Importance of the Aspergillus fumigatus Mismatch Repair Protein Msh6 in Antifungal Resistance Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Aspergillus Fumigatus Strains and Culture
2.2. Whole Genome Sequencing Analysis
2.3. Phylogenetic Analysis and Single-Nucleotide Variant Comparison
2.4. Generation of A. fumigatus Δmsh6 Strains
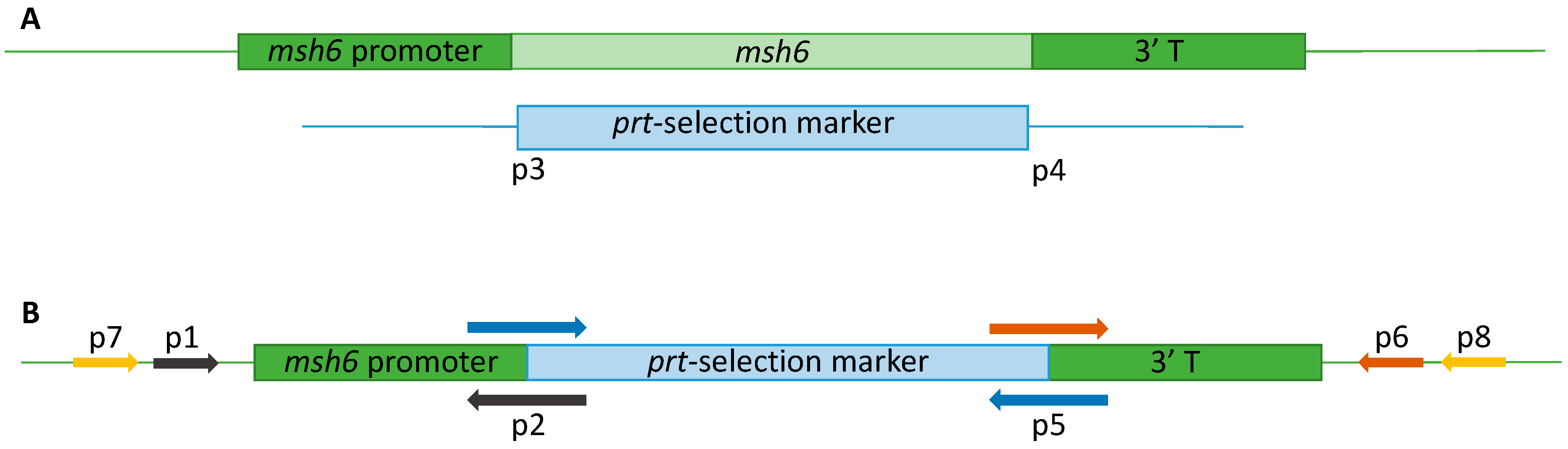
2.4.1. Vector Construction
2.4.2. Transformation
2.5. Phenotypic Characterization of Δmsh6 Mutant
2.6. Galleria Mellonella Survival Assay
2.7. Antifungal Susceptibility Testing
2.8. Mutagenesis Experiments in ∆msh6 Strains
2.9. Genetic Analysis of the Mutants
3. Results
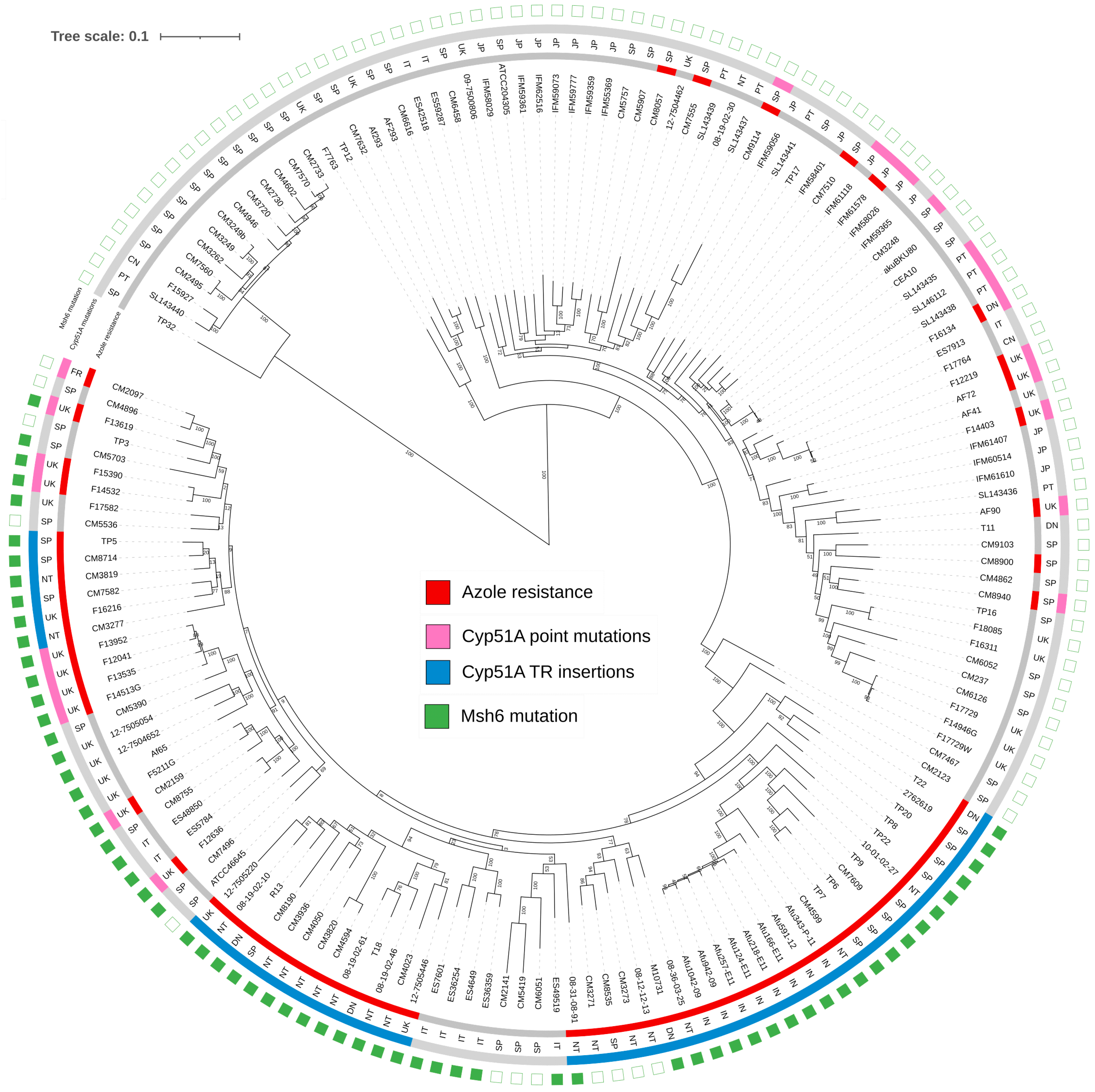
3.1. Analysis of Mutations in MMR Genes in a Collection of Isolates
3.2. Construction and Phenotypic Characterization of A. fumigatus Δmsh6 Strains
3.3. Deletion of msh6 Does Not Influence A. fumigatus Azole Susceptibility
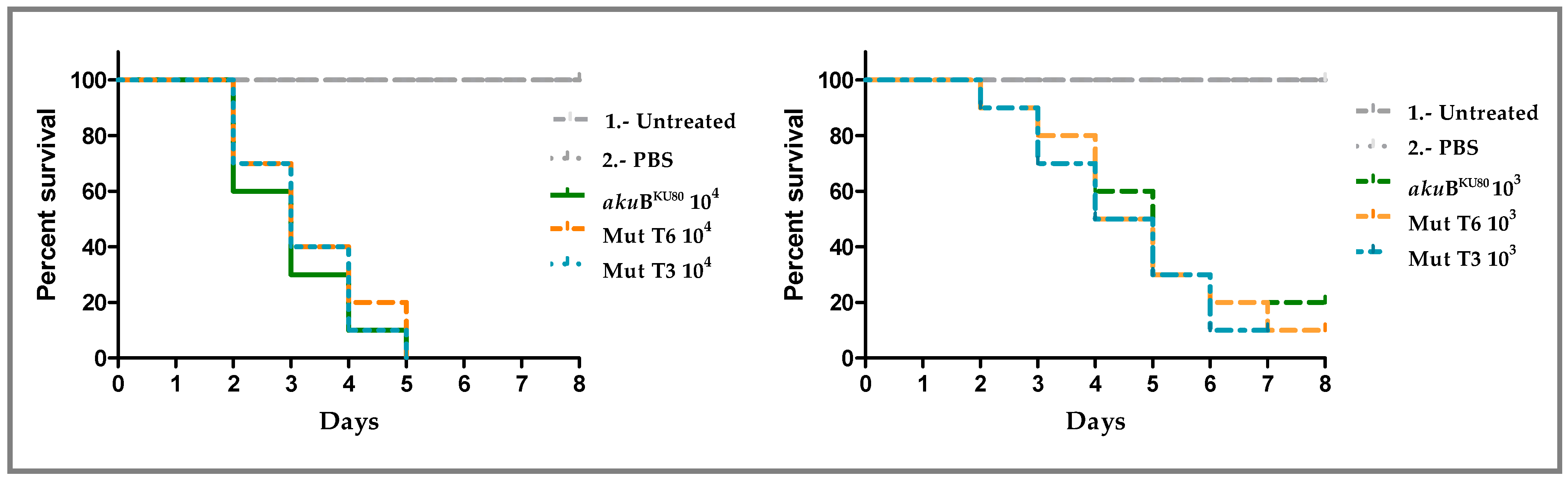
3.4. Deletion of msh6 Does Not Influence A. fumigatus Virulence
3.5. Deletion of msh6 Does Not Influence A. fumigatus Growth in Different Stress Conditions
3.6. Effects of Δmsh6 Deletion on A. fumigatus Antifungal Resistance Development
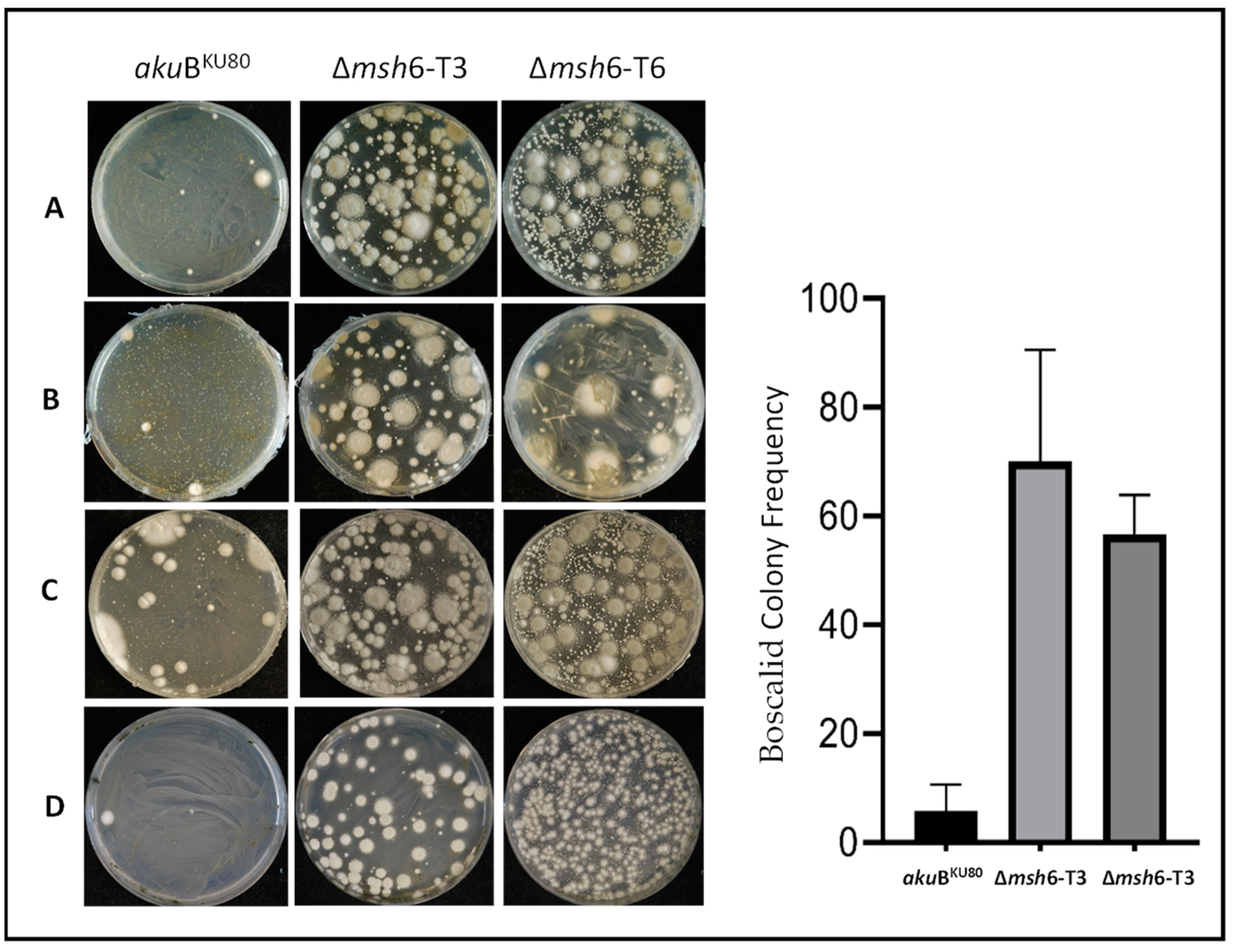
3.6.1. Development of Resistance to Azole Drugs
3.6.2. Development of Resistance to Non-Azole Fungicides
3.7. Genetic Analysis of Drug Target Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Patterson, T.F.; Thompson, G.R.; Denning, D.W.; Fishman, J.A.; Hadley, S.; Herbrecht, R.; Kontoyiannis, D.P.; Marr, K.A.; Morrison, V.A.; Nguyen, M.H.; et al. Practice guidelines for the diagnosis and management of aspergillosis: 2016 update by the infectious diseases society of America. Clin. Infect. Dis. 2016, 63, e1–e60. [Google Scholar] [CrossRef]
