Construction of a Cosmid-Based Ultraefficient Genomic Library System for Filamentous Fungi of the Genus Aspergillus
Abstract
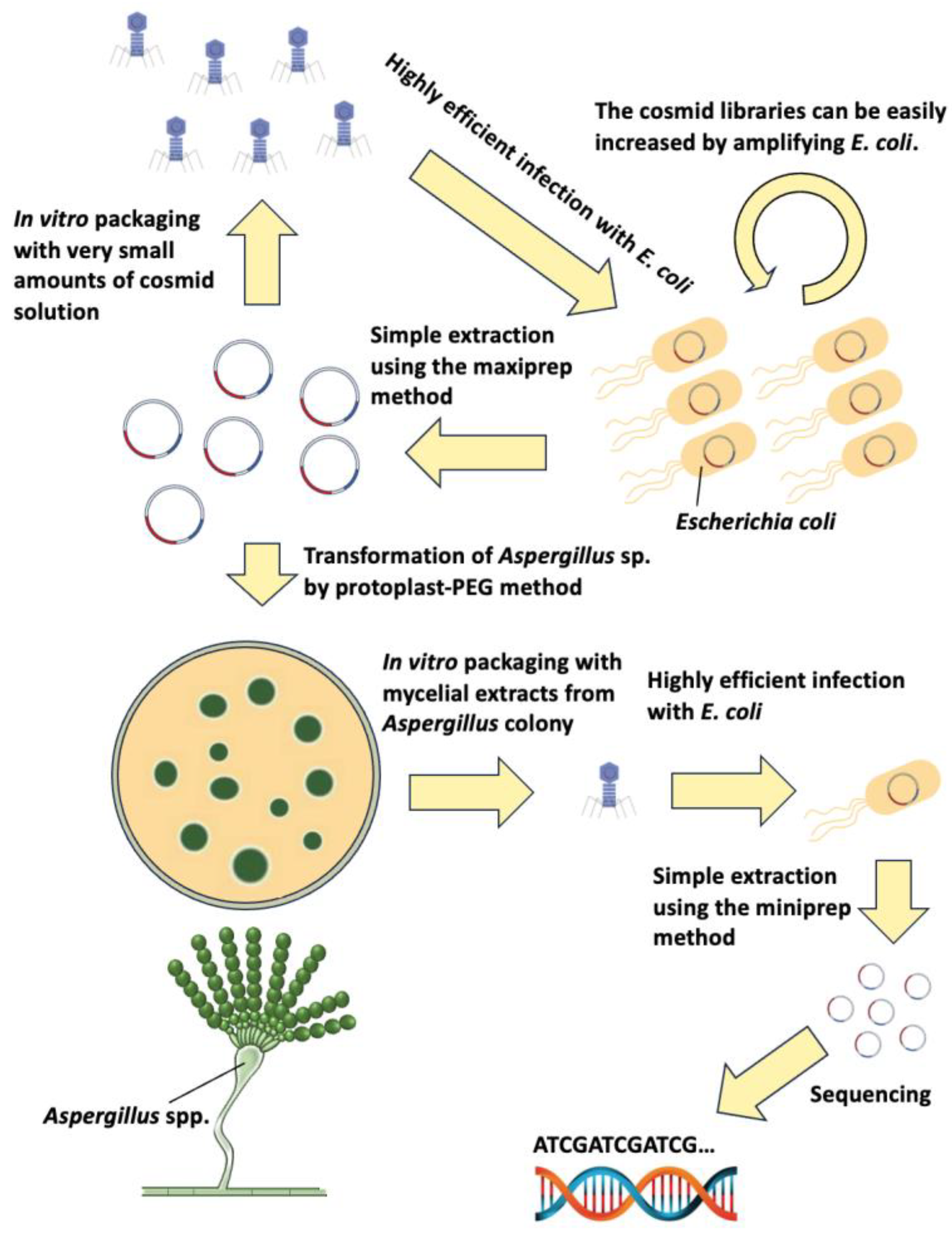
1. Introduction
2. Materials and Methods
2.1. Strains and Medium
2.2. Construction of the Shuttle Cosmid Vector pTOCK1
2.3. Construction of the A. nidulans Cosmid Library pTOCK1-gAKU89
2.4. Construction of AKU89P
2.5. Construction of AKU89PPAP
2.6. Construction of the gfsA Mutant Strain
2.7. Construction of pHSG396-nat1
2.8. Construction of AKU89PY
3. Results
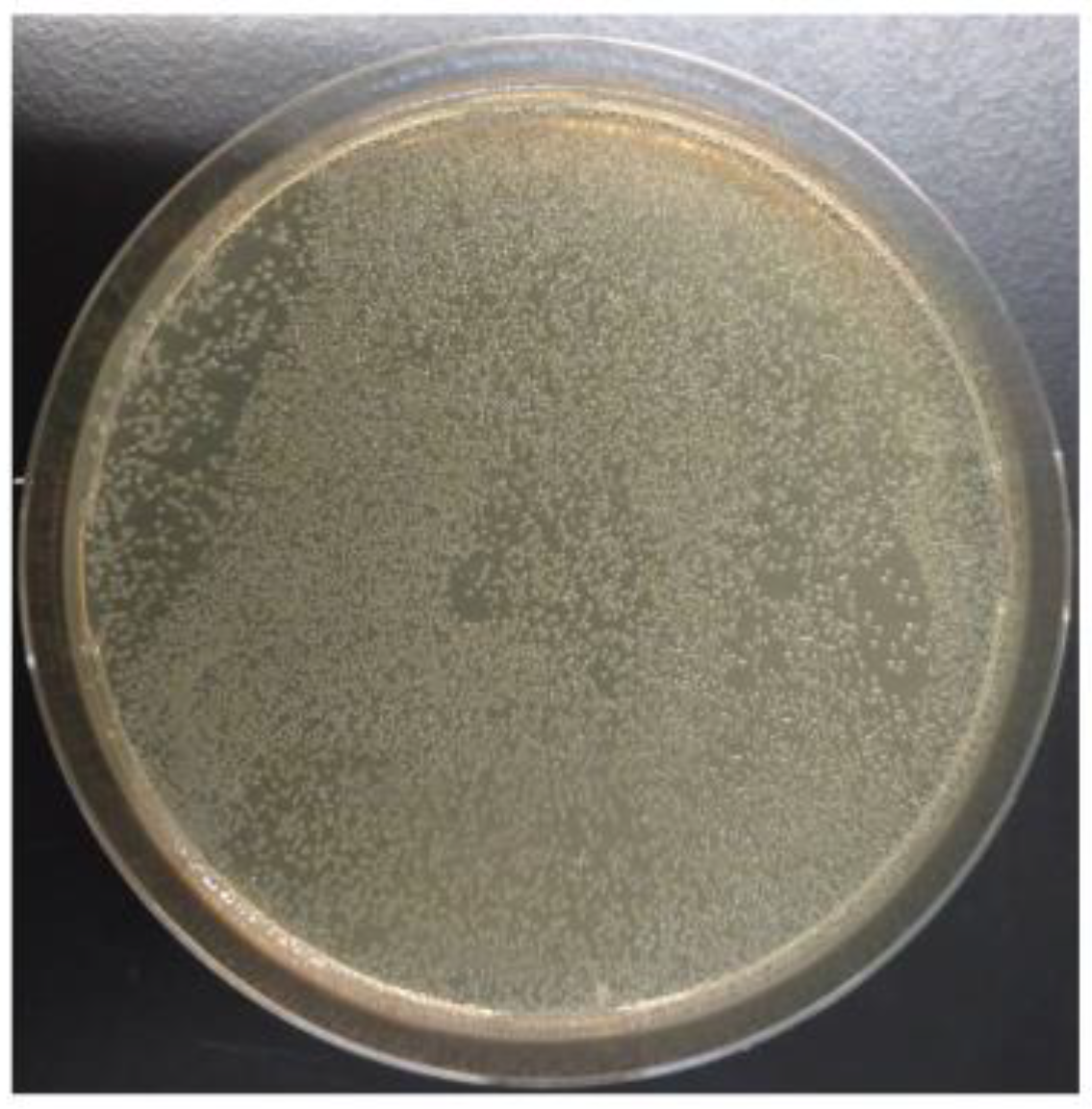
3.1. Construction of the A. nidulans Genomic Library pTOCK1-gAKU89
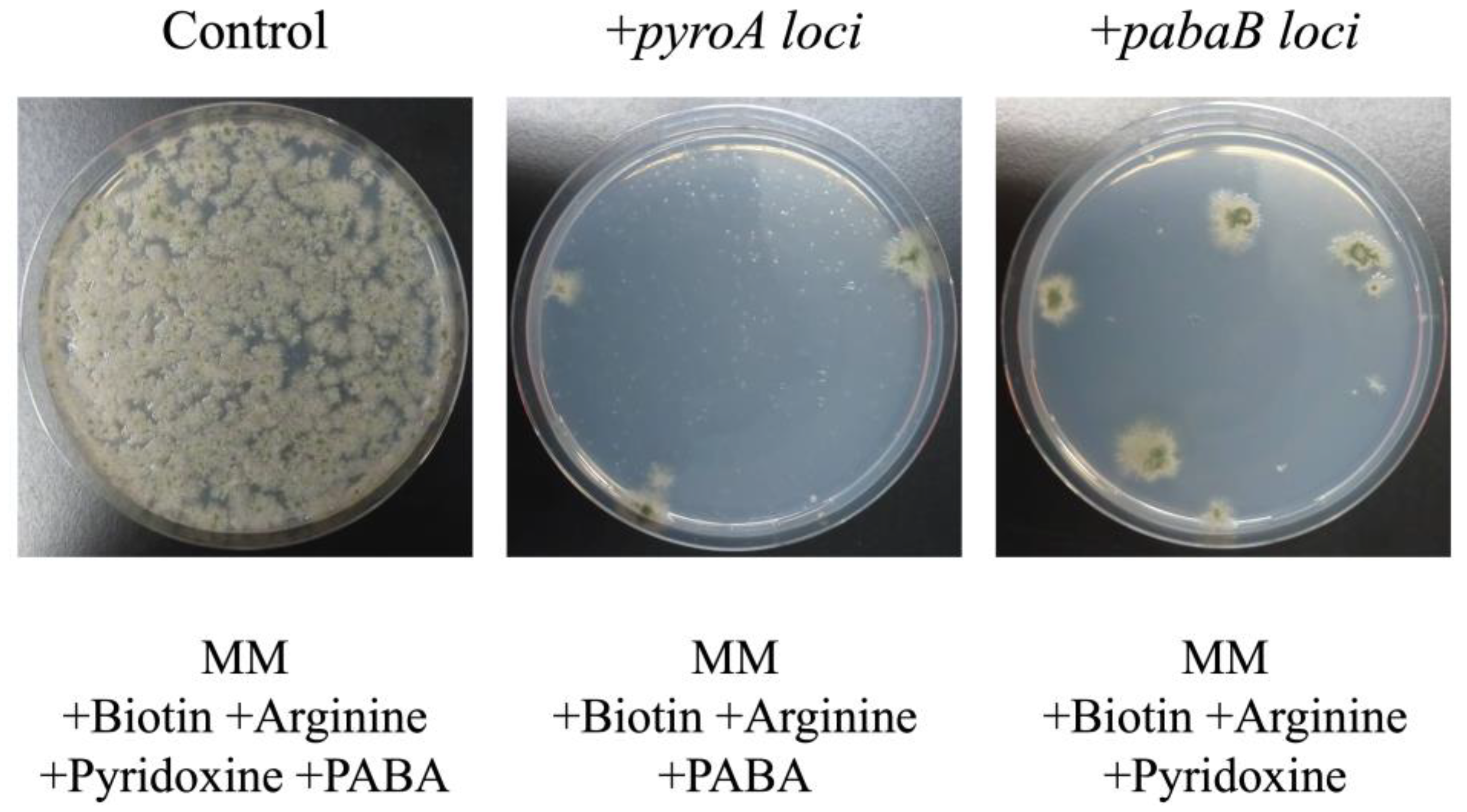
3.2. Complementation Analysis of Pyridoxine- and 4-Aminobenzoate-Auxotrophic Strains Using pTOCK1-gAKU89
3.3. Cosmid Rescue Techniques Using In Vitro Packaging
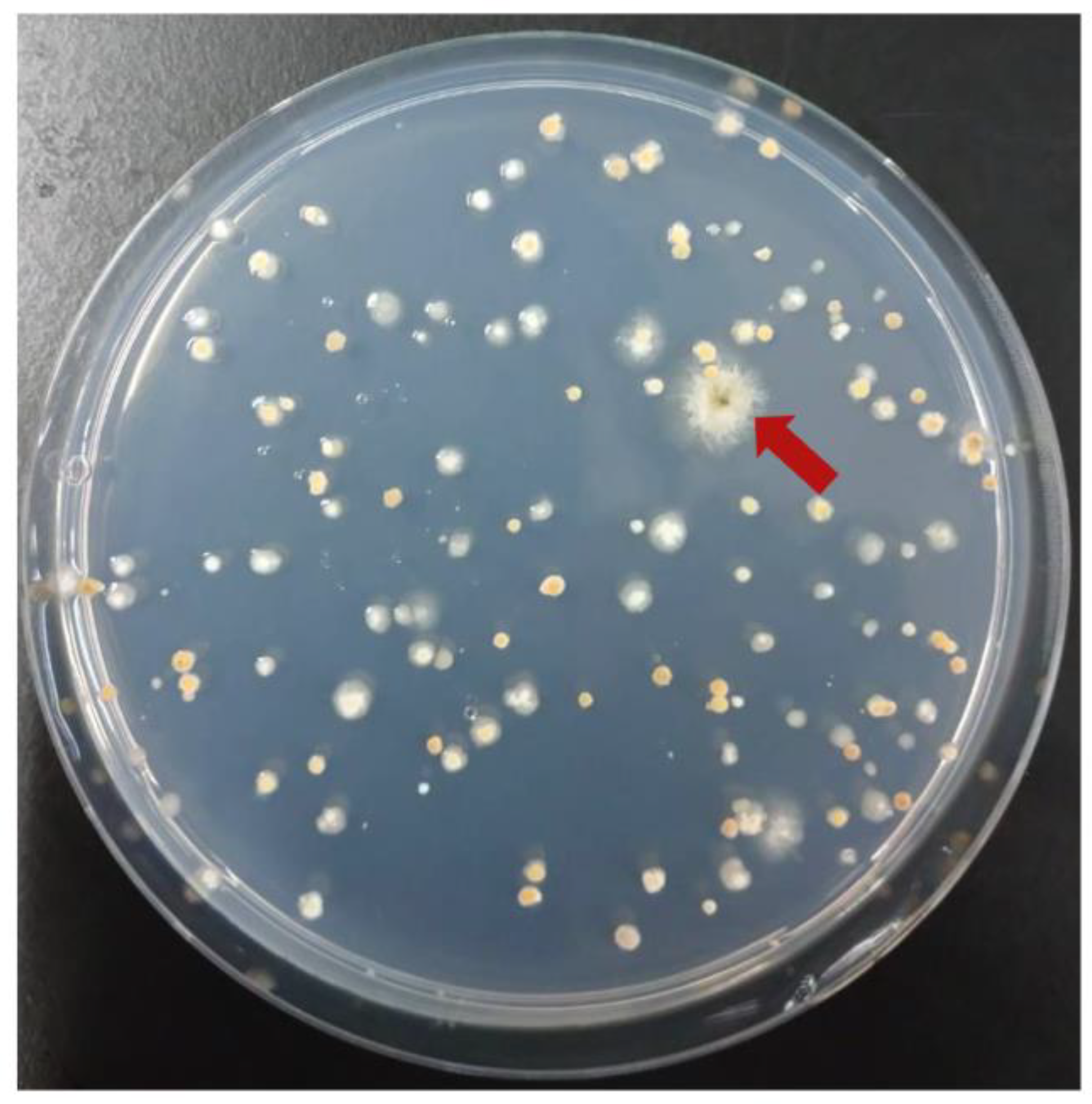
3.4. Complementation of gfsA with Cosmid Libraries
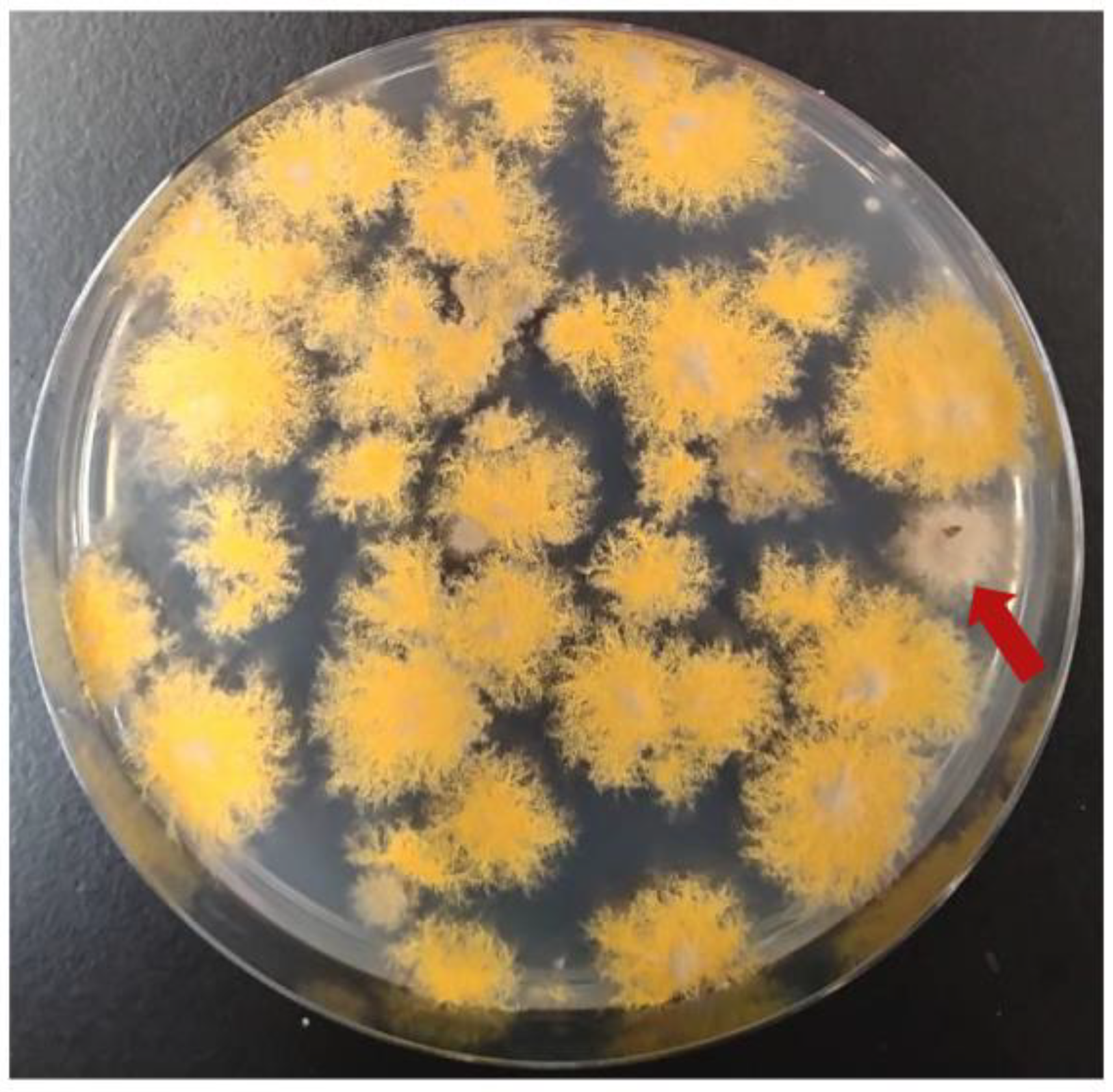
3.5. Complementation of yA with Cosmid Libraries
3.6. Reamplification of Cosmid Libraries
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Samson, R.A.; Visagie, C.M.; Houbraken, J.; Hong, S.B.; Hubka, V.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Susca, A.; Tanney, J.B.; et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 2014, 78, 141–173. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.C.; Larsen, T.O. Chemodiversity in the genus Aspergillus. Appl. Microbiol. Biotechnol. 2015, 99, 7859–7877. [Google Scholar] [CrossRef] [PubMed]
- De Vries, R.P.; Riley, R.; Wiebenga, A.; Aguilar-Osorio, G.; Amillis, S.; Uchima, C.A.; Anderluh, G.; Asadollahi, M.; Askin, M.; Barry, K.; et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017, 18, 28. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Sheng, Y.; Tang, Z.; Pan, J.; Wang, S.; Tang, B.; Zhou, T.; Shi, L.; Zhang, H. Polyketides as secondary metabolites from the genus Aspergillus. J. Fungi 2023, 9, 261. [Google Scholar] [CrossRef]
