Genome-Wide Identification of the ABC Gene Family and Its Expression in Response to the Wood Degradation of Poplar in Trametes gibbosa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Identification of ABC Genes in T. gibbosa
2.3. Phylogenetic Relationship and Motif Analyses of Tg-ABCs in T. gibbosa
2.4. Scaffold Localization and Collinearity Analysis of Tg-ABCs in T. gibbosa
2.5. Phylogenetic Relationship and Collinearity Analyses of ABC Gene Family
2.6. Other Characteristic Analyses in the Tg-ABCs
2.7. RT-qPCR Analysis of Tg-ABCs in Response to Wood Degradation by T. gibbosa
3. Results
3.1. Identification of ABC Genes in T. gibbosa
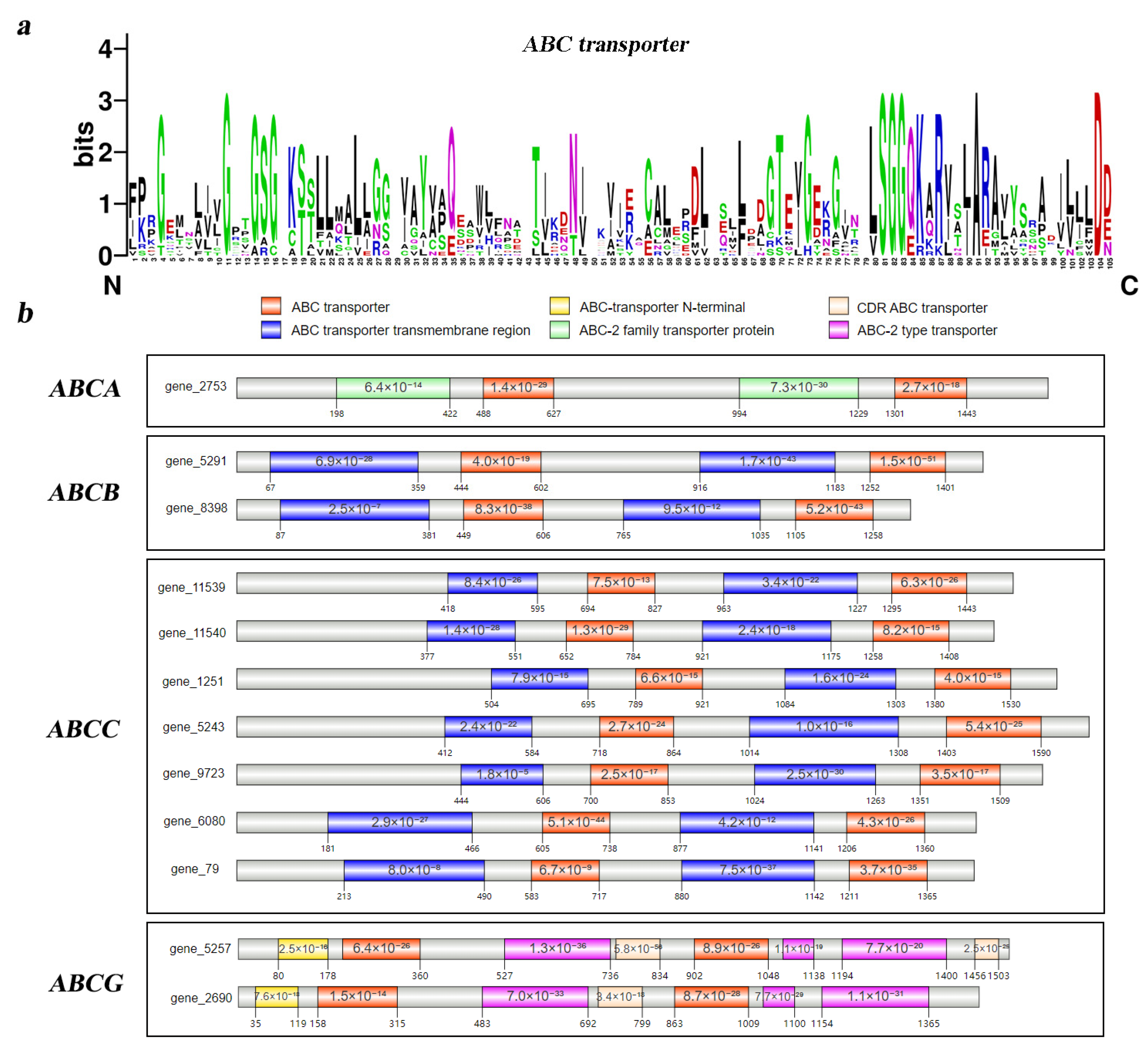
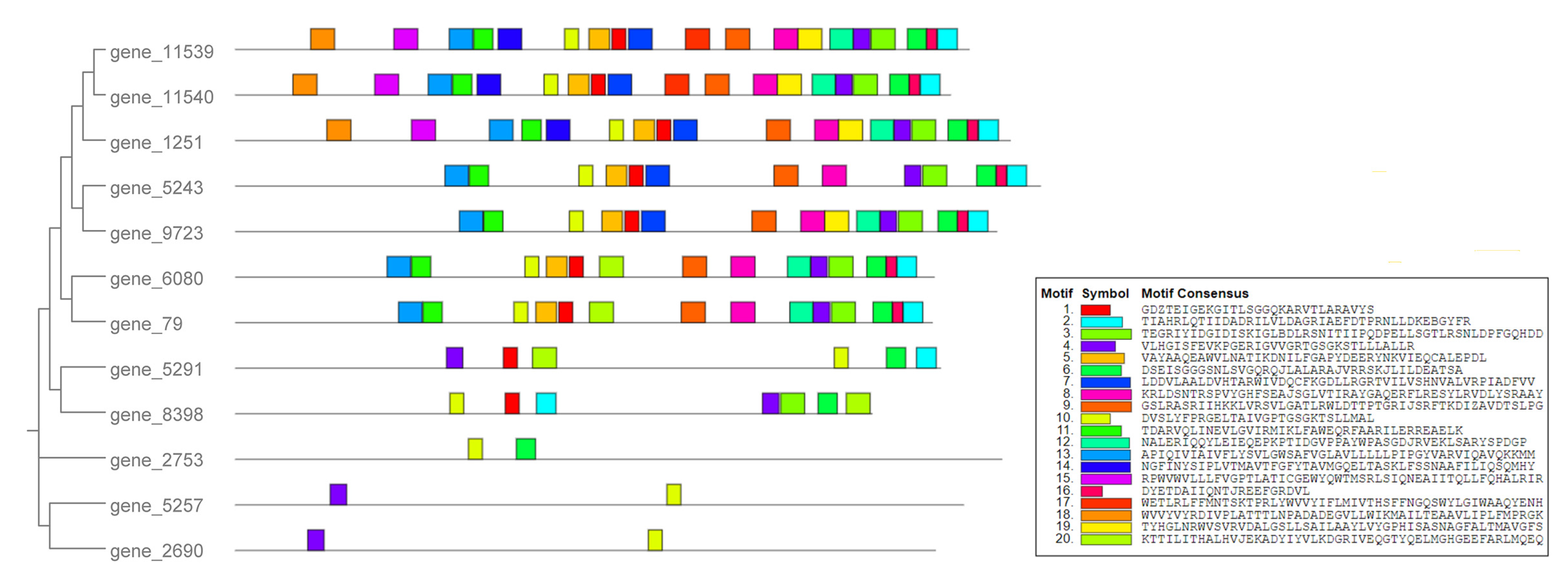
3.2. Phylogenetic Relationship and Motif Analyses of Tg-ABCs in T. gibbosa
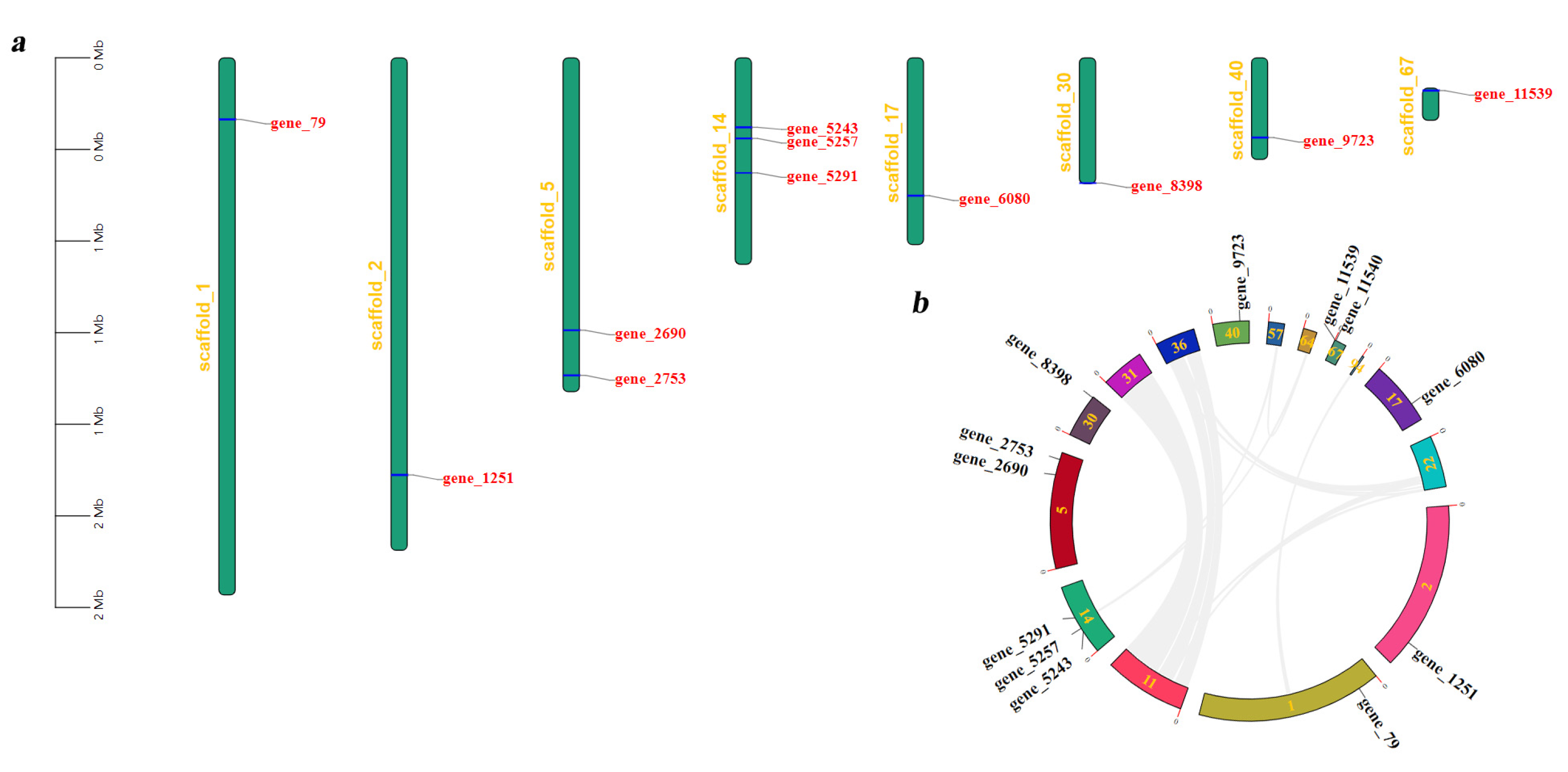
3.3. Scaffold Localization and Collinearity Analyses of Tg-ABCs in T. gibbosa
3.4. Physicochemical Analyses of Tg-ABCs in T. gibbosa
3.5. Phylogenetic Relationship and Collinearity Analyses of ABC Gene Family
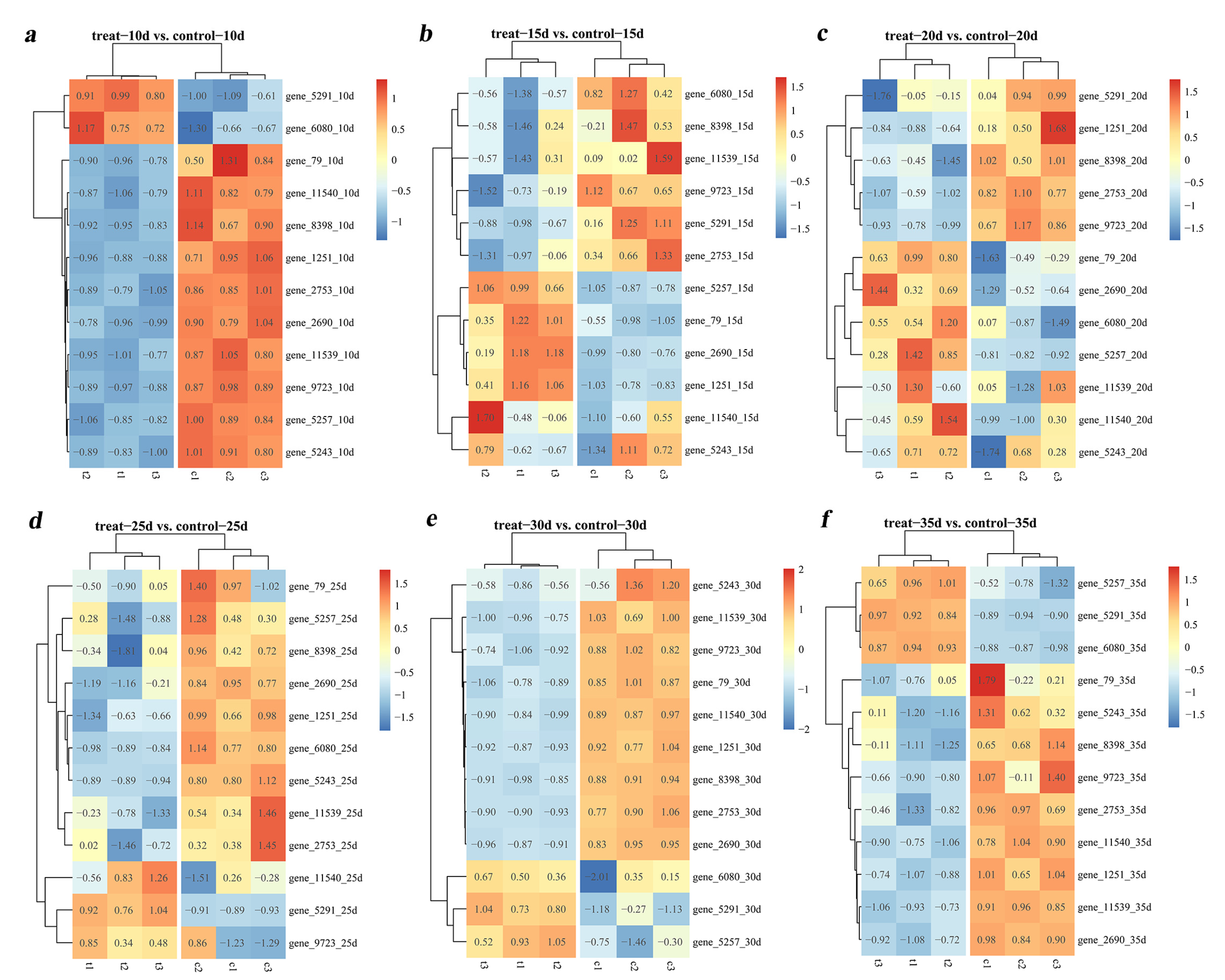
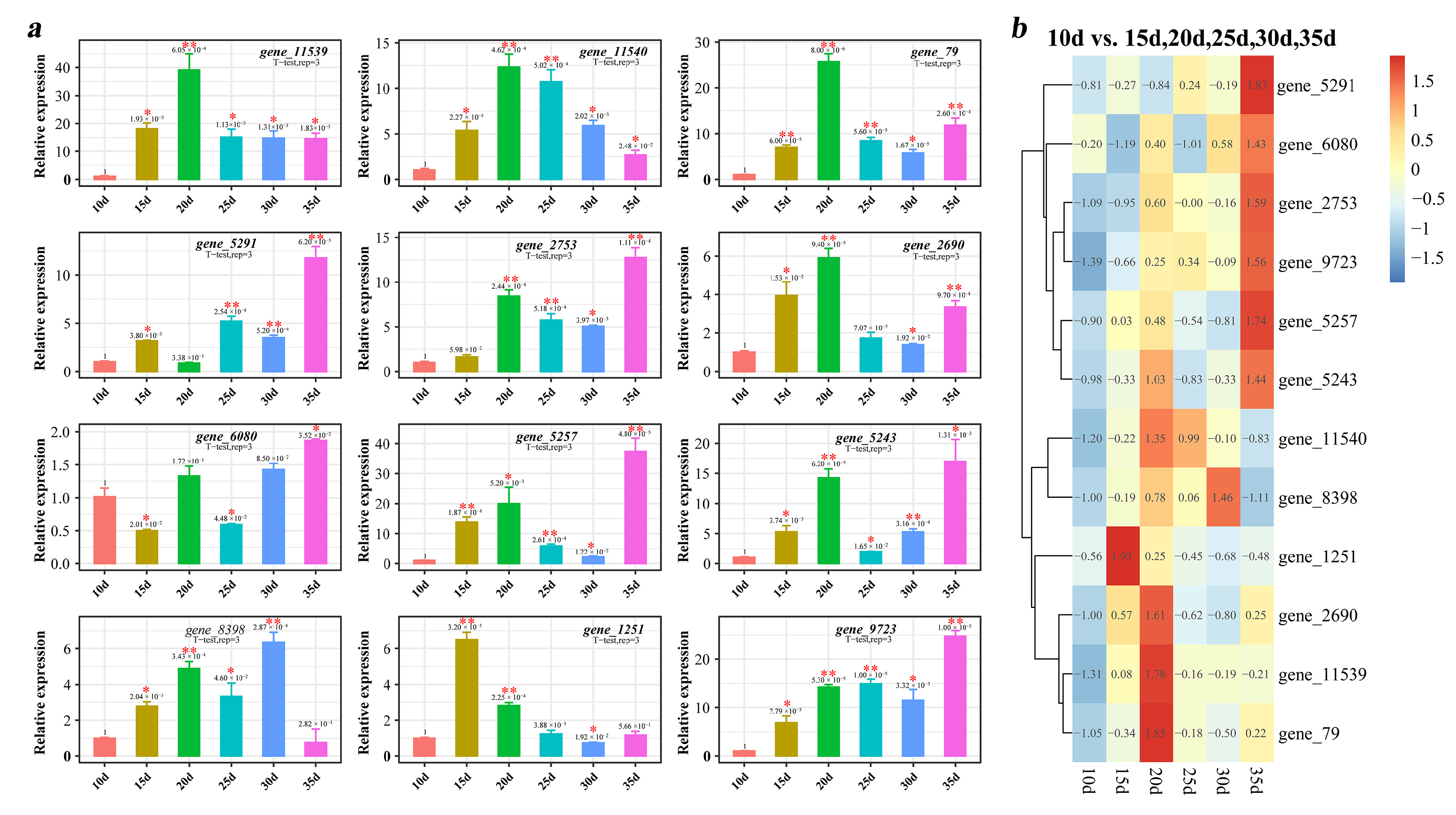
3.6. Quantitative Analysis of Tg-ABCs in T. gibbosa
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Thakur, A.K.; Kumar, P.; Parmar, N.; Shandil, R.K.; Aggarwal, G.; Gaur, A.; Srivastava, D.K. Achievements and prospects of genetic engineering in poplar: A review. New For. 2021, 52, 889–920. [Google Scholar] [CrossRef]