- Verweij, P.E.; Ananda-Rajah, M.; Andes, D.; Arendrup, M.C.; Brüggemann, R.J.; Chowdhary, A.; Cornely, O.A.; Denning, D.W.; Groll, A.H.; Izumikawa, K.; et al. International expert opinion on the management of infection caused by azole-resistant Aspergillus fumigatus. Drug Resist. Updates 2015, 21–22, 30–40. [Google Scholar] [CrossRef]
- Latgé, J.P.; Chamilos, G. Aspergillus fumigatus and aspergillosis in 2019. Clin. Microbiol. Rev. 2019, 33, e00140-18. [Google Scholar] [CrossRef]
- Lepesheva, G.I.; Waterman, M.R. Sterol 14alpha-demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms. Biochim. Biophys. Acta 2007, 1770, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rubio, R.; Gonzalez-Jimenez, I.; Lucio, J.; Mellado, E. Aspergillus fumigatus Cross-Resistance between Clinical and Demethylase Inhibitor Azole Drugs. Appl. Environ. Microbiol. 2021, 87, e02539-20. [Google Scholar] [CrossRef] [PubMed]
- Snelders, E.; Huis In ’t Veld, R.A.G.; Rijs, A.J.M.M.; Kema, G.H.J.; Melchers, W.J.G.; Verweij, P.E. Possible environmental origin of resistance of Aspergillus fumigatus to medical triazoles. Appl. Environ. Microbiol. 2009, 75, 4053–4057. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; van den Heuvel, J.; Debets, A.J.M.; Verweij, P.E.; Melchers, W.J.G.; Zwaan, B.J.; Schoustra, S.E. Evolution of cross-resistance to medical triazoles in Aspergillus fumigatus through selection pressure of environmental fungicides. Proc. R. Soc. B Biol. Sci. 2017, 284, 20170635. [Google Scholar] [CrossRef]
- Gonzalez-Jimenez, I.; Garcia-Rubio, R.; Monzon, S.; Lucio, J.; Cuesta, I.; Mellado, E. Multiresistance to nonazole fungicides in Aspergillus fumigatus TR34/L98H Azole-resistant isolates. Antimicrob. Agents Chemother. 2021, 65, AAC0064221. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rubio, R.; Cuenca-Estrella, M.; Mellado, E. Triazole Resistance in Aspergillus Species: An Emerging Problem. Drugs 2017, 77, 599–613. [Google Scholar] [CrossRef]
