Arthropods as Vectors of Grapevine Trunk Disease Pathogens: Quantification of Phaeomoniella chlamydospora on Arthropods and Mycobiome Analysis of Earwig Exoskeletons
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Site and Collection of Arthropods
2.2. Washing of Arthropod Exoskeletons and DNA Extraction
2.3. qPCR for the Quantification of Pa. chlamydospora on Arthropods
2.3.1. Selection of Arthropod Samples for qPCR Analysis
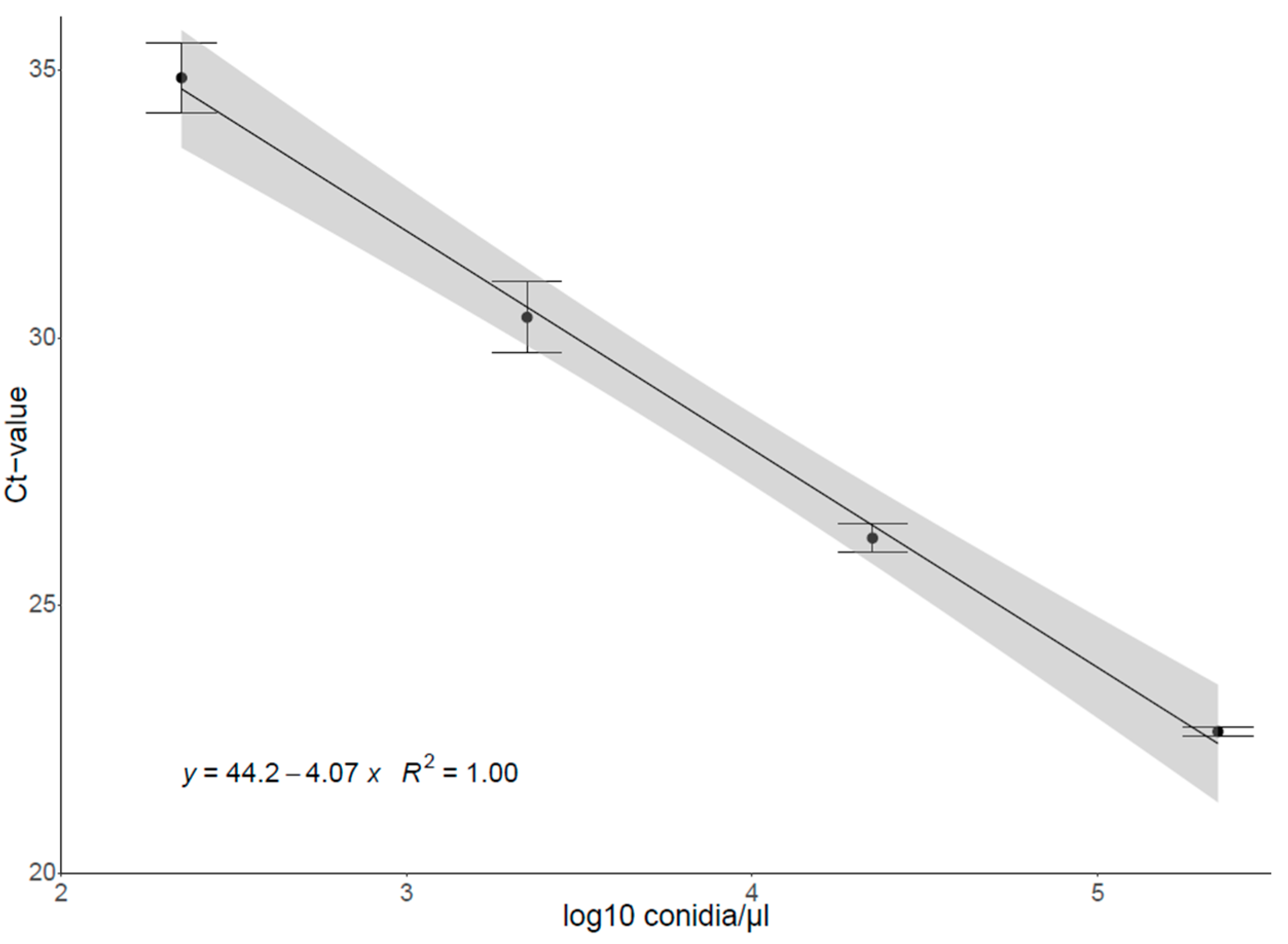
2.3.2. Construction of the Standard Curve
2.3.3. qPCR Set Up
2.3.4. qPCR Sample Processing
2.4. Mycobiome Analysis of Earwig Exoskeletons Using NGS
2.4.1. Selection of Earwig Samples for DNA Metabarcoding
2.4.2. DNA Metabarcoding
2.4.3. Bioinformatic Processing
3. Data Analysis
4. Results
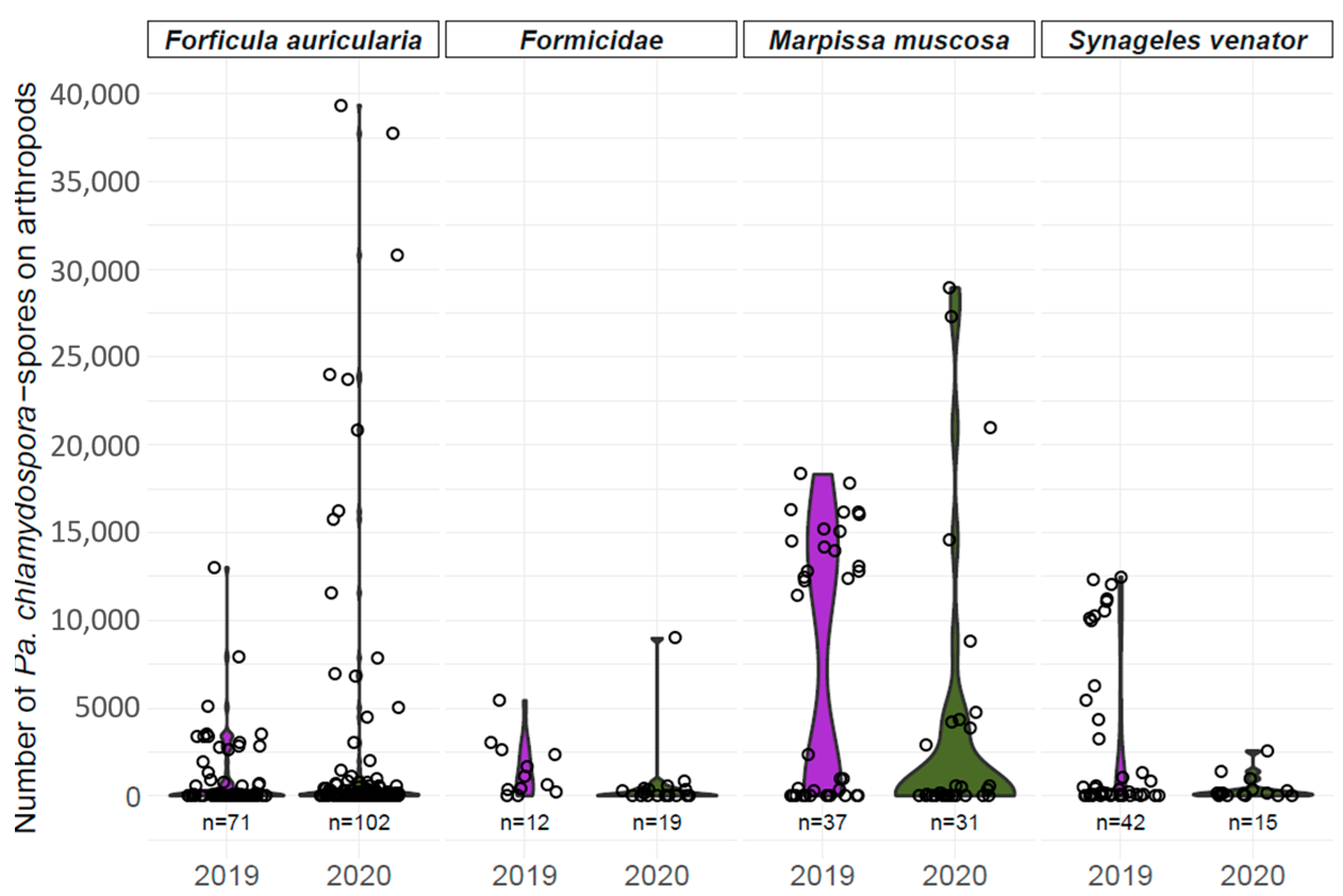
4.1. Spore Loads of Pa. chlamydospora on Arthropods
4.2. Mycobiome Analysis of Earwig Exoskeletons
4.2.1. Summary of Dataset
4.2.2. Alpha Diversity Metrics
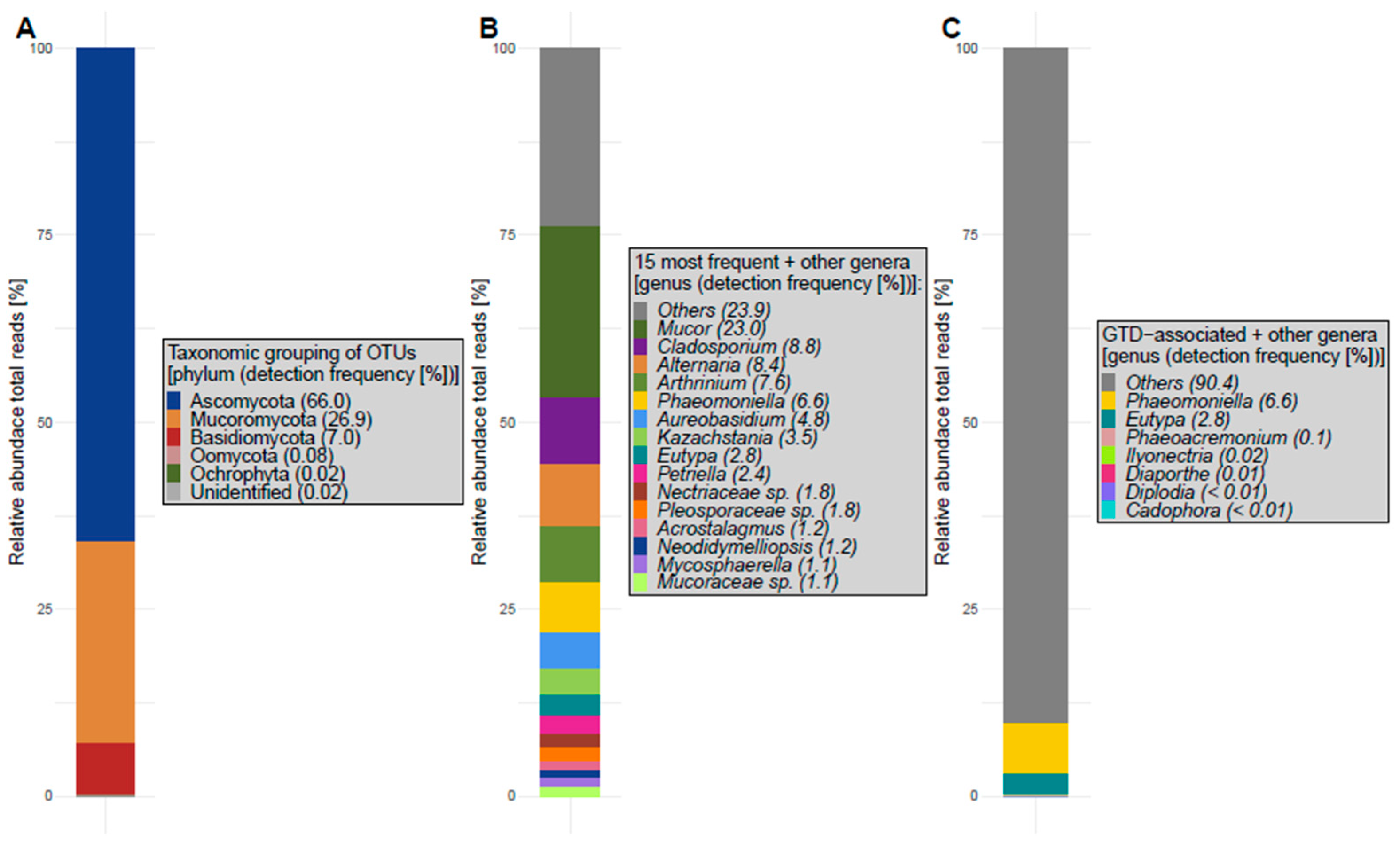
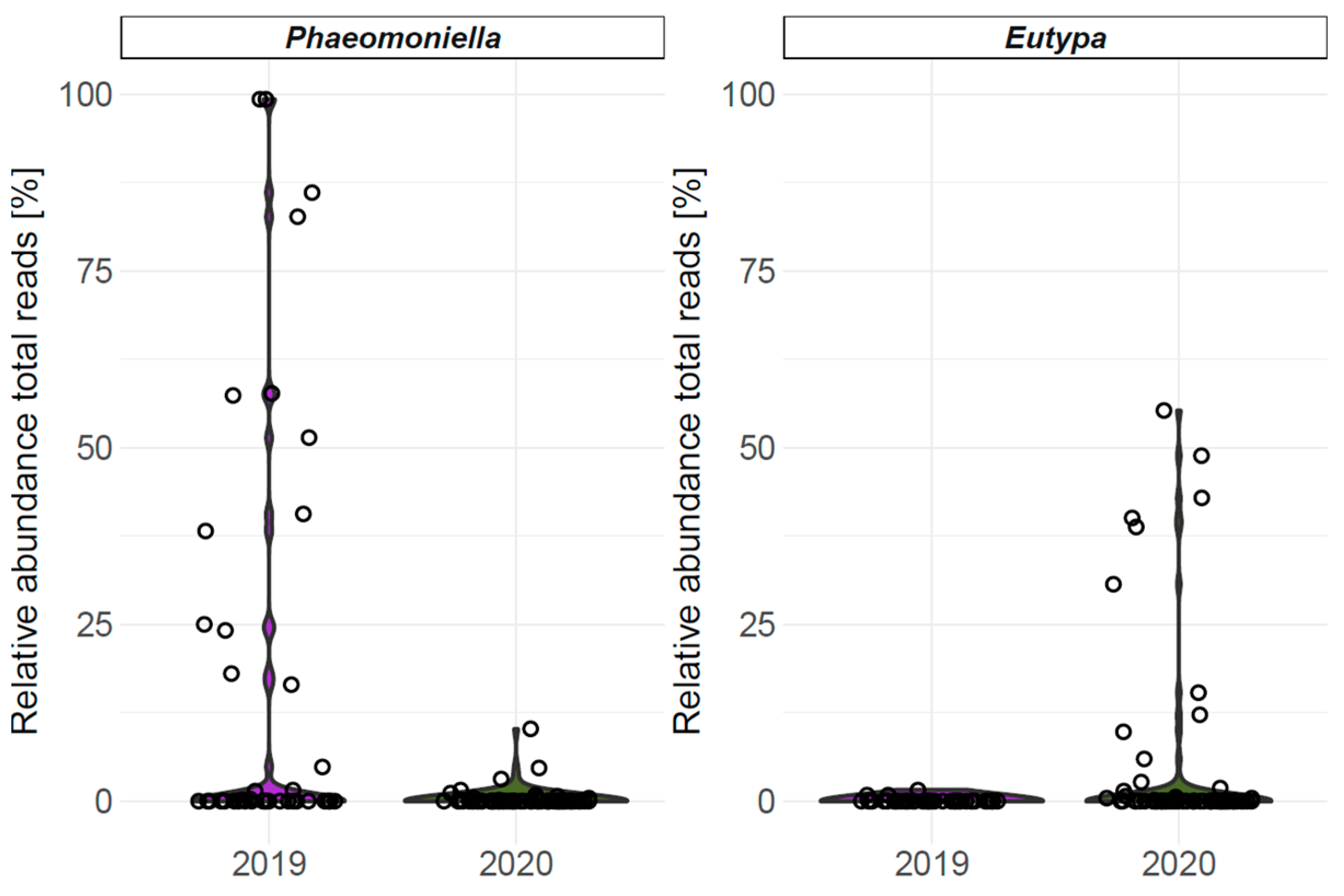
4.2.3. Fungal Abundance and Diversity
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Gramaje, D.; Úrbez-Torres, J.R.; Sosnowski, M.R. Managing grapevine trunk diseases with respect to etiology and epidemiology: Current strategies and future prospects. Plant Dis. 2018, 102, 12–39. [Google Scholar] [CrossRef]
- Larignon, P.; Fontaine, F.; Farine, S.; Clément, C.; Bertsch, C. Esca et black dead arm: Deux acteurs majeurs des maladies du bois chez la Vigne. C. R. Biol. 2009, 332, 765–783. [Google Scholar] [CrossRef]
- Bertsch, C.; Ramírez-Suero, M.; Magnin-Robert, M.; Larignon, P.; Chong, J.; Abou-Mansour, E.; Spagnolo, A.; Clément, C.; Fontaine, F. Grapevine trunk diseases: Complex and still poorly understood. Plant Pathol. 2013, 62, 243–265. [Google Scholar] [CrossRef]
- Mondello, V.; Songy, A.; Battiston, E.; Pinto, C.; Coppin, C.; Trotel-Aziz, P.; Clément, C.; Mugnai, L.; Fontaine, F. Grapevine trunk diseases: A review of fifteen years of trials for their control with chemicals and biocontrol agents. Plant Dis. 2018, 102, 1189–1217. [Google Scholar] [CrossRef] [PubMed]