- Gao, S.; Li, C.; Chen, X.; Li, S.; Liang, N.; Wang, H.; Zhan, Y.; Zeng, F. Basic helix-loop-helix transcription factor PxbHLH02 enhances drought tolerance in Populus (Populus simonii × P. nigra). Tree Physiol. 2023, 43, 185–202. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Dong, F.; Wang, Y.; Chen, H.; Tang, M. Arbuscular mycorrhizal fungi enhance photosynthesis and drought tolerance by regulating MAPK genes expressions of Populus simonii × P. nigra. Physiol. Plant. 2022, 174, e13829. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Kang, Y.; Leng, J.; Xu, Q. Genome-Wide Analysis of the miRNA-mRNAs Network Involved in Cold Tolerance in Populus simonii × P. nigra. Genes 2019, 10, 430. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.K.M.; Fu, Z.; Ding, C.; Jiang, L.; Han, X.; Yang, A.; Ma, Y.; Zhao, X. Growth and wood properties of a 38-year-old Populus simonii × P. nigra plantation established with different densities in semi-arid areas of northeastern China. J. For. Res. 2020, 31, 497–506. [Google Scholar] [CrossRef]
- Liu, D.; Liu, M.; Li, Z.; Wang, G.; Li, Y.; Zheng, M.; Liu, G.; Zhao, X. Variation Analysis of Growth Traits of Transgenic Populus simonii x P.nigra Clones Carrying TaLEA Gene. Bull. Bot. Res. 2015, 35, 540–546. [Google Scholar]
- de Vries, L.; Guevara-Rozo, S.; Cho, M.; Liu, L.Y.; Renneckar, S.; Mansfield, S.D. Tailoring renewable materials via plant biotechnology. Biotechnol. Biofuels 2021, 14, 167. [Google Scholar] [CrossRef]
- Vanholme, R.; De Meester, B.; Ralph, J.; Boerjan, W. Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 2019, 56, 230–239. [Google Scholar] [CrossRef]