- Galagan, J.E.; Calvo, S.E.; Cuomo, C.; Ma, L.J.; Wortman, J.R.; Batzoglou, S.; Lee, S.I.; Baştürkmen, M.; Spevak, C.C.; Clutterbuck, J.; et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 2005, 438, 1105–1115. [Google Scholar] [CrossRef]
- Nierman, W.C.; Pain, A.; Anderson, M.J.; Wortman, J.R.; Kim, H.S.; Arroyo, J.; Berriman, M.; Abe, K.; Archer, D.B.; Bermejo, C.; et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 2005, 438, 1151–1156. [Google Scholar] [CrossRef]
- Machida, M.; Asai, K.; Sano, M.; Tanaka, T.; Kumagai, T.; Terai, G.; Kusumoto, K.; Arima, T.; Akita, O.; Kashiwagi, Y.; et al. Genome sequencing and analysis of Aspergillus oryzae. Nature 2005, 438, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya, Y.; Suzuki, K.; Ishii, C.; Inoue, H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 2004, 101, 12248–12253. [Google Scholar] [CrossRef]
- Nayak, T.; Szewczyk, E.; Oakley, C.E.; Osmani, A.; Ukil, L.; Murray, S.L.; Hynes, M.J.; Osmani, S.A.; Oakley, B.R. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 2006, 172, 1557–1566. [Google Scholar] [CrossRef]
- Maruyama, J.; Kitamoto, K. Multiple gene disruptions by marker recycling with highly efficient gene-targeting background (DeltaligD) in Aspergillus oryzae. Biotechnol. Lett. 2008, 30, 1811–1817. [Google Scholar] [CrossRef]
- Mizutani, O.; Kudo, Y.; Saito, A.; Matsuura, T.; Inoue, H.; Abe, K.; Gomi, K. A defect of LigD (human Lig4 homolog) for nonhomologous end joining significantly improves efficiency of gene-targeting in Aspergillus oryzae. Fungal Genet. Biol. 2008, 45, 878–889. [Google Scholar] [CrossRef]