- Claverie, M.; Notaro, M.; Fontaine, F.; Wery, J. Current knowledge on Grapevine Trunk Diseases with complex etiology: A systematic approach. Phytopathol. Mediterr. 2020, 59, 29–53. [Google Scholar] [CrossRef]
- Vasquez, S.J.; Douglas, W.; Gubler, W.D.; Leavitt, G.M. Economic loss in California’s table grape vineyards due to measles. Phytopathol. Mediterr. 2007, 46, 103–104. [Google Scholar]
- Hofstetter, V.; Buyck, B.; Croll, D.; Viret, O.; Couloux, A.; Gindro, K. What if esca disease of grapevine were not a fungal disease? Fungal Divers. 2012, 54, 51–67. [Google Scholar] [CrossRef]
- Lecomte, P.; Darrieutort, G.; Liminana, J.-M.; Comont, G.; Muruamendiaraz, A.; Legorburu, F.-J.; Choueiri, E.; Jreijiri, F.; El Amil, R.; Fermaud, M. New insights into esca of grapevine: The development of foliar symptoms and their association with xylem discoloration. Plant Dis. 2012, 96, 924–934. [Google Scholar] [CrossRef]
- Pearson, R.C.; Burr, T.J. Eutypa Dieback; Disease Identification Sheet No. 1; New York State Agr. Exp. Stat.: Geneva, NY, USA, 1981; pp. 1–2. [Google Scholar]
- Rolshausen, P.E.; Mahoney, N.E.; Molyneux, R.J.; Gubler, W.D. A reassessment of the species concept in Eutypa lata, the causal agent of Eutypa dieback of grapevine. Phytopathology 2006, 96, 369–377. [Google Scholar] [CrossRef]
- Moyo, P.; Mostert, L.; Spies, C.F.J.; Damm, U.; Halleen, F. Diversity of Diatrypaceae species associated with dieback of grapevines in South Africa, with the description of Eutypa cremea sp. nov. Plant Dis. 2018, 102, 220–230. [Google Scholar] [CrossRef]
- Larignon, P.; Fulchic, R.; Cere, L.; Dubos, B. Observation on black dead arm in french vineyards. Phytopathol. Mediterr. 2001, 40, S336–S342. [Google Scholar]
- Ùrbez-Torres, J.R. The status of Botryosphaeriaceae species infecting grapevines. Phytopathol. Mediterr. 2011, 50, S5–S45. [Google Scholar]