- Gan, M.J.; Niu, Y.Q.; Qu, X.J.; Zhou, C.H. Lignin to value-added chemicals and advanced materials: Extraction, degradation, and functionalization. Green Chem. 2022, 24, 7705–7750. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, J.; Zhao, D.; Jia, L.; Qin, B.; Cao, X.; Zang, L.; Lu, F.; Liu, F. Biological degradation of lignin: A critical review on progress and perspectives. Ind. Crops Prod. 2022, 188, 115715. [Google Scholar] [CrossRef]
- Zhang, C.; Jin, Y.; Wang, F.; Shen, X.; Cheng, J.; Cai, C. Catalytic Strategies and Mechanism Analysis Orbiting the Center of Critical Intermediates in Lignin Depolymerization. Chem. Rev. 2023, 123, 4510–4601. [Google Scholar] [CrossRef]
- Weimer, P.J. Degradation of Cellulose and Hemicellulose by Ruminal Microorganisms. Microorganisms 2022, 10, 2345. [Google Scholar] [CrossRef] [PubMed]
- Long, L.; Hu, Y.; Sun, F.; Gao, W.; Hao, Z.; Yin, H. Advances in lytic polysaccharide monooxygenases with the cellulose-degrading auxiliary activity family 9 to facilitate cellulose degradation for biorefinery. Int. J. Biol. Macromol. 2022, 219, 68–83. [Google Scholar] [CrossRef] [PubMed]
- Nurul-Aliyaa, Y.A.; Awang, N.A.; Mohd, M.H. Characterisation of white rot fungi from wood decayed for lignin degradation. Lett. Appl. Microbiol. 2023, 76, ovad118. [Google Scholar] [CrossRef] [PubMed]
- Fackler, K.; Schwanninger, M. Polysaccharide degradation and lignin modification during brown rot of spruce wood: A polarised Fourier transform near infrared study. J. Near Infrared Spectrosc. 2010, 18, 403–416. [Google Scholar] [CrossRef]
- Chen, J.; Chi, Y.; Hao, X.; Ma, L. Metabolic regulation mechanism of Trametes gibbosa CB1 on lignin. Int. J. Biol. Macromol. 2023, 240, 124189. [Google Scholar] [CrossRef]
