Morpho-Molecular Characterization Reveals a New Genus, Three Novel Species and Two New Host Records in Xylariomycetidae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection, Isolation and Morphology
2.2. DNA Extraction, PCR Amplification and Sequencing
2.3. Phylogenetic Analyses
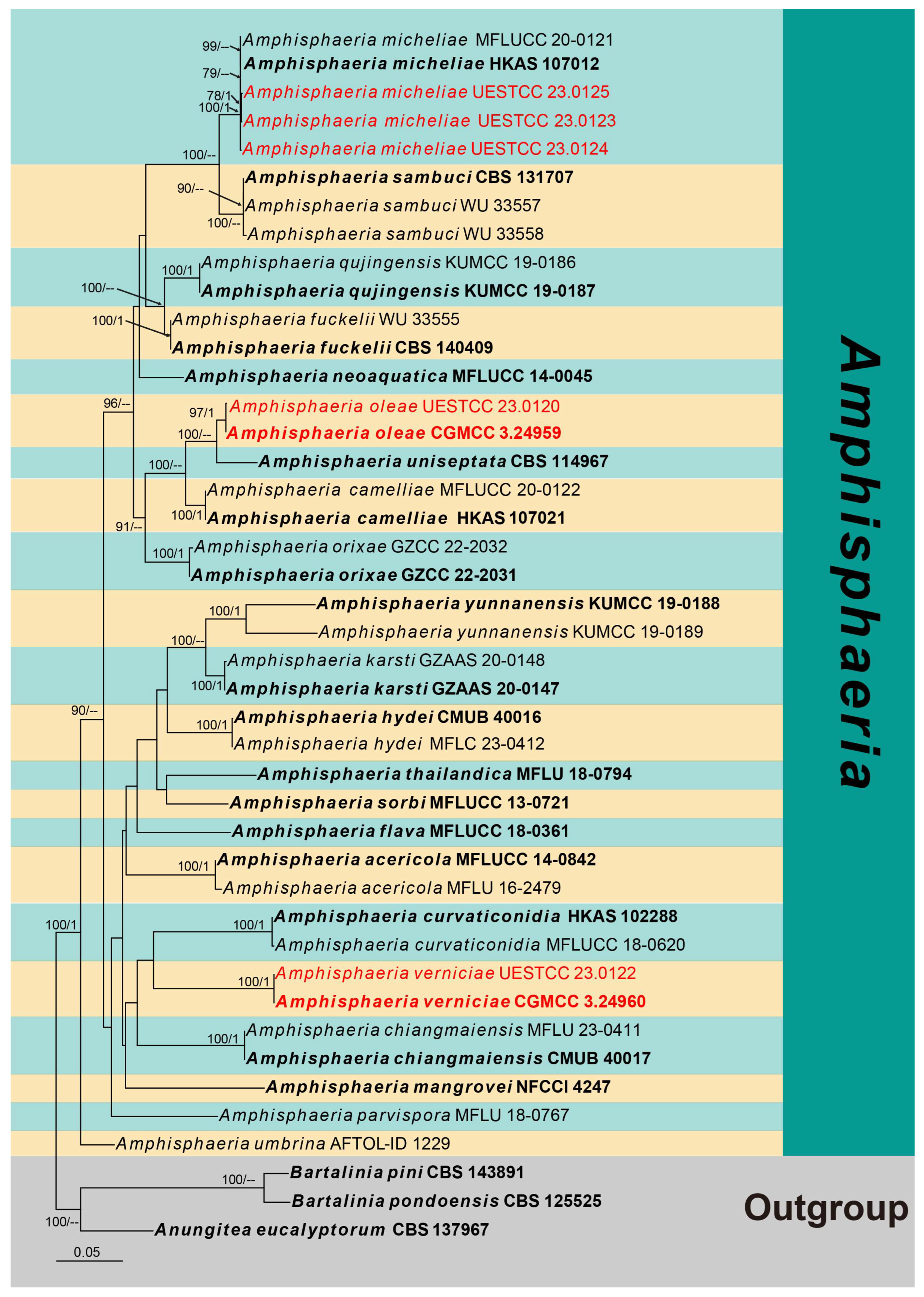
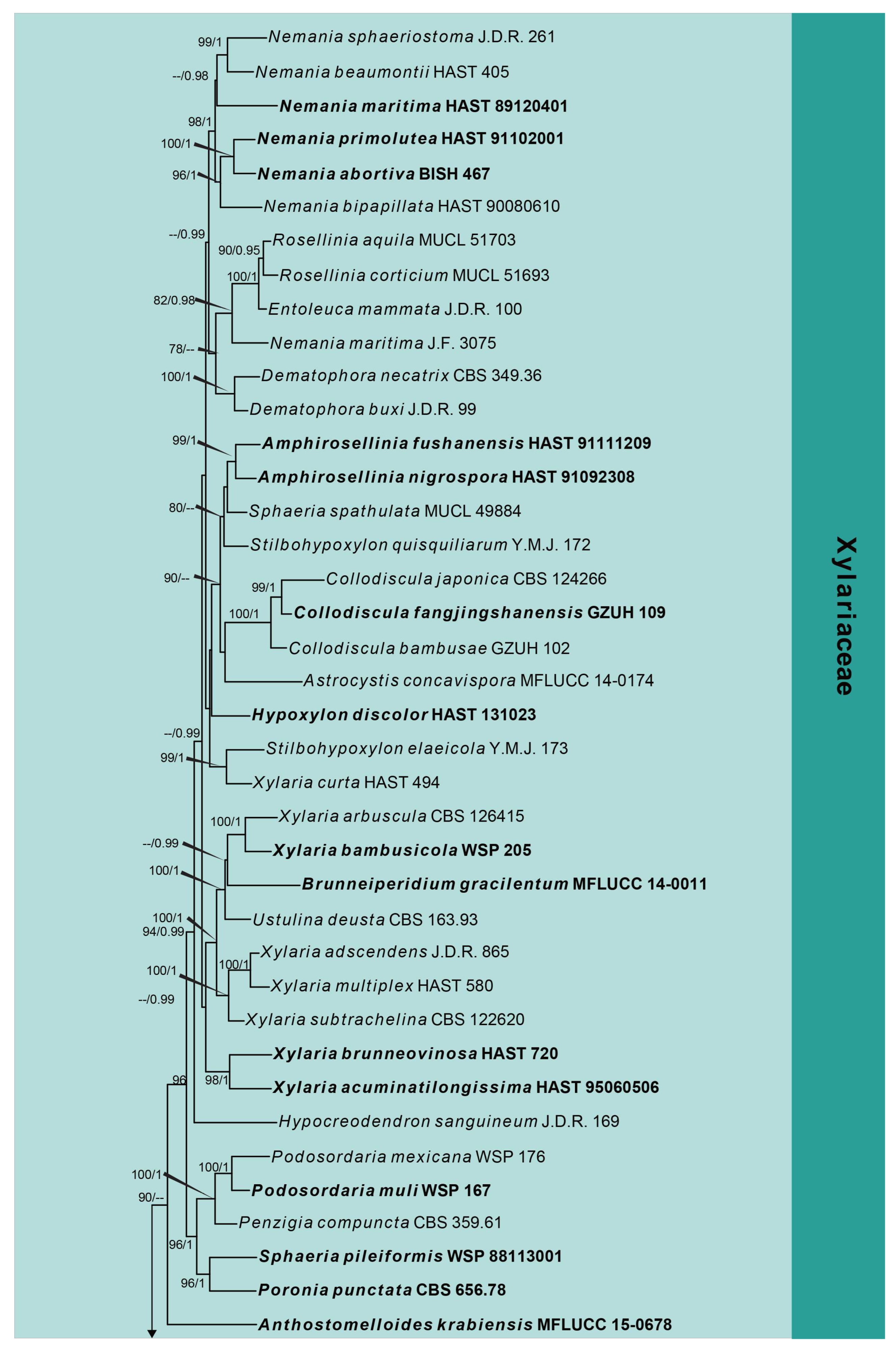
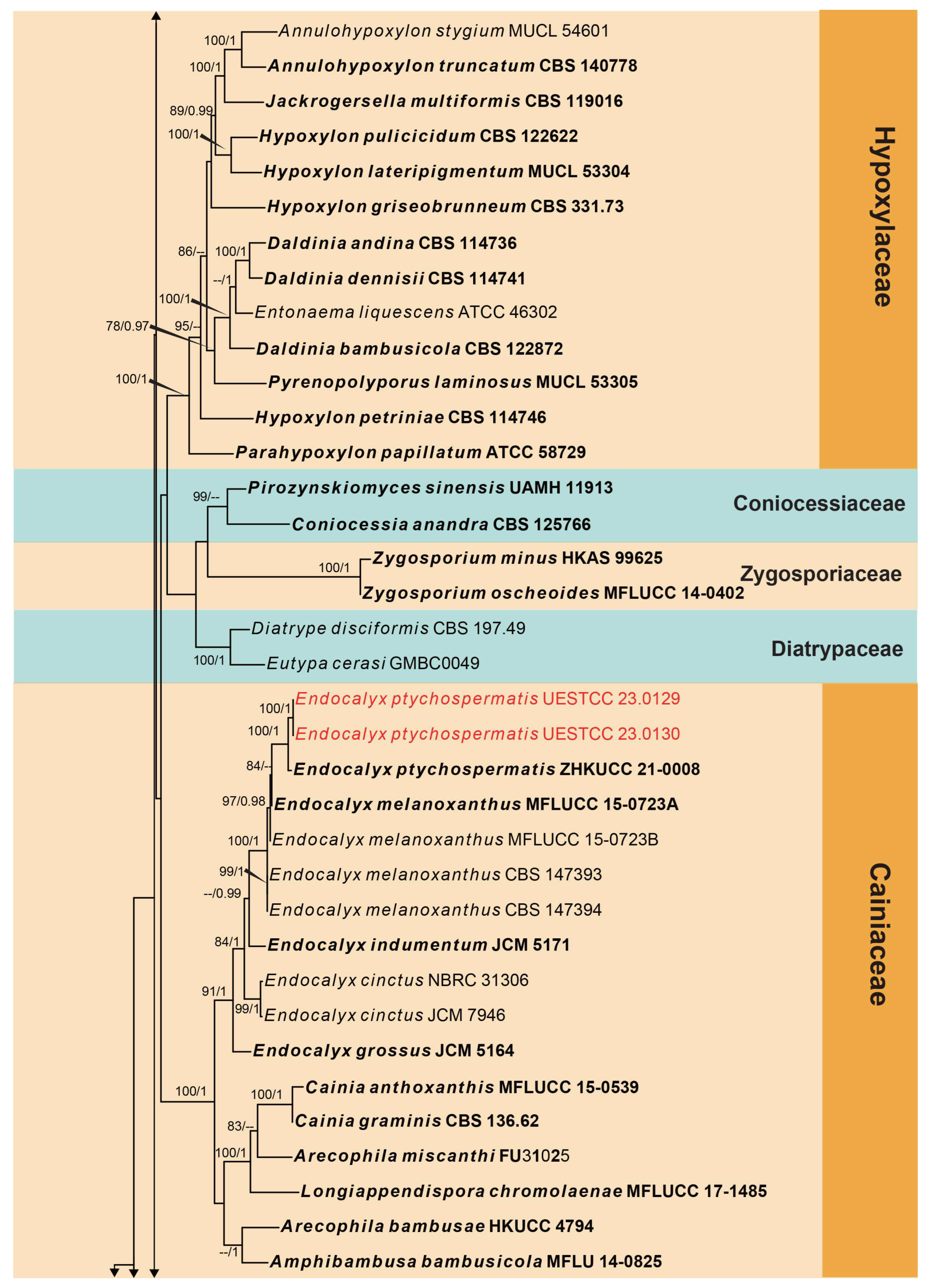
3. Results
3.1. Phylogenetic Analyses
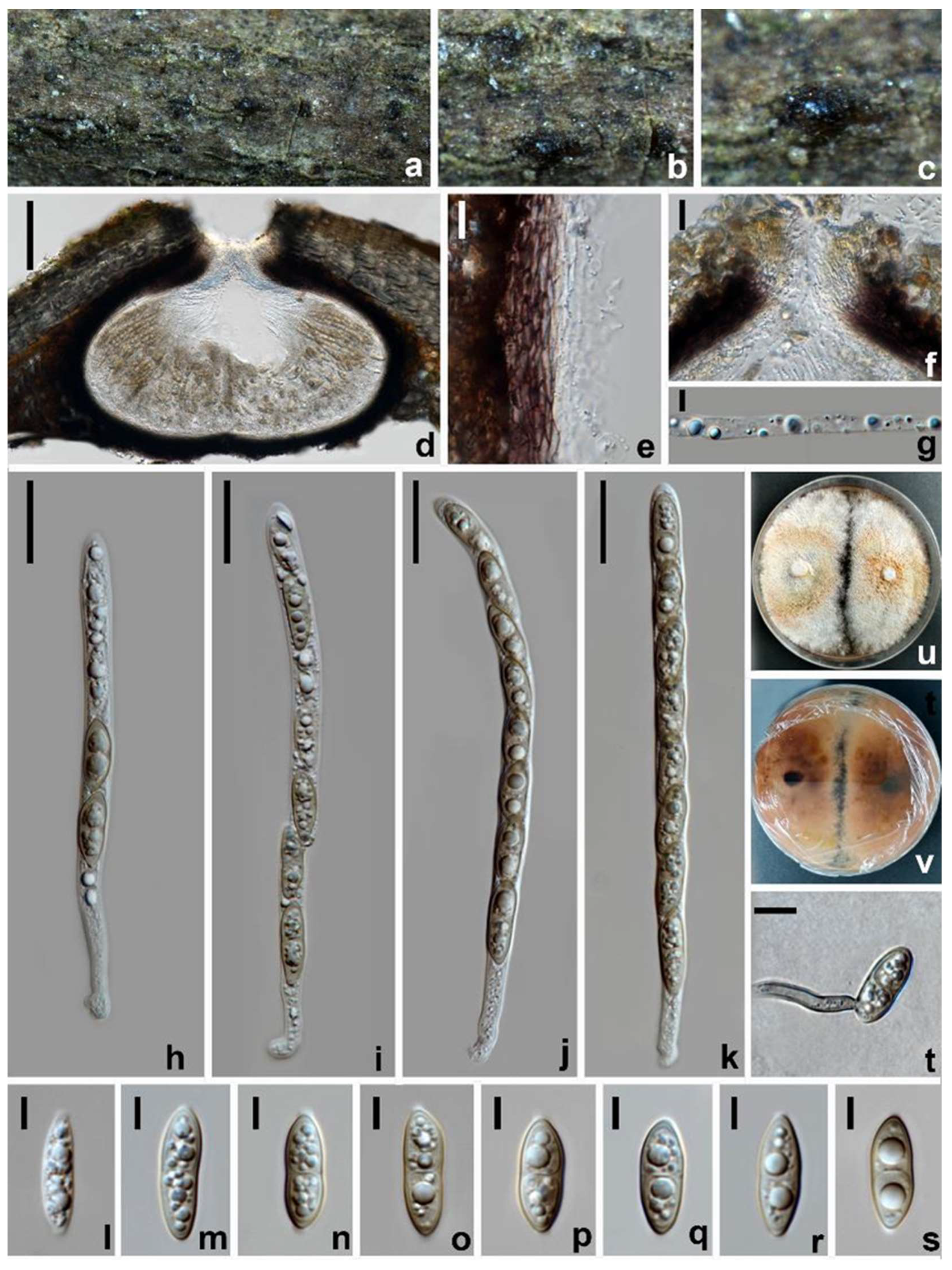
3.2. Taxonomy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Samarakoon, M.C.; Hyde, K.D.; Maharachchikumbura, S.S.N.; Stadler, M.; Gareth Jones, E.B.; Promputtha, I.; Suwannarach, N.; Camporesi, E.; Bulgakov, T.S.; Liu, J.K. Taxonomy, phylogeny, molecular dating and ancestral state reconstruction of Xylariomycetidae (Sordariomycetes). Fungal Divers. 2022, 112, 1–88. [Google Scholar] [CrossRef]
- Chen, Y.P.; Su, P.W.; Hyde, K.D.; Maharachchikumbura, S.S.N. Phylogenomics and diversification of Sordariomycetes. Mycosphere 2023, 14, 414–451. [Google Scholar] [CrossRef]
- Daranagama, D.A.; Hyde, K.D.; Sir, E.B.; Thambugala, K.M.; Tian, Q.; Samarakoon, M.C.; McKenzie, E.H.C.; Jayasiri, S.C.; Tibpromma, S.; Bhat, J.D.; et al. Towards a natural classification and backbone tree for Graphostromataceae, Hypoxylaceae, Lopadostomataceae and Xylariaceae. Fungal Divers. 2017, 88, 1–165. [Google Scholar] [CrossRef]
- Wendt, L.; Sir, E.B.; Kuhnert, E.; Heitkämper, S.; Lambert, C.; Hladki, A.I.; Romero, A.I.; Luangsa-ard, J.J.; Srikitikulchai, P.; Peršoh, D.; et al. Resurrection and emendation of the Hypoxylaceae, recognised from a multigene phylogeny of the Xylariales. Mycol. Prog. 2017, 17, 115–154. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Chen, Y.P.; Ariyawansa, H.A.; Hyde, K.D.; Haelewaters, D.; Perera, R.H.; Samarakoon, M.C.; Wanasinghe, D.N.; Bustamante, D.E.; Liu, J.K.; et al. Integrative approaches for species delimitation in Ascomycota. Fungal Divers. 2021, 109, 155–179. [Google Scholar] [CrossRef]
- Shenoy, B.D.; Jeewon, R.; Wu, W.P.; Bhat, D.J.; Hyde, K.D. Ribosomal and RPB2 DNA sequence analyses suggest that Sporidesmium and morphologically similar genera are polyphyletic. Mycol. Res. 2006, 110, 916–928. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.V.; Saxena, R.K.; et al. Outline of Fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Liu, J.K.; Chomnunti, P.; Cai, L.; Phookamsak, R. Phylogeny and morphology of Neodeightonia palmicola sp. nov. from palms. Sydowia 2010, 62, 261–276. [Google Scholar]
- Lu, B.S. A World Monograph of Anthostomella. Ph.D. Thesis, The University of Hong Kong, Pok Fu Lam, Hong Kong, 2000. [Google Scholar]
- Voglmayr, H.; Friebes, G.; Gardiennet, A.; Jaklitsch, W.M. Barrmaelia and Entosordaria in Barrmaeliaceae (fam. nov., Xylariales) and critical notes on Anthostomella-like genera based on multigene phylogenies. Mycol. Prog. 2018, 17, 155–177. [Google Scholar] [CrossRef]
- Krug, J. The genus Cainia and a new family, Cainiaceae. Sydowia 1978, 30, 122–133. [Google Scholar]
- Hyde, K.D.; Norphanphoun, C.; Maharachchikumbura, S.S.N.; Bhat, D.J.; Jones, E.B.G.; Bundhun, D.; Chen, Y.J.; Bao, D.F.; Boonmee, S.; Calabon, M.S.; et al. Refined families of Sordariomycetes. Mycosphere 2020, 11, 305–1059. [Google Scholar] [CrossRef]