- Arkam, M.; Alves, A.; Lopes, A.; Čechová, J.; Pokluda, R.; Eichmeier, A.; Zitouni, A.; Mahamedi, A.E.; Berraf-Tebbal, A. Diversity of Botryosphaeriaceae causing grapevine trunk diseases and their spatial distribution under different climatic conditions in Algeria. Eur. J. Plant Pathol. 2021, 161, 933–952. [Google Scholar] [CrossRef]
- Mugnai, L.; Graniti, A.; Surico, G. Esca (black measles) and brown wood-streaking: Two old and elusive diseases of grapevines. Plant Dis. 1999, 83, 404–418. [Google Scholar] [CrossRef]
- Graniti, A.; Surico, G.; Mugnai, L. Esca of grapevine: A disease complex or a complex of diseases? Phytopathol. Mediterr. 2000, 39, 16–20. [Google Scholar]
- Surico, G. Towards a redefinition of the diseases within the esca complex of grapevine. Phytopathol. Mediterr. 2009, 48, 5–10. [Google Scholar]
- Crous, P.W.; Gams, W. Phaeomoniella chlamydospora gen. et comb. nov., a causal organism of Petri grapevine decline and esca. Phytopathol. Mediterr. 2000, 39, 112–118. [Google Scholar]
- Mostert, L.; Halleen, F.; Fourie, P.H.; Crous, P.W. A review of Phaeoacremonium species involved in Petri disease and esca of grapevine. Phytopathol. Mediterr. 2006, 45, S12–S29. [Google Scholar]
- Gramaje, D.; Mostert, L.; Groenewald, J.Z.; Crous, P.W. Phaeoacremonium: From esca disease to phaeohyphomycosis. Fungal Biol. 2015, 119, 759–783. [Google Scholar] [CrossRef]
- Baloyi, M.A.; Mostert, L.; Halleen, F. Pathogenicity of ten Phaeoacremonium species associated with esca and Petri disease of grapevine. Phytopathol. Mediterr. 2018, 57, 538–546. [Google Scholar] [CrossRef]
- Halleen, F.; Mostert, L.; Crous, P.W. Pathogenicity testing of lesser-known vascular fungi of grapevines. Austral. Plant Pathol. 2007, 36, 277. [Google Scholar] [CrossRef]
- Gramaje, D.; Armengol, J. Fungal trunk pathogens in the grapevine propagation process: Potential inoculum sources, detection, identification, and management strategies. Plant Dis. 2011, 95, 1040–1055. [Google Scholar] [CrossRef]
- Travadon, R.; Lawrence, D.P.; Rooney-Latham, S.; Gubler, W.D.; Wilcox, W.F.; Rolshausen, P.E.; Baumgartner, K. Cadophora species associated with wood-decay of grapevine in North America. Fungal Biol. 2015, 119, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M. A new wood-decaying basidiomycete species associated with esca of grapevine: Fomitiporia mediterranea (Hymenochaetales). Mycol. Prog. 2002, 1, 315–324. [Google Scholar] [CrossRef]