- Chowdhary, A.; Sharma, C.; Meis, J.F. Azole-Resistant Aspergillosis: Epidemiology, Molecular Mechanisms, and Treatment. J. Infect. Dis. 2017, 216 (Suppl. S3), S436–S444. [Google Scholar] [CrossRef]
- Fraaije, B.; Atkins, S.; Hanley, S.; Macdonald, A.; Lucas, J. The Multi-Fungicide Resistance Status of Aspergillus fumigatus Populations in Arable Soils and the Wider European Environment. Front. Microbiol. 2020, 11, 599233. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.E.; Sumabat, L.G.; Melie, T.; Mangum, B.; Momany, M.; Brewer, M.T. Evidence for the agricultural origin of resistance to multiple antimicrobials in Aspergillus fumigatus, a fungal pathogen of humans. G3 Genes Genomes Genet. 2022, 12, jkab427. [Google Scholar] [CrossRef]
- Hahn, M. The rising threat of fungicide resistance in plant Pathogenic fungi: Botrytis as a case study. J. Chem. Biol. 2014, 7, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Grasso, V.; Palermo, S.; Sierotzki, H.; Garibaldi, A.; Gisi, U. Cytochrome b gene structure and consequences for resistance to Qo inhibitor fungicides in plant pathogens. Pest Manag. Sci. 2006, 62, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Veloukas, T.; Kalogeropoulou, P.; Markoglou, A.N.; Karaoglanidis, G.S. Fitness and competitive ability of Botrytis cinerea field isolates with dual resistance to SDHI and QoI fungicides, associated with several sdhB and the cytb G143A mutations. Phytopathology 2014, 104, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Sierotzki, H.; Scalliet, G. A review of current knowledge of resistance aspects for the next-generation succinate dehydrogenase inhibitor fungicides. Phytopathology 2013, 103, 880–887. [Google Scholar] [CrossRef]