- Knezevic, A.; Stajic, M.; Milovanovic, I.; Vukojevic, J. Degradation of beech wood and wheat straw by Trametes gibbosa. Wood Sci. Technol. 2017, 51, 1227–1247. [Google Scholar] [CrossRef]
- Knezevic, A.; Stajic, M.; Milovanovic, I.; Vukojevic, J. Wheat Straw Degradation by Trametes gibbosa: The Effect of Calcium Ions. Waste Biomass Valorization 2018, 9, 1903–1908. [Google Scholar] [CrossRef]
- Han, M.-L.; Lin, L.; Guo, X.-X.; An, M.; Geng, Y.-J.; Xin, C.; Ma, L.-C.; Mi, Q.; Ping, A.-Q.; Yang, Q.-Y. Comparative Analysis of the Laccase Secretion Ability of Five White-rot Fungi in Submerged Fermentation with Lignocellulosic Biomass. Bioresources 2023, 18, 584–598. [Google Scholar] [CrossRef]
- Yi, X.; Lin, L.; Mei, J.; Wang, W. Transporter proteins in Zymomonas mobilis contribute to the tolerance of lignocellulose-derived phenolic aldehyde inhibitors. Bioprocess Biosyst. Eng. 2021, 44, 1875–1882. [Google Scholar] [CrossRef]
- Takeuchi, M.; Watanabe, A.; Tamura, M.; Tsutsumi, Y. The gene expression analysis of Arabidopsis thaliana ABC transporters by real-time PCR for screening monolignol-transporter candidates. J. Wood Sci. 2018, 64, 477–484. [Google Scholar] [CrossRef]
- Sibout, R.; Höfte, H. Plant cell biology: The ABC of monolignol transport. Curr. Biol. 2012, 22, R533–R535. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Zhong, Z.; Liu, H.; Lin, L.; Guo, M.; Guo, W.; Wang, Z.; Zhang, Q.; Feng, L.; Lu, G. Whole genome and transcriptome analysis reveal adaptive strategies and pathogenesis of Calonectria pseudoreteaudii to Eucalyptus. BMC Genom. 2018, 19, 358. [Google Scholar] [CrossRef] [PubMed]