- Konta, S.; Hyde, K.D.; Eungwanichayapant, P.D.; Karunarathna, S.C.; Samarakoon, M.C.; Xu, J.; Dauner, L.A.; Aluthwattha, S.T.; Lumyong, S.; Tibpromma, S. Multigene phylogeny reveals Haploanthostomella elaeidis gen. et sp. nov. and familial replacement of Endocalyx (Xylariales, Sordariomycetes, Ascomycota). Life 2021, 11, 486. [Google Scholar] [CrossRef] [PubMed]
- Delgado, G.; Miller, A.N.; Hashimoto, A.; Iida, T.; Ohkuma, M.; Okada, G. A phylogenetic assessment of Endocalyx (Cainiaceae, Xylariales) with E. grossus comb. et stat. nov. Mycol. Prog. 2022, 21, 221–242. [Google Scholar] [CrossRef]
- Phukhamsakda, C.; Nilsson, R.H.; Bhunjun, C.S.; de Farias, A.R.G.; Sun, Y.R.; Wijesinghe, S.N.; Raza, M.; Bao, D.F.; Lu, L.; Tibpromma, S.; et al. The numbers of fungi: Contributions from traditional taxonomic studies and challenges of metabarcoding. Fungal Divers. 2022, 114, 327–386. [Google Scholar] [CrossRef]
- Hughes, S.J. Fungi from the Gold Coast II. Mycol. Pap. 1953, 50, 1–104. [Google Scholar]
- Hughes, S.J. New Zealand fungi 25. Miscellaneous species. N. Z. J. Bot. 1978, 16, 311–370. [Google Scholar] [CrossRef]
- Okada, G.; Tubaki, K.J.M. A new species and a new variety of Endocalyx (Deuteromycotina) from Japan. Mycologia 1984, 76, 300–313. [Google Scholar] [CrossRef]
- Eriksson, O.; Hawksworth, D.L. Notes on ascomycete systematics, Nos. 1-224. Syst. Ascomycetum 1986, 5, 113–174. [Google Scholar]
- Senanayake, I.C.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Bhat, J.D.; Jones, E.B.G.; McKenzie, E.H.C.; Dai, D.Q.; Daranagama, D.A.; Dayarathne, M.C.; Goonasekara, I.D.; et al. Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Divers. 2015, 73, 73–144. [Google Scholar] [CrossRef]
