.; Puccia, R. The human pathogen Paracoccidioides brasiliensis has a unique 1-cys peroxiredoxin that localizes both intracellularly and at the cell surface. Front. Cell Infect. Microbiol. 2020, 10, 394. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Escobedo, G.; Hernández-Carreón, O.; Morales-Rojano, B.; Revuelta-Rodríguez, B.; Vázquez-Franco, N.; Castaño, I.; De Las Peñas, A. Candida glabrata peroxiredoxins, Tsa1 and Tsa2, and sulfiredoxin, Srx1, protect against oxidative damage and are necessary for virulence. Fungal Genet. Biol. 2020, 135, 103287. [Google Scholar] [CrossRef] [PubMed]
- Rocha, M.C.; de Godoy, K.F.; Bannitz-Fernandes, R.; Fabri, J.; Barbosa, M.M.F.; de Castro, P.A.; Almeida, F.; Goldman, G.H.; da Cunha, A.F.; Netto, L.E.S.; et al. Analyses of the three 1-cys peroxiredoxins from Aspergillus fumigatus reveal that cytosolic Prx1 is central to H2O2 metabolism and virulence. Sci. Rep. 2018, 8, 12314. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Xiao, J.; Chen, Y.; Yu, Y.; Xiao, X.; Yu, Y.; Wu, H. Secretory expression and characterization of a novel peroxiredoxin for zearalenone detoxification in Saccharomyces cerevisiae. Microbiol. Res. 2013, 168, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Yaakoub, H.; Mina, S.; Calenda, A.; Bouchara, J.P.; Papon, N. Oxidative stress response pathways in fungi. Cell. Mol. Life Sci. 2022, 79, 333. [Google Scholar] [CrossRef]
- Li, X.; Kang, Y.Q.; Luo, Y.L.; Zhang, K.Q.; Zou, C.G.; Liang, L.M. The NADPH oxidase AoNoxA in Arthrobotrys oligospora functions as an initial factor in the infection of Caenorhabditis elegans. J. Microbiol. 2017, 55, 885–891. [Google Scholar] [CrossRef]
- Park, G.; Colot, H.V.; Collopy, P.D.; Krystofova, S.; Crew, C.; Ringelberg, C.; Litvinkova, L.; Altamirano, L.; Li, L.; Curilla, S.; et al. High-throughput production of gene replacement mutants in Neurospora crassa. Methods Mol. Biol. 2011, 722, 179–189. [Google Scholar]
- Yang, X.; Ma, N.; Yang, L.; Zheng, Y.; Zhen, Z.; Li, Q.; Xie, M.; Li, J.; Zhang, K.Q.; Yang, J. Two Rab GTPases play different roles in conidiation, trap formation, stress resistance, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Microbiol. Biotechnol. 2018, 102, 4601–4613. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Yang, J.; Wang, W.; Liu, Y.; Xie, M.; Yang, J. The MADS-box transcription factor AoRlmA is involved in the regulation of mycelium development, conidiation, cell-wall integrity, stress response, and trap formation of Arthrobotrys oligospora. Microbiol. Res. 2023, 268, 127299. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, X. Tools and basic procedures of gene manipulation in nematode-trapping fungi. Mycology 2023, 14, 75–90. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Bai, N.; Yang, X.; Liu, Y.; Zhang, K.Q.; Yang, J. Fus3 regulates asexual development and trap morphogenesis in the nematode-trapping fungus Arthrobotrys oligospora. iScience 2023, 26, 107404. [Google Scholar] [CrossRef]
- Zhu, M.C.; Zhao, N.; Liu, Y.K.; Li, X.M.; Zhen, Z.Y.; Zheng, Y.Q.; Zhang, K.Q.; Yang, J.K. The cAMP-PKA signalling pathway regulates hyphal growth, conidiation, trap morphogenesis, stress tolerance, and autophagy in Arthrobotrys oligospora. Environ. Microbiol. 2022, 24, 6524–6538. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, X.; Xie, M.; Bai, N.; Yang, J.; Jiang, K.; Zhang, K.Q.; Yang, J. Pleiotropic roles of Ras GTPases in the nematode-trapping fungus Arthrobotrys oligospora identified through multi-omics analyses. iScience 2021, 24, 102820. [Google Scholar] [CrossRef]