- Fischer, M.; Gonzálz Garcíca, V. An annotated checklist of European basidiomycetes related to white rot of grapevine (Vitis vinifera). Phytopathol. Mediterr. 2015, 54, 281–298. [Google Scholar] [CrossRef]
- Brown, A.A.; Lawrence, D.P.; Baumgartner, K. Role of basidiomycete fungi in the grapevine trunk disease esca. Plant Pathol. 2020, 69, 205–220. [Google Scholar] [CrossRef]
- Larignon, P.; Dubos, B. Preliminary studies on the biology of Phaeoacremonium. Phytopathol. Mediterr. 2000, 39, 184–189. [Google Scholar]
- Eskalen, A.; Gubler, D.W. Association of spores of Phaeomoniella chlamydospora, Phaeoacremonium inflatipes, and Pm. aleophilum with grapevine cordons in California. Phytopathol. Mediterr. 2001, 40, S429–S432. [Google Scholar]
- Munkvold, G.P.; Marois, J.J. Factors associated with variation in susceptibility of grapevine pruning wounds to infection by Eutypa lata. Phytopathology 1995, 85, 249–256. [Google Scholar] [CrossRef]
- Larignon, P.; Dubos, B. Fungi associated with esca disease in grapevine. Eur. J. Plant Pathol. 1997, 103, 147–157. [Google Scholar] [CrossRef]
- van Niekerk, J.M.; Halleen, F.; Fourie, P.H. Temporal susceptibility of grapevine pruning wounds to trunk pathogen infection in South African grapevines. Phytopathol. Mediterr. 2011, 50, S139–S150. [Google Scholar]
- Makatini, G.J.; Halleen, F.; Mutawila, C.; Moyo, P.; Mostert, L. Susceptibility of grapevine sucker and green shoot wounds to trunk disease pathogens. S. Afr. J. Enol. Vitic. 2023, 44, 55–63. [Google Scholar] [CrossRef]
- Rosace, M.C.; Legler, S.E.; Salotti, I.; Rossi, V. Susceptibility of pruning wounds to grapevine trunk diseases: A quantitative analysis of literature data. Front. Plant Sci. 2023, 14, 1063932. [Google Scholar] [CrossRef] [PubMed]
- Leach, J.G. Insect Transmission of Plant Diseases; McGraw-Hill Book Co.: New York, NY, USA, 1940. [Google Scholar]