- Pilze, W.H., II. Abtheilung. Ascomyceten: Gymnoasceen und Pyrenomyceten. In Rabenhorst’s Kryptogamen-Flora von Deutschland, Oesterreich und der Schweiz, 2nd ed.; Rabenhorst, G.L., Ed.; Kummer: Leipzig, Germany, 1885; Volume 21, pp. 203–238. [Google Scholar]
- Wijayawardene, N.N.; Hyde, K.D.; Lumbsch, H.T.; Liu, J.K.; Maharachchikumbura, S.S.N.; Ekanayaka, A.H.; Tian, Q.; Phookamsak, R. Outline of ascomycota: 2017. Fungal Divers. 2018, 88, 167–263. [Google Scholar] [CrossRef]
- Samarakoon, M.C.; Maharachchikumbura, S.S.N.; Liu, J.K.; Hyde, K.D.; Promputtha, I.; Stadler, M. Molecular Phylogeny and Morphology of Amphisphaeria (= Lepteutypa) (Amphisphaeriaceae). J. Fungi 2020, 6, 174. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Lian, T.T.; Mai, X.M.; Camporesi, E.; Zeng, Y.J.; Tian, S.L.; Xie, N. Taxonomy and phylogeny of Amphisphaeria acericola sp. nov. from Italy. Phytotaxa 2019, 403, 285–292. [Google Scholar] [CrossRef]
- Wang, Y.Z.; Aptroot, A.; Hyde, K.D. Revision of the genus Amphisphaeria. Hong Kong SAR, China. Fungal Divers. Res. Ser. 2004, 13, 1–168. [Google Scholar]
- Phookamsak, R.; Lu, Y.Z.; Hyde, K.D.; Jeewon, R.; Li, J.F.; Doilom, M.; Boonmee, S.; Promputtha, I. Phylogenetic characterization of two novel Kamalomyces species in Tubeufiaceae (Tubeufiales). Mycol. Prog. 2018, 17, 647–660. [Google Scholar] [CrossRef]
- Samarakoon, M.C.; Liu, J.K.; Hyde, K.D.; Promputtha, I. Two new species of Amphisphaeria (Amphisphaeriaceae) from northern Thailand. Phytotaxa 2019, 391, 207–217. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Rathnayaka, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dissanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid Genetic Identification and Mapping of Enzymatically Amplified Ribosomal DNA from Several Cryptococcus Species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Cigelnik, E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungusfusariumare nonorthologous. Mol. Phylogenet. Evol. 1997, 7, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Rehner, S.A.; Samuels, G.J. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 1994, 98, 625–634. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press, Inc.: New York, NY, USA, 1990; Volume 18, pp. 315–322. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Nylander, J. MrModeltest v25; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Rannala, B.; Yang, Z. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996, 43, 304–311. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree, a Graphical Viewer of Phylogenetic Trees; Institute of Evolutionary Biology University of Edinburgh: Edinburgh, UK, 2009. [Google Scholar]
- Tsui, C.K.M.; Hyde, K.D.; Hodgkiss, I.J. Paraniesslia tuberculata gen. et sp. nov., and new records or species of Clypeosphaeria, Leptosphaeria and Astrosphaeriella in Hong Kong freshwater habitats. Mycologia 2001, 93, 1002–1009. [Google Scholar] [CrossRef]
- Jaklitsch, W.M.; Gardiennet, A.; Voglmayr, H. Resolution of morphology-based taxonomic delusions: Acrocordiella, Basiseptospora, Blogiascospora, Clypeosphaeria, Hymenopleella, Lepteutypa, Pseudapiospora, Requienella, Seiridium and Strickeria. Persoonia 2016, 37, 82–105. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Abreu, V.P.; Bazzicalupo, A.; Thilini Chethana, K.W.; Clericuzio, M.; Dayarathne, M.C.; Dissanayake, A.J.; Ekanayaka, A.H.; He, M.Q.; et al. Fungal diversity notes 603–708: Taxonomic and phylogenetic notes on genera and species. Fungal Divers. 2017, 87, 1–235. [Google Scholar] [CrossRef]