- Wang, W.; Liu, Y.; Duan, S.; Bai, N.; Zhu, M.; Yang, J. Cellular communication and fusion regulate cell fusion, trap morphogenesis, conidiation, and secondary metabolism in Arthrobotrys oligospora. Microbiol. Res. 2024, 278, 127516. [Google Scholar] [CrossRef]
- Zhu, Y.; Yang, X.; Bai, N.; Liu, Q.; Yang, J. AoRab7A interacts with AoVps35 and AoVps41 to regulate vacuole assembly, trap formation, conidiation, and functions of proteasomes and ribosomes in Arthrobotrys oligospora. Microbiol. Res. 2024, 280, 127573. [Google Scholar] [CrossRef]
- Zhen, Z.; Zhang, G.; Yang, L.; Ma, N.; Li, Q.; Ma, Y.; Niu, X.; Zhang, K.Q.; Yang, J. Characterization and functional analysis of calcium/calmodulin-dependent protein kinases (CaMKs) in the nematode-trapping fungus Arthrobotrys oligospora. Appl. Microbiol. Biotechnol. 2019, 103, 819–832. [Google Scholar] [CrossRef]
- Xie, M.; Ma, N.; Bai, N.; Yang, L.; Yang, X.; Zhang, K.Q.; Yang, J. PKC-SWI6 signaling regulates asexual development, cell wall integrity, stress response, and lifestyle transition in the nematode-trapping fungus Arthrobotrys oligospora. Sci. China Life Sci. 2022, 65, 2455–2471. [Google Scholar] [CrossRef]
- Zhen, Z.; Xing, X.; Xie, M.; Yang, L.; Yang, X.; Zheng, Y.; Chen, Y.; Ma, N.; Li, Q.; Zhang, K.Q.; et al. MAP kinase Slt2 orthologs play similar roles in conidiation, trap formation, and pathogenicity in two nematode-trapping fungi. Fungal Genet. Biol. 2018, 116, 42–50. [Google Scholar] [CrossRef]
- Zhou, D.; Zhu, Y.; Bai, N.; Yang, L.; Xie, M.; Yang, J.; Zhu, M.; Zhang, K.Q.; Yang, J. AoATG5 plays pleiotropic roles in vegetative growth, cell nucleus development, conidiation, and virulence in the nematode-trapping fungus Arthrobotrys oligospora. Sci. China Life Sci. 2022, 65, 412–425. [Google Scholar] [CrossRef]
- Li, X.; Zhu, M.; Liu, Y.; Yang, L.; Yang, J. Aoatg11 and Aoatg33 are indispensable for mitophagy, and contribute to conidiation, the stress response, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Microbiol. Res. 2023, 266, 127252. [Google Scholar] [CrossRef]
- Yang, L.; Li, X.; Ma, Y.; Zhang, K.; Yang, J. The Arf-Gap proteins AoGcs1 and AoGts1 regulate mycelial development, endocytosis, and pathogenicity in Arthrobotrys oligospora. J. Fungi 2022, 8, 463. [Google Scholar] [CrossRef]
- Kong, X.; Zhang, H.; Wang, X.; van der Lee, T.; Waalwijk, C.; van Diepeningen, A.; Brankovics, B.; Xu, J.; Xu, J.; Chen, W.; et al. FgPex3, a peroxisome biogenesis factor, is involved in regulating vegetative growth, conidiation, sexual development, and virulence in Fusarium graminearum. Front. Microbiol. 2019, 10, 2088. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhou, D.; Bai, N.; Liu, Q.; Zhao, N.; Yang, J. SNARE protein AoSec22 orchestrates mycelial growth, vacuole assembly, trap formation, stress response, and secondary metabolism in Arthrobotrys oligospora. J. Fungi 2023, 9, 75. [Google Scholar] [CrossRef]
- Waters, E.C.T.; Baark, F.; Yu, Z.; Mota, F.; Eykyn, T.R.; Yan, R.; Southworth, R. Detecting validated intracellular ROS generation with 18F-dihydroethidine-based PET. Mol. Imaging Biol. 2022, 24, 377–383. [Google Scholar] [CrossRef]
- Rhee, S.G. Overview on peroxiredoxin. Mol. Cells 2016, 39, 1–5. [Google Scholar] [CrossRef]