- Yazaki, K. ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett. 2006, 580, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- de Lima, L.G.A.; Ferreira, S.S.; Simoes, M.S.; Cunha, L.X.; Fernie, A.R.; Cesarino, I. Comprehensive expression analyses of the ABCG subfamily reveal SvABCG17 as a potential transporter of lignin monomers in the model C4 grass Setaria viridis. J. Plant Physiol. 2023, 280, 153900. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Li, J.; Qin, G.; Liu, C.; Cao, Z.; Jia, B.; Xu, Y.; Li, G.; Yang, Y.; Su, Y. Characterization of the ABC Transporter G Subfamily in Pomegranate and Function Analysis of PgrABCG14. Int. J. Mol. Sci. 2022, 23, 11661. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Gong, W.; Zhu, Z.; Zhou, Y.; Xu, C.; Yan, L.; Hu, Z.; Ai, L.; Peng, Y. Comparative secretome of white-rot fungi reveals co-regulated carbohydrate-active enzymes associated with selective ligninolysis of ramie stalks. Microb. Biotechnol. 2021, 14, 911–922. [Google Scholar] [CrossRef]
- Kirk, T.K.; Schultz, E.; Connors, W.J.; Lorenz, L.F.; Zeikus, J.G. Influence of culture parameters on lignin metabolism byPhanerochaete chrysosporium. Arch. Microbiol. 1978, 117, 277–285. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Hage, H.; Miyauchi, S.; Viragh, M.; Drula, E.; Min, B.; Chaduli, D.; Navarro, D.; Favel, A.; Norest, M.; Lesage-Meessen, L. Gene family expansions and transcriptome signatures uncover fungal adaptations to wood decay. Environ. Microbiol. 