- Jin, F.J.; Wang, B.T.; Wang, Z.D.; Jin, L.; Han, P. CRISPR/Cas9-based genome editing and its application in Aspergillus species. J. Fungi 2022, 8, 467. [Google Scholar] [CrossRef] [PubMed]
- Beadle, G.W.; Tatum, E.L. Genetic control of biochemical reactions in Neurospora. Proc. Natl. Acad. Sci. USA 1941, 27, 499–506. [Google Scholar] [CrossRef]
- Tatum, E.L.; Beadle, G.W. Genetic control of biochemical reactions in Neurospora: An “Aminobenzoicless” mutant. Proc. Natl. Acad. Sci. USA 1942, 28, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, N.H. The one gene-one enzyme hypothesis. Genetics 1948, 33, 612. [Google Scholar]
- Pontecorvo, G.; Roper, J.A.; Hemmons, L.M.; Macdonald, K.D.; Bufton, A.W. The genetics of Aspergillus nidulans. Adv. Genet. 1953, 5, 141–238. [Google Scholar] [CrossRef]
- Todd, R.B.; Davis, M.A.; Hynes, M.J. Genetic manipulation of Aspergillus nidulans: Meiotic progeny for genetic analysis and strain construction. Nat. Protoc. 2007, 2, 811–821. [Google Scholar] [CrossRef]
- Ramer, S.W.; Elledge, S.J.; Davis, R.W. Dominant genetics using a yeast genomic library under the control of a strong inducible promoter. Proc. Natl. Acad. Sci. USA 1992, 89, 11589–11593. [Google Scholar] [CrossRef]
- Adams, T.H.; Wieser, J.K.; Yu, J.H. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 1998, 62, 35–54. [Google Scholar] [CrossRef]
- Punt, P.J.; Schuren, F.H.; Lehmbeck, J.; Christensen, T.; Hjort, C.; van den Hondel, C.A. Characterization of the Aspergillus niger prtT, a unique regulator of extracellular protease encoding genes. Fungal Genet. Biol. 2008, 45, 1591–1599. [Google Scholar] [CrossRef] [PubMed]
- Yelton, M.M.; Timberlake, W.E.; Hondel, C.A. A cosmid for selecting genes by complementation in Aspergillus nidulans: Selection of the developmentally regulated yA locus. Proc. Natl. Acad. Sci. USA 1985, 82, 834–838. [Google Scholar] [CrossRef]