- Evans, C. Invertebrate vectors of Phytophthora palmivora, causing black pod disease of cocoa in Ghana. Ann. Appl. Biol. 1973, 75, 331–345. [Google Scholar] [CrossRef]
- Brasier, C.M. Ophiostoma novo-ulmi sp. nov., causative agent of current Dutsch elm disease pandemics. Mycopathologia 1991, 115, 151–161. [Google Scholar] [CrossRef]
- El-Hamalawi, Z.A.; Menge, J.A. The role of snails and ants in transmitting the avocado stem canker pathogen, Phytophthora citricola. J. Am. Soc. Hortic. Sci. 1996, 121, 973–977. [Google Scholar] [CrossRef]
- Kluth, S.; Kruess, A.; Tscharntke, T. Insects as vectors of plant pathogens: Mutualistic and antagonistic interactions. Oecologia 2002, 133, 193–199. [Google Scholar] [CrossRef]
- Chen, G.; Zhang, R.-R.; Liu, Y.; Sun, W.-B. Spore dispersal of fetid Lysurus mokusin by feces of mycophagous insects. J. Chem. Ecol. 2014, 40, 893–899. [Google Scholar] [CrossRef]
- Moyo, P.; Allsopp, E.; Roets, F.; Mostert, L.; Halleen, F. Arthropods vector grapevine trunk disease pathogens. Phytopathology 2014, 104, 1063–1069. [Google Scholar] [CrossRef]
- Kalvelage, E.M.; Voegele, R.T.; Fischer, M. Dissemination of esca-related pathogens in German vineyards: Do arthropods play roles in vectoring spores? Phytopathol. Mediterr. 2021, 60, 467–478. [Google Scholar] [CrossRef]
- Kalvelage, E.M.; Behrens, F.H.; Rauch, C.; Voegele, R.T.; Fischer, M. Arthropods as vectors of esca-related pathogens: Transmission efficiency of ants and earwigs and the potential of earwig feces as inoculum source in vineyards. Vitis 2022, 61, 77–85. [Google Scholar] [CrossRef]
- Moyo, P. The Role of Arthropods in the Dispersal of Trunk Disease Pathogens Associated with Petri Disease and Esca. Available online: https://core.ac.uk/download/pdf/37411221.pdf (accessed on 17 March 2024).
- Huth, C.D. Untersuchungen zur Lebensweise und zur Populationskontrolle des Gemeinen Ohrwurms Forficula auricularia L. (Insecta, Dermaptera) in Rebanlagen. Available online: https://openscience.ub.uni-mainz.de/handle/20.500.12030/3141 (accessed on 17 March 2024).