- Vela-Corcía, D.; Romero, D.; de Vicente, A.; Pérez-García, A. Analysis of β-tubulin-carbendazim interaction reveals that binding site for MBC fungicides does not include residues involved in fungicide resistance. Sci. Rep. 2018, 8, 7161. [Google Scholar] [CrossRef]
- Garcia-Rubio, R.; Monzon, S.; Alcazar-Fuoli, L.; Cuesta, I.; Mellado, E. Genome-wide comparative analysis of Aspergillus fumigatus strains: The reference genome as a matter of concern. Genes 2018, 9, 363. [Google Scholar] [CrossRef]
- dos Reis, T.F.; Silva, L.P.; de Castro, P.A.; Almeida de Lima, P.B.; do Carmo, R.A.; Marini, M.M.; da Silveira, J.F.; Ferreira, B.H.; Rodrigues, F.; Malavazi, I.; et al. The Influence of Genetic Stability on Aspergillus fumigatus Virulence and Azole Resistance. G3 Genes Genomes Genet. 2018, 8, 265–278. [Google Scholar] [CrossRef]
- dos Reis, T.F.; Silva, L.P.; de Castro, P.A.; do Carmo, R.A.; Marini, M.M.; da Silveira, J.F.; Ferreira, B.H.; Rodrigues, F.; Lind, A.L.; Rokas, A.; et al. The Aspergillus fumigatus Mismatch Repair MSH2 Homolog Is Important for Virulence and Azole Resistance. mSphere 2019, 4, e00416-19. [Google Scholar] [CrossRef]
- Boyce, K.J.; Wang, Y.; Verma, S.; Shakya VP, S.; Xue, C.; Idnurm, A. Mismatch repair of DNA replication errors contributes to microevolution in the pathogenic fungus Cryptococcus neoformans. mBio 2017, 8, 10–1128. [Google Scholar] [CrossRef]
- Byun, S.A.; Won, E.J.; Kim, M.N.; Lee, W.G.; Lee, K.; Lee, H.S.; Uh, Y.; Healey, K.R.; Perlin, D.S.; Choi, M.J.; et al. Multilocus sequence typing (MLST) genotypes of Candida glabrata bloodstream isolates in Korea: Association with antifungal resistance, mutations in mismatch repair gene (Msh2), and clinical outcomes. Front. Microbiol. 2018, 9, 1523. [Google Scholar] [CrossRef]