- Bok, J.W.; Keller, N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell. 2004, 3, 527–535. [Google Scholar] [CrossRef]
- Borgia, P.T.; Dodge, C.L.; Eagleton, L.E.; Adams, T.H. Bidirectional gene transfer between Aspergillus fumigatus and Aspergillus nidulans. FEMS Microbiol. Lett. 1994, 122, 227–231. [Google Scholar] [CrossRef][Green Version]
- Gems, D.H.; Clutterbuck, A.J. Co-transformation with autonomously-replicating helper plasmids facilitates gene cloning from an Aspergillus nidulans gene library. Curr. Genet. 1993, 24, 520–524. [Google Scholar] [CrossRef]
- Goto, M.; Harada, Y.; Oka, T.; Matsumoto, S.; Takegawa, K.; Furukawa, K. Protein O-mannosyltransferases B and C support hyphal development and differentiation in Aspergillus nidulans. Eukaryot. Cell. 2009, 8, 1465–1474. [Google Scholar] [CrossRef]
- Komachi, Y.; Hatakeyama, S.; Motomatsu, H.; Futagami, T.; Kizjakina, K.; Sobrado, P.; Ekino, K.; Takegawa, K.; Goto, M.; Nomura, Y.; et al. GfsA encodes a novel galactofuranosyltransferase involved in biosynthesis of galactofuranose antigen of O-glycan in Aspergillus nidulans and Aspergillus fumigatus. Mol. Microbiol. 2013, 90, 1054–1073. [Google Scholar] [CrossRef]
- Ling, S.O.; Storms, R.; Zheng, Y.; Rodzi, M.R.; Mahadi, N.M.; Illias, R.M.; Abdul Murad, A.M.; Abu Bakar, F.D. Development of a pyrG mutant of Aspergillus oryzae strain S1 as a host for the production of heterologous proteins. Sci. World J. 2013, 2013, 634317. [Google Scholar] [CrossRef]
- Kück, U.; Hoff, B. Application of the nourseothricin acetyltransferase gene (nat1) as dominant marker for the transformation of filamentous fungi. Fungal Genet. Rep. 2006, 53, 3. [Google Scholar] [CrossRef]