- Molnar, M.; Voegele, R.T.; Fischer, M. Grapevine trunk disease in German viticulture IV. Spreading of spores of Phaeomoniella chlamydospora in Esca-affected vineyards. Vitis 2020, 59, 63–69. [Google Scholar] [CrossRef]
- Schäfer, M. (Ed.) Brohmer—Fauna von Deutschland, 24th ed.; Quelle & Meyer Verlag GmbH & Co.: Wiebelsheim, Germany, 2009. [Google Scholar]
- Roberts, M.J. (Ed.) Spiders of Britain and Northern Europe; Collins Field Guide; Harper Collins Publishers: London, UK, 1995. [Google Scholar]
- Haag, N. Grapevine Trunk Diseases: Epidemiologie und Molekulardiagnose wichtiger Esca-Erreger während der Pflanzenguterzeugung. Available online: http://opus.uni-hohenheim.de/volltexte/2018/1552/ (accessed on 17 March 2024). [CrossRef]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Pauvert, C.; Buée, M.; Laval, V.; Edel-Hermann, V.; Fauchery, L.; Gautier, A.; Lesur, I.; Vallance, J.; Vacher, C. Bioinformatics matters: The accuracy of plant and soil fungal community data is highly dependent on the metabarcoding pipeline. Fungal Ecol. 2019, 41, 23–33. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high throughput sequencing reads. EMBnet.journal 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Morgan, M.; Anders, S.; Lawrence, M.; Aboyoun, P.; Pages, H.; Gentleman, R. ShortRead: A bioconductor package for input, quality assessment and exploration of high-throughput sequence data. J. Bioinform. 2009, 25, 2607–2608. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample interference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.-H.; Alexander, J.J.; Eberhardt, U.; Erland, S.; Hoiland, K.; Kjoller, R.; Larsson, E.; Pennanen, T.; et al. The UNITE database for molecular identification of fungi—Recent updates and future perspectives. New Phytol. 2020, 186, 281–285. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Kassambara, A. ggpubr: “ggplot” Based Publication Ready Plots. R Package Version 0.4.0. 2020. Available online: https://rpkgs.datanovia.com/ggpubr/ (accessed on 17 March 2024).
- Revelle, W. psych: Procedures for Personality and Psychological Research; Northwestern University: Evanston, IL, USA, 2022. [Google Scholar]
- Bates, D.; Maechler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Lenth, R.V. emmeans: Estimated Marginal Means, Aka Least-Squares Means. R Package Version 1.7.4-1. 2022. Available online: https://cran.r-project.org/web/packages/emmeans/emmeans.pdf (accessed on 17 March 2024).
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous inference in general parametric models. Biom. J. 2008, 50, 346–363. [Google Scholar] [CrossRef]
- Fox, J.; Weisberg, S. An {R} Companion to Applied Regression, 3rd ed.; Sage: Thousand Oaks, CA, USA, 2019. [Google Scholar]
- McMurdie, P.J.; Holmes, S. An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. vegan: Community Ecology Package. R Package Version 2.5-7. 2020. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 17 March 2024).
- Wilke, C.O. cowplot: Streamlined Plot Theme and Plot. 2020. Available online: https://wilkelab.org/cowplot/ (accessed on 17 March 2024).
- Hrycan, J.; Hart, M.; Bowen, P.; Forge, T.; Úrbez-Torres, J.R. Grapevine trunk disease fungi: Their roles as latent pathogens and stress factors that favour disease development and symptom expression. Phytopathol. Mediterr. 2020, 3, 395–424. [Google Scholar] [CrossRef]
- Kenfaoui, J.; Radouane, N.; Mennani, M.; Tahiri, A.; El Ghadraoui, L.; Belabess, Z.; Fontaine, F.; El Hamss, H.; Amiri, S.; Lahlali, R.; et al. A panoramic view on grapevine trunk diseases threats: Case of Eutypa dieback, Botryosphaeria dieback, and Esca disease. J. Fungi 2022, 8, 595. [Google Scholar] [CrossRef]
- Edwards, J.; Constable, F.; Wiechel, T.; Salib, S. Comparison of the molecular tests—Single PCR, nested PCR and quantitative PCR (SYBR Green and TaqMan)—For detection of Phaemoniella chlamydospora during grapevine nursery propagation. Phytopathol. Mediterr. 2007, 46, 58–72. [Google Scholar]
- Edwards, J.; Laukart, N.; Pascoe, I.G. In situ sporulation of Phaemoniella chlamydospora in the vineyard. Phytopathol. Mediterr. 2001, 40, 61–66. [Google Scholar]
- Eskalen, A.; Feliciano, A.J.; Gubler, W.D. Susceptibility of grapevine pruning wounds and symptom development in response to infection by Phaeoacremonium aleophilum and Phaeomoniella chlamydospora. Plant Dis. 2007, 91, 1100–1104. [Google Scholar] [CrossRef]
- Elena, G.; Luque, J. Seasonal susceptibility of grapevine pruning wounds and cane colonization in Catalonia, Spain following artificial infection with Diplodia seriata and Phaeomoniella chlamydospora. Plant Dis. 2016, 100, 1651–1659. [Google Scholar] [CrossRef] [PubMed]