- Legrand, M.; Chan, C.L.; Jauert, P.A.; Kirkpatrick, D.T. Role of DNA mismatch repair and double-strand break repair in genome stability and antifungal drug resistance in Candida albicans. Eukaryot. Cell 2007, 6, 2194–2205. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Walker, G.-C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Feng, Y.; Zhang, Y.; Feng, J. DNA damage checkpoint and repair: From the budding yeast Saccharomyces cerevisiae to the pathogenic fungus Candida albicans. Comput. Struct. Biotechnol. J. 2021, 19, 6343–6354. [Google Scholar] [CrossRef]
- Iyer, R.R.; Pluciennik, A.; Burdett, V.; Modrich, P.L. DNA Mismatch Repair: Functions and Mechanisms. Chem. Rev. 2006, 106, 302–323. [Google Scholar] [CrossRef] [PubMed]
- Larrea, A.A.; Lujan, S.A.; Kunkel, T.A. SnapShot: DNA Mismatch Repair. Cell 2010, 141, 730. [Google Scholar] [CrossRef]
- Boyce, K.J. Mutators Enhance Adaptive Micro-Evolution in Pathogenic Microbes. Microorganisms 2022, 10, 442. [Google Scholar] [CrossRef]
- Phillips, M.A.; Steenwyk, J.L.; Shen, X.X.; Rokas, A. Examination of Gene Loss in the DNA Mismatch Repair Pathway and Its Mutational Consequences in a Fungal Phylum. Genome Biol. Evol. 2021, 13, evab219. [Google Scholar] [CrossRef]
- Gambhir, N.; Harris, S.D.; Everhart, S.E. Evolutionary Significance of Fungal Hypermutators: Lessons Learned from Clinical Strains and Implications for Fungal Plant Pathogens. mSphere 2022, 7, e0008722. [Google Scholar] [CrossRef]
- Alcazar-Fuoli, L.; Mellado, E.; Alastruey-Izquierdo, A.; Cuenca-Estrella, M.; Rodriguez-Tudela, J.L. Aspergillus section Fumigati: Antifungal susceptibility patterns and sequence-based identification. Antimicrob. Agents Chemother. 2008, 52, 1244–1251. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.Y. ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Alcazar-Fuoli, L.; Clavaud, C.; Lamarre, C.; Aimanianda, V.; Seidl-Seiboth, V.; Mellado, E.; Latgé, J.P. Functional analysis of the fungal/plant class chitinase family in Aspergillus fumigatus. Fungal Genet. Biol. 2011, 48, 418–429. [Google Scholar] [CrossRef]
- Szewczyk, E.; Nayak, T.; Oakley, C.E.; Edgerton, H.; Xiong, Y.; Taheri-Talesh, N.; Osmani, S.A.; Oakley, B.R. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 2006, 1, 3111–3120. [Google Scholar] [CrossRef]
- Gomez-Lopez, A.; Forastiero, A.; Cendejas-Bueno, E.; Gregson, L.; Mellado, E.; Howard, S.J.; Livermore, J.L.; Hope, W.W.; Cuenca-Estrella, M. An invertebrate model to evaluate virulence in Aspergillus fumigatus: The role of azole resistance. Med. Mycol. 2014, 52, 311–319. [Google Scholar] [CrossRef]
- Pfaller, J.B.; Messer, S.A.; Hollis, R.J.; Diekema, D.J.; Pfaller, M.A. In vitro susceptibility testing of Aspergillus spp.: Comparison of Etest and reference microdilution methods for determining voriconazole and itraconazole MICs. J. Clin. Microbiol. 2003, 41, 1126–1129. [Google Scholar] [CrossRef] [PubMed]
- Guinea, J.; Meletiadis, J.; Arikan-Akdagli, S.; Kahlmeter, G.; Arendrup, M.C.; The Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST). EUCAST DEFINITIVE DOCUMENT E.DEF 9.4 Method for the Determination of Broth Dilution Minimum Inhibitory Concentrations of Antifungal Agents for Conidia Forming Moulds. 2022. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Files/EUCAST_EDef_9.4_method_for_susceptibility_testing_of_moulds.pdf (accessed on 1 April 2022).
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs for Antifungal Agents, Version 10.0. 2020. Available online: http://www.eucast.org/astoffungi/clinicalbreakpointsforantifungals (accessed on 4 February 2020).