- Kadooka, C.; Hira, D.; Tanaka, Y.; Chihara, Y.; Goto, M.; Oka, T. Mnt1, an α-(1→2)-mannosyltransferase responsible for the elongation of N-glycans and O-glycans in Aspergillus fumigatus. Glycobiology 2020, 32, 1137–1152. [Google Scholar] [CrossRef] [PubMed]
- Gems, D.; Johnstone, I.L.; Clutterbuck, A.J. An autonomously replicating plasmid transforms Aspergillus nidulans at high frequency. Gene 1991, 98, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Katafuchi, Y.; Li, Q.; Tanaka, Y.; Shinozuka, S.; Kawamitsu, Y.; Izumi, M.; Ekino, K.; Mizuki, K.; Takegawa, K.; Shibata, N.; et al. GfsA is a β1,5-galactofuranosyltransferase involved in the biosynthesis of the galactofuran side chain of fungal-type galactomannan in Aspergillus fumigatus. Glycobiology 2017, 27, 568–581. [Google Scholar] [CrossRef]
- Chihara, Y.; Tanaka, Y.; Izumi, M.; Hagiwara, D.; Watanabe, A.; Takegawa, K.; Kamei, K.; Shibata, N.; Ohta, K.; Oka, T. Biosynthesis of β-(1→5)-galactofuranosyl chains of fungal-type and O-mannose-type galactomannans within the invasive pathogen Aspergillus fumigatus. mSphere 2020, 5, e00770-19. [Google Scholar] [CrossRef]
- Osherov, N.; Mathew, J.; May, G.S. Polarity-defective mutants of Aspergillus nidulans. Fungal Genet. Biol. 2000, 31, 181–188. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Osherov, N.; May, G. Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics 2000, 155, 647–656. [Google Scholar] [CrossRef]
- Zhou, S.; Narukami, T.; Nameki, M.; Ozawa, T.; Kamimura, Y.; Hoshino, T.; Takaya, N. Heme-biosynthetic porphobilinogen deaminase protects Aspergillus nidulans from nitrosative stress. Appl. Environ. Microbiol. 2012, 78, 103–109. [Google Scholar] [CrossRef]
- Zhou, S.; Narukami, T.; Masuo, S.; Shimizu, M.; Fujita, T.; Doi, Y.; Kamimura, Y.; Takaya, N. NO-inducible nitrosothionein mediates NO removal in tandem with thioredoxin. Nat. Chem. Biol. 2013, 9, 657–663. [Google Scholar] [CrossRef]