- Daranagama, D.A.; Camporesi, E.; Tian, Q.; Liu, X.; Chamyuang, S.; Stadler, M.; Hyde, K.D. Anthostomella is polyphyletic comprising several genera in Xylariaceae. Fungal Divers. 2015, 73, 203–238. [Google Scholar] [CrossRef]
- Minter, D.W.; Cannon, P.F. Anthostomella tomicoides. In Descriptions of Fungi and Bacteria; Sheet 1990; CABI: Wallingford, UK, 2014. [Google Scholar]
- Seonju, L.; Crous, P.W. New species of Anthostomella on fynbos, with a key to the genus in South Africa. Mycol. Res. 2003, 107, 360–370. [Google Scholar]
- Ellis, J.B.; Everhart, B.M. New West American Fungi; University of California: Los Angeles, CA, USA, 1893. [Google Scholar]
- Jepson, W.L. Erythea: A Journal of Botany, West American and General; University of California: Los Angeles, CA, USA, 1893; Volume 1. [Google Scholar]
- Barr, M.E. Prodromus to Class Loculoascomycetes; Hamilton I. Newell, Inc.: Amherst, MA, USA, 2004. [Google Scholar]
- Hsieh, H.M.; Lin, C.R.; Fang, M.J.; Rogers, J.D.; Fournier, J.; Lechat, C.; Ju, Y.M. Phylogenetic status of Xylaria subgenus Pseudoxylaria among taxa of the subfamily Xylarioideae (Xylariaceae) and phylogeny of the taxa involved in the subfamily. Mol. Phylogenet. Evol. 2010, 54, 957–969. [Google Scholar] [CrossRef]
- Jaklitsch, W.M.; Fournier, J.; Rogers, J.D.; Voglmayr, H. Phylogenetic and taxonomic revision of Lopadostoma. Persoonia 2014, 32, 52–82. [Google Scholar] [CrossRef] [PubMed]