- Hillmann, F.; Bagramyan, K.; Straßburger, M.; Heinekamp, T.; Hong, T.B.; Bzymek, K.P.; Williams, J.C.; Brakhage, A.A.; Kalkum, M. The crystal structure of peroxiredoxin ASP f3 provides mechanistic insight into oxidative stress resistance and virulence of Aspergillus fumigatus. Sci. Rep. 2016, 6, 33396. [Google Scholar] [CrossRef] [PubMed]
- Schröter, C.; Hipler, U.C.; Wilmer, A.; Künkel, W.; Wollina, U. Generation of reactive oxygen species by Candida albicans in relation to morphogenesis. Arch. Dermatol. Res. 2000, 292, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.H.; Jung, S.; Park, S.J.; Kim, Y.J.; Ahn, J.M.; Kim, W.; Choi, W. Characterization of thiol-specific antioxidant 1 (TSAI) of Candida albicans. Yeast 2005, 22, 907–918. [Google Scholar] [CrossRef] [PubMed]
- Iraqui, I.; Kienda, G.; Soeur, J.; Faye, G.; Baldacci, G.; Kolodner, R.D.; Huang, M.E. Peroxiredoxin Tsa1 is the key peroxidase suppressing genome instability and protecting against cell death in Saccharomyces cerevisiae. PLoS Genet. 2009, 5, e1000524. [Google Scholar] [CrossRef]
- Kuo, T.H.; Yang, C.T.; Chang, H.Y.; Hsueh, Y.P.; Hsu, C.C. Nematode-trapping fungi produce diverse metabolites during predator-prey interaction. Metabolites 2020, 10, 117. [Google Scholar] [CrossRef]
- Xu, Z.F.; Wang, B.L.; Sun, H.K.; Yan, N.; Zeng, Z.J.; Zhang, K.Q.; Niu, X.M. High trap formation and low metabolite production by disruption of the polyketide synthase gene involved in the biosynthesis of arthrosporols from nematode-trapping fungus Arthrobotrys oligospora. J. Agric. Food Chem. 2015, 63, 9076–9082. [Google Scholar] [CrossRef]
- Bahn, Y.S.; Xue, C.; Idnurm, A.; Rutherford, J.C.; Heitman, J.; Cardenas, M.E. Sensing the environment: Lessons from fungi. Nat. Rev. Microbiol. 2007, 5, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.; Priest, E.; Naglik, J.R.; Richardson, J.P. Fungal toxins and host immune responses. Front. Microbiol. 2021, 12, 643639. [Google Scholar] [CrossRef] [PubMed]
- Finotti, E.; Parroni, A.; Zaccaria, M.; Domin, M.; Momeni, B.; Fanelli, C.; Reverberi, M. Aflatoxins are natural scavengers of reactive oxygen species. Sci. Rep. 2021, 11, 16024. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.J.; Yu, Y.M.; Kim, G.B.; Lee, G.W.; Maeng, P.J.; Kim, S.; Floyd, A.; Heitman, J.; Bahn, Y.S. Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot. Cell 2009, 8, 1197–1217. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Hu, X.; Pop, M.; Wernet, N.; Kirschhöfer, F.; Brenner-Weiß, G.; Keller, J.; Bunzel, M.; Fischer, R. Fatal attraction of Caenorhabditis elegans to predatory fungi through 6-methyl-salicylic acid. Nat. Commun. 2021, 12, 5462. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, N.; Zhu, M.; Liu, Q.; Shen, Y.; Duan, S.; Zhu, L.; Yang, J. AoPrdx2 Regulates Oxidative Stress, Reactive Oxygen Species, Trap Formation, and Secondary Metabolism in Arthrobotrys oligospora. J. Fungi 2024, 10, 110. https://doi.org/10.3390/jof10020110
Zhao N, Zhu M, Liu Q, Shen Y, Duan S, Zhu L, Yang J. AoPrdx2 Regulates Oxidative Stress, Reactive Oxygen Species, Trap Formation, and Secondary Metabolism in Arthrobotrys oligospora. Journal of Fungi. 2024; 10(2):110. https://doi.org/10.3390/jof10020110
Chicago/Turabian StyleZhao, Na, Meichen Zhu, Qianqian Liu, Yanmei Shen, Shipeng Duan, Lirong Zhu, and Jinkui Yang. 2024. "AoPrdx2 Regulates Oxidative Stress, Reactive Oxygen Species, Trap Formation, and Secondary Metabolism in Arthrobotrys oligospora" Journal of Fungi 10, no. 2: 110. https://doi.org/10.3390/jof10020110