- Amahisa, M.; Tsukagoshi, M.; Kadooka, C.; Masuo, S.; Takeshita, N.; Doi, Y.; Takagi, H.; Takaya, N. The Metabolic regulation of amino acid synthesis counteracts reactive nitrogen stress via Aspergillus nidulans cross-pathway control. J. Fungi 2024, 10, 58. [Google Scholar] [CrossRef]
- Shaaban, M.I.; Bok, J.W.; Lauer, C.; Keller, N.P. Suppressor mutagenesis identifies a velvet complex remediator of Aspergillus nidulans secondary metabolism. Eukaryot. Cell. 2010, 9, 1816–1824. [Google Scholar] [CrossRef]
- Soukup, A.A.; Chiang, Y.M.; Bok, J.W.; Reyes-Dominguez, Y.; Oakley, B.R.; Wang, C.C.; Strauss, J.; Keller, N.P. Overexpression of the Aspergillus nidulans histone 4 acetyltransferase EsaA increases activation of secondary metabolite production. Mol. Microbiol. 2012, 86, 314–330. [Google Scholar] [CrossRef]
- Otamendi, A.; Espeso, E.A.; Etxebeste, O. Identification and characterization of Aspergillus nidulans mutants impaired in asexual development under phosphate stress. Cells 2019, 8, 1520. [Google Scholar] [CrossRef]
- Damveld, R.A.; Franken, A.; Arentshorst, M.; Punt, P.J.; Klis, F.M.; van den Hondel, C.A.; Ram, A.F. A novel screening method for cell wall mutants in Aspergillus niger identifies UDP-galactopyranose mutase as an important protein in fungal cell wall biosynthesis. Genetics 2008, 178, 873–881. [Google Scholar] [CrossRef]
- Smith, D.J.; Burnham, M.K.; Edwards, J.; Earl, A.J.; Turner, G. Cloning and heterologous expression of the penicillin biosynthetic gene cluster from Penicillum chrysogenum. Biotechnology 1990, 8, 39–41. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Kinoshita, H.; Shimizu, T.; Nihira, T. Construction of a citrinin gene cluster expression system in heterologous Aspergillus oryzae. J. Biosci. Bioeng. 