2021, 23, 5716–5732. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.U.; Ahring, B.K. Lignin degradation under anaerobic digestion: Influence of lignin modifications—A review. Biomass Bioenergy 2019, 128, 105325. [Google Scholar] [CrossRef]
- Jiang, B.; Jiao, H.; Guo, X.; Chen, G.; Guo, J.; Wu, W.; Jin, Y.; Cao, G.; Liang, Z. Lignin-Based Materials for Additive Manufacturing: Chemistry, Processing, Structures, Properties, and Applications. Adv. Sci. 2023, 10, 2206055. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Kong, W.; Ren, K.; Cheng, H. Advance of Research in Function of Plant ABC Transporters. Acat Agric. Boreali-Occident. Sin. 2023, 32, 1–10. [Google Scholar]
- Chi, Y.; Gu, X. Advances in the Research of Lenzites gibbosa (Pers.) Hemmi. J. Jilin Agric. Univ. 2021, 43, 275–282. [Google Scholar]
- Viglas, J.; Olejnikova, P. An update on ABC transporters of filamentous fungi-from physiological substrates to xenobiotics. Microbiol. Res. 2021, 246, 126684. [Google Scholar] [CrossRef]
- Chi, Y.; Yan, H.; Wu, S. Phylogenetic Analysis and Cloning of the Gene Lg-mnp2 and Lg-mnp3 of Lenzites gibbosa CB1. J. North-East For. Univ. 2019, 47, 64–87. [Google Scholar]
- Kovalchuk, A.; Driessen, A.J.M. Phylogenetic analysis of fungal ABC transporters. BMC Genom. 2010, 11, 177. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, M.; Schuetz, M.; Lin, B.S.P.; Chanis, C.; Hamberger, B.; Western, T.L.; Ehlting, J.; Samuels, A.L. ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. J. Exp. Bot. 2011, 62, 2063–2077. [Google Scholar] [CrossRef] [PubMed]