- Elena, G.; Sosnowski, M.R.; Ayres, M.R.; Lecomte, P.; Benetreau, C.; Garcia-Figueres, F.; Luque, J. Effect of the inoculum dose of thee grapevine trunk pathogens on the infection of artificially inoculated pruning wounds. Phytopathol. Mediterr. 2015, 54, 345–354. [Google Scholar] [CrossRef]
- Hong, T.D.; Ellis, R.H.; Moore, D. Development of a model to predict the effect of temperature and moisture on fungal spore longevity. Ann. Bot. 1997, 79, 121–128. [Google Scholar] [CrossRef]
- Perazzolli, M.; Antonielli, L.; Storari, M.; Puopolo, G.; Pancher, M.; Giovannini, O.; Pindo, M.; Pertot, I. Resilience of the natural phyllospjere microbiota of the grapevine to chemical and biological pesticides. AMS J. 2014, 80, 3589–3596. [Google Scholar] [CrossRef]
- Behrens, F.H.; Fischer, M. Evaluation of different phyllosphere sample types for parallel metabacroding of Fungi and Oomycetes in Vitis vinifera. Phytobiomes J. 2022, 6, 207–213. [Google Scholar] [CrossRef]
- Castañeda, L.E.; Miura, T.; Sánchez, R.; Barbosa, O. Effects of agricultural management on phyllosphere fungal diversity in vineyards and the association with adjacent native forests. PeerJ 2018, 6, e5715. [Google Scholar] [CrossRef] [PubMed]
- Kraus, C.; Rauch, C.; Kalvelage, E.M.; Behrens, F.H.; d’Aguiar, D.; Dubois, C.; Fischer, M. Minimal versus intensive: How the pruning intensity affects occurrence of grapevine leaf stripe disease, wood integrity, and the mycobiome in grapevine trunks. J. Fungi 2022, 8, 247. [Google Scholar] [CrossRef] [PubMed]
- Bruez, E.; Vallance, J.; Gautier, A.; Laval, V.; Compant, S.; Maurer, W.; Sessitsch, A.; Lebrun, M.-H.; Rey, P. Major changes in grapevine wood microbiota are associated with the onset of esca, a devastating trunk disease. Environ. Microbiol. 2020, 22, 5189–5206. [Google Scholar] [CrossRef] [PubMed]
- Vanga, B.R.; Panda, P.; Shah, A.S.; Thompson, S.; Woolley, R.H.; Ridgway, H.J.; Mundy, D.C.; Bulman, S. DNA metabarcoding reveals high relative abundance of trunk disease fungi in grapevines from Marlborough, New Zealand. BMC Microbiol. 2022, 22, 126. [Google Scholar] [CrossRef] [PubMed]
- Gobbi, A.; Kyrkou, I.; Filippi, E.; Ellegaard-Jensen, L.; Hestbjerg Hansen, L. Seasonal epiphytic microbial dynamics on grapevine leaves under biocontrol and copper fungicide treatments. Sci. Rep. 2020, 10, 681. [Google Scholar] [CrossRef] [PubMed]
- Mundy, D.C.; Vanga, B.R.; Thompson, S.; Bulman, S. Assessment of sampling and DNA extraction methods for identification of grapevine trunk microorganisms using metabarcoding. NZPP 2018, 71, 10–18. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Purahong, W.; Wubet, T.; Hyde, K.D.; Zhang, W.; Xu, H.; Zhang, G.; Fu, C.; Liu, M.; Xing, Q.; et al. Direct comparison of culture-dependent and culture-independent molecular approaches reveal the diversity of fungal endophytic communities in stems of grapevine (Vitis vinifera). Fungal Divers. 2018, 90, 85–107. [Google Scholar] [CrossRef]
- Eichmeier, A.; Pečenka, J.; Peňázová, E.; Baránek, M.; Català-García, S.; León, M.; Armengol, J.; Gramaje, D. High-throughput amplicon sequencing-based analysis of active fungal communities inhabiting grapevine after hot-water treatments reveals unexpectedly high fungal diversity. Fungal Ecol. 2018, 36, 26–38. [Google Scholar] [CrossRef]
- Lin, M.M.-H.; Boss, P.K.; Walker, M.E.; Sumby, K.M.; Grbin, P.R.; Jiranek, V. Evaluation of indigenous non-Saccharomyces yeasts isolated from a South Australian vineyard for their potential as wine starter cultures. Int. J. Food Microbiol. 2020, 312, 108373. [Google Scholar] [CrossRef] [PubMed]
- Setati, M.E.; Jacobson, D.; Bauer, F. Sequence-based analysis of the Vitis vinifera L. cv Cabernet Sauvignon grape must mycobiome in three South African vineyards employing distinct agronomic systems. Front. Micobiol. 2015, 6, 1358. [Google Scholar] [CrossRef]
- Shi, T.; Wang, H.; Li, Y.-J.; Wang, Y.-F.; Pan, Q.; Wang, B.; Shang, E.-L. Genus Acrostalagmus: A prolific producer of natural products. Biomolecules 2023, 13, 1191. [Google Scholar] [CrossRef] [PubMed]
- Cueva, C.; García-Ruiz, A.; González-Rompinelli, E.; Bartolome, B.; Martín-Álvarez, P.J.; Salazar, O.; Vicente, M.F.; Bills, G.F.; Moreno-Arribas, M.V. Degradation of biogenic amines by vineyard ecosystem fungi. Potential use in winemaking. J. Appl. Microbiol. 2012, 112, 672–682. [Google Scholar] [CrossRef]
- Ettinger, C.L.; Wu-Woods, J.; Kurbessoian, T.; Brown, D.J.; Souza Pacheco, I.d.; Vindiola, B.G.; Walling, L.L.; Atkinson, P.W.; Byrne, F.J.; Redak, R.; et al. Geographical survey of the mycobiome and microbiome of Southern California glassy-winged sharpshooters. mSphere 2023, 8, e0026723. [Google Scholar] [CrossRef] [PubMed]