2008, 106, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Kinoshita, H.; Nihira, T. Heterologous expression system in Aspergillus oryzae for fungal biosynthetic gene clusters of secondary metabolites. Appl. Microbiol. Biotechnol. 2012, 93, 2011–2022. [Google Scholar] [CrossRef] [PubMed]
- Anyaogu, D.C.; Mortensen, U.H. Heterologous production of fungal secondary metabolites in Aspergilli. Front. Microbiol. 2015, 6, 77. [Google Scholar] [CrossRef]


| Strains | Genotype | Source |
|---|---|---|
| Aspergillus nidulans | ||
| A26 | biA1 | obtained from FGSC |
| A426 | yA2, pabaB22, pyroA4 | obtained from FGSC |
| AKU89 | biA1, argB2, nkuB::aurA+ | Goto et al., 2009 [25] |
| AKU89P | biA1, argB2, nkuB::aurA+, ΔpyrG | This study |
| AKU89PPAP | biA1, argB2, nkuB::aurA+, ΔpyrG, pabaB22, pyroA4 | This study |
| ΔgfsA | biA1, argB2, nkuB::aurA+, gfsA::argB | Komachi et al., 2013 [26] |
| ΔgfsA ΔpyrG | biA1, argB2, nkuB::aurA+, ΔpyrG, gfsA::argB | This study |
| AKU89PY | biA1, argB2, nkuB::aurA+, ΔpyrG yA::nat1 | This study |
| SuperCos1-PmaCI-F2 | AAAACACGTGTACGTCTGCTTTTTGTTGACTTCC | This study |
| SuperCos1-PmaCI-R2 | AAAACACGTGCACGTCTGAAGCTAGCTTCGA | This study |
| AoPyrG-Scos1-infusion(AatII)-F | GAAAAGTGCCACCTGTATGGATCTCAGAACAATATACC | This study |
| AoPyrG-Scos1-infusion(AatII)-R | ATAATGGTTTCTTAGTGTACGATAGTGACCGACTG | This study |
| pyrG-del-1 | AGCTTAAAGAAAACGCGCAGC | This study |
| pyrG-del-2 | ACTAGAGCCGTCAGTGAGGCGATGGCGGTTCTCCAATGAT | This study |
| pyrG-del-3 | ATCATTGGAGAACCGCCATCGCCTCACTGACGGCTCTAGT | This study |
| pyrG-del-4 | GATGGTGAGGATAGGAATTGCC | This study |
| pyrG-FC | GTATCTTTCCCCCTTCAACGC | This study |
| pyrG-RC | GATCAAGGAAAGGGGCGAGG | This study |
| AN8677-7 | CGCGTAGATCACACAGGGTC | Komachi et al., 2013 [26] |