- Michalska, K.; Chang, C.; Mack, J.C.; Zerbs, S.; Joachimiak, A.; Collart, F.R. Characterization of Transport Proteins for Aromatic Compounds Derived from Lignin: Benzoate Derivative Binding Proteins. J. Mol. Biol. 2012, 423, 555–575. [Google Scholar] [CrossRef] [PubMed]
- Dao, A.T.N.; Loenen, S.J.; Swart, K.; Dang, H.T.C.; Brouwer, A.; de Boer, T.E. Characterization of 2,3,7,8-tetrachlorodibenzo-p-dioxin biodegradation by extracellular lignin-modifying enzymes from ligninolytic fungus. Chemosphere 2021, 263, 128280. [Google Scholar] [CrossRef]
- Leriche-Grandchamp, M.; Flourat, A.; Shen, H.; Picard, F.; Giordana, H.; Allais, F.; Fayeulle, A. Inhibition of Phenolics Uptake by Ligninolytic Fungal Cells and Its Potential as a Tool for the Production of Lignin-Derived Aromatic Building Blocks. J. Fungi 2020, 6, 362. [Google Scholar] [CrossRef]

| ID | Number of Amino Acids | Molecular Weight | Theoretical pI | Aliphatic Index | Grand Average of Hydropathicity | Formula | Total Number of Atoms | Instability Index | Stability |
|---|---|---|---|---|---|---|---|---|---|
| gene_11539 | 1536 | 169,596.99 | 6.28 | 107.98 | 0.186 | C7664H12155N2041O2200S47 | 24,107 | 37.98 | stable |
| gene_11540 | 1497 | 165,312.94 | 5.92 | 106.73 | 0.117 | C7435H11789N1999O2176S41 | 23,440 | 40.53 | unstable |
| gene_79 | 1459 | 161,918.99 | 7.99 | 95.42 | −0.030 | C7256H11479N1993O2107S49 | 22,884 | 35.49 | stable |
| gene_5291 | 1476 | 161,417.17 | 6.84 | 97.24 | 0.104 | C7183H11477N1997O2095S65 | 22,817 | 48.30 | unstable |
| gene_2753 | 1604 | 174,825.02 | 8.45 | 105.57 | 0.262 | C7940H12497N2113O2241S44 | 24,835 | 35.36 | stable |