- Munkvold, G.P.; Duthie, J.A.; Marois, J.J. Reduction in yield and vegetative growth of grapevine due to Eutypa dieback. Phytopathology 1994, 84, 186–192. [Google Scholar] [CrossRef]
- Úrbez-Torres, J.R.; Peduto, F.; Smith, R.J.; Gubler, W.D. Phomopsis dieback: A grapevine trunk disease caused by Phomopsis viticola in California. Plant Dis. 2013, 97, 1571–1579. [Google Scholar] [CrossRef]
- Cabral, A.; Rego, C.; Nascimento, T.; Oliveira, H.; Groenewald, J.Z.; Crous, P.W. Multi-gene analysis and morphology reveal novel Ilyonectria species associated with black foot disease of grapevines. Fungal Biol. 2012, 116, 62–80. [Google Scholar] [CrossRef]
- Pinto, C.; Pinho, D.; Sousa, S.; Pinheiro, M.; Egas, C.; Gomes, A.C. Unravelling the diversity of grapevine microbiome. PLoS ONE 2014, 9, e85622. [Google Scholar] [CrossRef]
- Del Frari, G.; Gobbi, A.; Aggerbeck, M.R.; Oliveira, H.; Hansen, L.H.; Ferreira, R.B. Characterization of the wood mycobiome of Vitis vinifera in a vineyard affected by esca. Spatial distribution of fungal communities and their putative relation with leaf symptoms. Front. Plant Sci. 2019, 10, 910. [Google Scholar] [CrossRef]
- Fischer, M. Grapevine trunk diseases in German viticulture: III. Biodiversity and spatial distribution of fungal pathogens in rootstock mother plants and possible relation to leaf symptoms. Vitis 2019, 58, 141–149. [Google Scholar] [CrossRef]
- Fischer, M.; Kassemeyer, H.-H. Fungi associated with Esca disease of grapevine in Germany. Vitis 2003, 42, 109–116. [Google Scholar]
- Moretti, S.; Pacetti, A.; Pierron, R.; Kassemeyer, H.-H.; Fischer, M.; Péros, J.-P.; Perez-Gonzales, G.; Bieler, E.; Schilling, M.; Di Marco, S.; et al. Fomitiporia mediterranea M. Fisch., the historical Esca agent: A comprehensive review on the main grapevine wood rot agent in Europe. Phytopathol. Mediterr. 2021, 60, 351–379. [Google Scholar] [CrossRef]
- Pinto, C.; Custódio, V.; Nunes, M.; Songy, A.; Rabenoelina, F.; Courteaux, B.; Clément, C.; Gomes, A.C.; Fontaine, F. Understand the potential role of Aureobasidium pullulans, a resident microorganism from grapevine, to prevent the infection caused by Diplodia seriata. Front. Micobiol. 2018, 9, 3047. [Google Scholar] [CrossRef] [PubMed]
- Karácsony, Z.; Mondello, V.; Fontaine, F.; Váczy, K.Z. The potential role of Aureobasidium pullulans in the development of foliar symptoms of Esca disease in grapevine. OENO One 2023, 57, 189–203. [Google Scholar] [CrossRef]
- Travadon, R.; Lecomte, P.; Diarra, B.; Lawrence, D.P.; Renault, D.; Ojeda, H.; Rey, P.; Baumgartner, K. Grapevine pruning systems and cultivars influence the diversity of wood-colonizing fungi. Fungal Ecol. 2016, 24, 82–93. [Google Scholar] [CrossRef]
- Fontaine, F.; Armengol, J.; Smart, R.; Nagy, Z.A.; Borgo, M.; Rego, C.; Corio-Costet, M.-F. Grapevine Trunk Diseases. A Review. OIV Publications. 2016. Available online: https://www.oiv.int/public/medias/4650/trunk-diseases-oiv-2016.pdf (accessed on 17 March 2024).
- Berbegal, M.; Ramón-Albalat, A.; León, M.; Armengol, J. Evaluation of long-term protection from nursery to vineyard provided by Trichoderma atroviride SC1 against fungal grapevine trunk pathogens. Pest Manag. Sci. 2020, 76, 967–977. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brandenburg, E.M.; Voegele, R.T.; Fischer, M.; Behrens, F.H. Arthropods as Vectors of Grapevine Trunk Disease Pathogens: Quantification of Phaeomoniella chlamydospora on Arthropods and Mycobiome Analysis of Earwig Exoskeletons. J. Fungi 2024, 10, 237. https://doi.org/10.3390/jof10040237
Brandenburg EM, Voegele RT, Fischer M, Behrens FH. Arthropods as Vectors of Grapevine Trunk Disease Pathogens: Quantification of Phaeomoniella chlamydospora on Arthropods and Mycobiome Analysis of Earwig Exoskeletons. Journal of Fungi. 2024; 10(4):237. https://doi.org/10.3390/jof10040237
Chicago/Turabian StyleBrandenburg, Elisa Maria, Ralf Thomas Voegele, Michael Fischer, and Falk Hubertus Behrens. 2024. "Arthropods as Vectors of Grapevine Trunk Disease Pathogens: Quantification of Phaeomoniella chlamydospora on Arthropods and Mycobiome Analysis of Earwig Exoskeletons" Journal of Fungi 10, no. 4: 237. https://doi.org/10.3390/jof10040237
APA StyleBrandenburg, E. M., Voegele, R. T., Fischer, M., & Behrens, F. H. (2024). Arthropods as Vectors of Grapevine Trunk Disease Pathogens: Quantification of Phaeomoniella chlamydospora on Arthropods and Mycobiome Analysis of Earwig Exoskeletons. Journal of Fungi, 10(4), 237. https://doi.org/10.3390/jof10040237