- Mellado, E.; Garcia-Effron, G.; Alcázar-Fuoli, L.; Melchers WJ, G.; Verweij, P.E.; Cuenca-Estrella, M.; Rodríguez-Tudela, J.L. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 2007, 51, 1897–1904. [Google Scholar] [CrossRef]
- Mann, P.A.; Parmegiani, R.M.; Wei, S.Q.; Mendrick, C.A.; Li, X.; Loebenberg, D.; DiDomenico, B.; Hare, R.S.; Walker, S.S.; McNicholas, P.M. Mutations in Aspergillus fumigatus resulting in reduced susceptibility to posaconazole appear to be restricted to a single amino acid in the cytochrome P450 14alpha-demethylase. Antimicrob. Agents Chemother. 2003, 47, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Espinel-Ingroff, A.; Turnidge, J.; Alastruey-Izquierdo, A.; Dannaoui, E.; Garcia-Effron, G.; Guinea, J.; Kidd, S.; Pelaez, T.; Sanguinetti, M.; Meletiadis, J.; et al. Posaconazole MIC Distributions for Aspergillus fumigatus Species Complex by Four Methods: Impact of cyp51A Mutations on Estimation of Epidemiological Cutoff Values. Antimicrob. Agents Chemother. 2018, 62, e01916-17. [Google Scholar] [CrossRef]
- Warris, A. Azole-resistant aspergillosis. J. Infect. 2015, 71, S121–S125. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.; Heinen, C.D. The mismatch repair-dependent DNA damage response: Mechanisms and implications. DNA Repair 2019, 78, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Healey, K.R.; Zhao, Y.; Perez, W.B.; Lockhart, S.R.; Sobel, J.D.; Farmakiotis, D.; Kontoyiannis, D.P.; Sanglard, D.; Taj-Aldeen, S.J.; Alexander, B.D.; et al. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat. Commun. 2016, 7, 11128. [Google Scholar] [CrossRef]
- Bordallo-Cardona, M.Á.; Agnelli, C.; Gómez-Nuñez, A.; Sánchez-Carrillo, C.; Bouza, E.; Muñoz, P.; Escribano, P.; Guinea, J. MSH2 Gene Point Mutations Are Not Antifungal Resistance Markers in Candida glabrata. Antimicrob. Agents Chemother. 2019, 63, e01876-18. [Google Scholar] [CrossRef]
- Billmyre, R.B.; Clancey, S.A.; Heitman, J. Natural mismatch repair mutations mediate phenotypic diversity and drug resistance in Cryptococcus deuterogattii. eLife 2017, 6, e28802. [Google Scholar] [CrossRef]
- Buscaino, A. Chromatin-mediated regulation of genome plasticity in human fungal pathogens. Genes 2019, 10, 855. [Google Scholar] [CrossRef]
- Sundin, G.W.; Weigand, M.R. The microbiology of mutability. FEMS Microbiol. Lett. 2007, 277, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Verweij, P.E.; Zhang, J.; Debets, A.J.M.; Meis, J.F.; van de Veerdonk, F.L.; Schoustra, S.E.; Zwaan, B.J.; Melchers, W.J.G. In-host adaptation and acquired triazole resistance in Aspergillus fumigatus: A dilemma for clinical management. Lancet Infect. Dis. 2016, 16, e251–e260. [Google Scholar] [CrossRef] [PubMed]
- Hokken, M.W.J.; Zwaan, B.J.; Melchers, W.J.G.; Verweij, P.E. Facilitators of adaptation and antifungal resistance mechanisms in clinically relevant fungi. Fungal Genet. Biol. 2019, 132, 103254. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Healey, K.R.; Yadav, P.; Upadhyaya, G.; Sachdeva, N.; Sarma, S.; Kumar, A.; Tarai, B.; Perlin, D.S.; Chowdhary, A. Absence of azole or echinocandin resistance in Candida glabrata isolates in India despite background prevalence of strains with defects in the dna mismatch repair pathway. Antimicrob. Agents Chemother. 2018, 62, e00195-18. [Google Scholar] [CrossRef]