- Willis, K.J. State of the World’s Fungi 2018. Report; CABI: Wallingford, UK, 2018; pp. 1–92. [Google Scholar]
- Rogers, J.D. The Xylariaceae: Systematic, biological and evolutionary aspects. Mycologia 1979, 71, 1–42. [Google Scholar] [CrossRef]
- Suwannasai, N.; Whalley, M.A.; Whalley, A.J.; Thienhirun, S.; Sihanonth, P. Ascus apical apparatus and ascospore characters in Xylariaceae. IMA Fungus 2012, 3, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Barr, M.E. Prodromus to nonlichenized, pyrenomycetous members of class Hymenoascomycetes. Mycotax 1990, 39, 43–184. [Google Scholar]
- Maharachchikumbura, S.S.M.; Hyde, K.D.; Groenewald, J.Z.; Xu, J.; Crous, P.W. Pestalotiopsis revisited. Stud. Mycol. 2014, 79, 121–186. [Google Scholar] [CrossRef]
- Jones, E.; Moss, S. Ascospore appendages of marine ascomycetes: An evaluation of appendages as taxonomic criteria. Mar. Biol. 1978, 49, 11–26. [Google Scholar] [CrossRef]
- Berkeley, M.J.; Broome, C.E. Supplement to the enumeration of fungi of Ceylon. Bot. J. Linn. Soc. 1876, 15, 82–86. [Google Scholar] [CrossRef]






| Species | Clypeus | Apical Ring | Ascospores | Germ Slit | Sheath | References | |||
|---|---|---|---|---|---|---|---|---|---|
| Shape | Size (μm) | Color of Dwarf Cells | Size of Dwarf Cells (μm) | Larger Cell | |||||
| Anthostomella castnopsis | Absent | Wedge-shaped | 5–8 × 5–6.5 | Hyaline | 5–8 × 5–6.5 | Hyaline | Absent | Absent | [49] |
| A. clypeata | Not dense, black or sometimes with a halo around the central pore, subglobose | Wedge-shaped | / | Hyaline | 2–3 × 2–2.5 | Brown | Longitudinal, very fine | Absent | [47] |