| gene_2690 | 1465 | 164,291.53 | 6.96 | 86.22 | −0.065 | C7446H11457N1969O2109S62 | 23,043 | 39.54 | stable |
| gene_6080 | 1463 | 160,933.00 | 8.51 | 97.58 | −0.019 | C7214H11504N1958O2122S41 | 22,839 | 36.87 | stable |
| gene_5257 | 1524 | 168,164.73 | 7.00 | 86.71 | −0.018 | C7560H11723N2059O2194S50 | 23,586 | 37.30 | stable |
| gene_5243 | 1685 | 185,461.04 | 5.89 | 101.38 | 0.082 | C8390H13216N2228O2432S39 | 26,305 | 36.75 | stable |
| gene_1251 | 1622 | 178,758.07 | 5.85 | 104.06 | 0.156 | C8094H12776N2108O2350S48 | 25,376 | 39.28 | stable |
| gene_9723 | 1594 | 175,020.65 | 5.90 | 102.61 | 0.169 | C7909H12477N2083O2292S50 | 24,811 | 41.41 | unstable |
| gene_8398 | 1333 | 145,732.30 | 6.24 | 96.05 | −0.051 | C6518H10338N1774O1951S30 | 20,611 | 39.62 | stable |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Wang, A.; Wang, Q. Genome-Wide Identification of the ABC Gene Family and Its Expression in Response to the Wood Degradation of Poplar in Trametes gibbosa. J. Fungi 2024, 10, 96. https://doi.org/10.3390/jof10020096
Zhao J, Wang A, Wang Q. Genome-Wide Identification of the ABC Gene Family and Its Expression in Response to the Wood Degradation of Poplar in Trametes gibbosa. Journal of Fungi. 2024; 10(2):96. https://doi.org/10.3390/jof10020096
Chicago/Turabian StyleZhao, Jia, Achuan Wang, and Qian Wang. 2024. "Genome-Wide Identification of the ABC Gene Family and Its Expression in Response to the Wood Degradation of Poplar in Trametes gibbosa" Journal of Fungi 10, no. 2: 96. https://doi.org/10.3390/jof10020096