- Peng, G.; Lin, S.Y. Exploiting the homologous recombination DNA repair network for targeted cancer therapy. World J. Clin. Oncol. 2011, 2, 73–79. [Google Scholar] [CrossRef]
- Hawkins, N.J.; Fraaije, B.A. Predicting Resistance by Mutagenesis: Lessons from 45 Years of MBC Resistance. Front. Microbiol. 2016, 7, 1814. [Google Scholar] [CrossRef]
- Snelders, E.; Camps, S.M.; Karawajczyk, A.; Rijs, A.J.; Zoll, J.; Verweij, P.E.; Melchers, W.J. Genotype-phenotype complexity of the TR46/Y121F/T289A cyp51A azole resistance mechanism in Aspergillus fumigatus. Fungal Genet. Biol. 2015, 82, 129–135. [Google Scholar] [CrossRef]
| Gene (Gene Code) | Mutations | % of Strains |
|---|---|---|
| msh6 (Afu4g08300) | A55V | 0.62 |
| V118A | 0.62 | |
| D121E | 0.62 | |
| G178A | 1.86 | |
| I183R | 10.56 | |
| G240A | 42.86 | |
| N289S | 2.48 | |
| msh2 (Afu3g09850) | A45T | 3.73 |
| P329T | 3.73 | |
| E467D | 0.62 | |
| E812G | 1.24 | |
| A889E | 0.62 | |
| pms1 (Afu2g13410) | G286C | 0.62 |
| P401A, V438A, K464R, Q611E, E87K, E760K | 4.35 | |
| E444G | 2.48 | |
| S758Y | 1.24 | |
| D1013Y | 0.62 | |
| mlh1 (Afu5g11700) | K310R | 4.35 |
| S368N | 4.35 | |
| I510T | 1.86 | |
| A641S | 4.35 |
| Strains | MIC Ranges (mg/L) | |||
|---|---|---|---|---|
| ITC | VCZ | PSZ | IVZ | |
| akuBKU80 | 1–1 | 0.5–0.5 | 0.125–0.25 | 1–1 |
| ∆msh6 T3 | 0.5–1 | 0.5–1 | 0.125–0.25 | 0.5–1 |
| ∆msh6 T6 | 0.5–1 | 0.5–0.5 | 0.25–0.25 | 1–1 |
| Gene | Nucleotide Position (cDNA) | Codon | Amino Acid Change | akuBKU80 | ∆msh6-T3 | ∆msh6-T6 |
|---|---|---|---|---|---|---|
| cyp51A | g160t | Ggg/Tgg | G54W | 16 | 31 | 80 |
| g160a | Ggg/Agg | G54R | 0 | 1 | 0 | |
| benA | a593c | gAg/gCg | E198A | 0 | 0 | 1 |
| g592c | Gag/Cag | E198Q | 0 | 0 | 0 | |
| g592a | Gag/Aag | E198K | 0 | 8 | 0 | |
| g594c | gag/gaC | E198D | 0 | 3 | 3 | |
| a593c | gAg/gTg | E198V | 0 | 1 | 0 | |
| a593g | gAg/gGg | E198G | 2 | 0 | 0 | |
| t599a | tTc/tCc | F200S | 0 | 10 | 2 | |
| t599a | tTc/tAc | F200Y | 0 | 0 | 0 | |
| sdhB | a809t | cAc/cTc | H270L | 0 | 9 | 3 |
| c808t | Cac/Tac | H270Y | 3 | 7 | 7 | |
| a809g | cAc/cGc | H270R | 0 | 8 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lucio, J.; Gonzalez-Jimenez, I.; Roldan, A.; Amich, J.; Alcazar-Fuoli, L.; Mellado, E. Importance of the Aspergillus fumigatus Mismatch Repair Protein Msh6 in Antifungal Resistance Development. J. Fungi 2024, 10, 210. https://doi.org/10.3390/jof10030210
Lucio J, Gonzalez-Jimenez I, Roldan A, Amich J, Alcazar-Fuoli L, Mellado E. Importance of the Aspergillus fumigatus Mismatch Repair Protein Msh6 in Antifungal Resistance Development. Journal of Fungi. 2024; 10(3):210. https://doi.org/10.3390/jof10030210
Chicago/Turabian StyleLucio, Jose, Irene Gonzalez-Jimenez, Alejandra Roldan, Jorge Amich, Laura Alcazar-Fuoli, and Emilia Mellado. 2024. "Importance of the Aspergillus fumigatus Mismatch Repair Protein Msh6 in Antifungal Resistance Development" Journal of Fungi 10, no. 3: 210. https://doi.org/10.3390/jof10030210