| AN8677-8 | GCATTGTTCGATCGCTCGTC | Komachi et al., 2013 [26] |
| AN8677-FC | GCTCGAATCTTCCGTATGCC | This study |
| AN8677-RC | GTCCCGCCAATAATTTTAGCCATA | This study |
| pyroA-F | AGTAGAGGCAATATAGTCCCTGCG | This study |
| pyroA-R | CAGATGTAAATGTCAAAAGGTCCGTC | This study |
| nat1-F | ATGGCCACCCTCGACGACACGGC | This study |
| nat1-R | TCAGGGGCAGGGCATGCTCATG | This study |
| AnGpdA-P-TrpC-T-F(nat1) | ATGCCCTGCCCCTGAGGATCCACTTAACGTTACTGAAATC | This study |
| AnGpdA-P-TrpC-T-R(nat1) | GTCGAGGGTGGCCATGGTGATGTCTGCTCAAGCGG | This study |
| yA-1 | CGGGCTGCAGGAATTCCCCGACAAGTTTCTCAGTGG | This study |
| yA-2 | AGAGTCGACCCCTCGGCTGGATCCCGGAGGAATCA | This study |
| yA-3 | CCCATCGATGGGGTAAAATGGGAGGAATGGCGCTG | This study |
| yA-4 | CCCATCGATGGGGTAATTGCCCTCCCTGGCGTATA | This study |
| yA-FC | GGACCTGACAATTCTCGACGT | This study |
| yA-RC | GCCGGTCAAAACTGCATCGG | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kadooka, C.; Oka, T. Construction of a Cosmid-Based Ultraefficient Genomic Library System for Filamentous Fungi of the Genus Aspergillus. J. Fungi 2024, 10, 188. https://doi.org/10.3390/jof10030188
Kadooka C, Oka T. Construction of a Cosmid-Based Ultraefficient Genomic Library System for Filamentous Fungi of the Genus Aspergillus. Journal of Fungi. 2024; 10(3):188. https://doi.org/10.3390/jof10030188
Chicago/Turabian StyleKadooka, Chihiro, and Takuji Oka. 2024. "Construction of a Cosmid-Based Ultraefficient Genomic Library System for Filamentous Fungi of the Genus Aspergillus" Journal of Fungi 10, no. 3: 188. https://doi.org/10.3390/jof10030188
APA StyleKadooka, C., & Oka, T. (2024). Construction of a Cosmid-Based Ultraefficient Genomic Library System for Filamentous Fungi of the Genus Aspergillus. Journal of Fungi, 10(3), 188. https://doi.org/10.3390/jof10030188