| A. cynaroides | Black, ellipsoidal, margin indistinct | Wedge-shaped | 4.5–5 × 3.5–4 | Hyaline | 1.5–2 × 3 | Brown | Absent | Absent | [47] |
| A. formosa | Absent | Absent | / | Hyaline | 1.5–2 | Dark brown | Longitudinal | Narrow hyaline | [45] |
| A. proteae | Absent | Wedge-shaped | 4–5 × (2–)3 | Hyaline | 1.5–2 × 2–3 | Dark brown | Longitudinal, indistinct | Gelatinous | [47] |
| A. tomicoides | Dense, with conspicuous blackened patches | / | 2 × 3 | Hyaline | 1.5–2 | Brown | Longitudinal, indistinct | Present | [46] |
| Bicellulospora elaeidis | Black, subglobose | Discoid | 0.8–1.6 × 1.4–2 | Hyaline, olivaceous green | 3.3–4.4 × 3.3–4.2 µm | Hyaline, yellow, dark brown | Absent | Absent | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.-L.; Liang, R.-R.; Yang, J.; Liu, J.-K. Morpho-Molecular Characterization Reveals a New Genus, Three Novel Species and Two New Host Records in Xylariomycetidae. J. Fungi 2024, 10, 189. https://doi.org/10.3390/jof10030189
Li W-L, Liang R-R, Yang J, Liu J-K. Morpho-Molecular Characterization Reveals a New Genus, Three Novel Species and Two New Host Records in Xylariomycetidae. Journal of Fungi. 2024; 10(3):189. https://doi.org/10.3390/jof10030189
Chicago/Turabian StyleLi, Wen-Li, Rui-Ru Liang, Jing Yang, and Jian-Kui Liu. 2024. "Morpho-Molecular Characterization Reveals a New Genus, Three Novel Species and Two New Host Records in Xylariomycetidae" Journal of Fungi 10, no. 3: 189. https://doi.org/10.3390/jof10030189
APA StyleLi, W.-L., Liang, R.-R., Yang, J., & Liu, J.-K. (2024). Morpho-Molecular Characterization Reveals a New Genus, Three Novel Species and Two New Host Records in Xylariomycetidae. Journal of Fungi, 10(3), 189. https://doi.org/10.3390/jof10030189







