Characterization of Culturable Mycobiome of Newly Excavated Ancient Wooden Vessels from the Archeological Site of Viminacium, Serbia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Sampling Points
2.2. Indoor Microclimate and Moisture Content Measurements
2.3. Isolation and Identification of Culturable Fungi
2.4. Biodegradation Plate Assays
3. Results
3.1. Microclimate of the Mammoth Park Ambient
3.2. Diversity of Cultivable Mycobiomes
3.3. Biodegradative Profile of Isolated Fungi
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blanchette, R.A. A review of microbial deterioration found in archaeological wood from different environments. Int. Biodeterior. Biodegrad. 2000, 46, 189–204. [Google Scholar] [CrossRef]
- Colombini, M.P.; Lucejko, J.J.; Modugno, F.; Orlandi, M.; Tolppa, E.-L.; Zoia, L. A multi-analytical study of degradation of lignin in archaeological waterlogged wood. Talanta 2009, 80, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Singh, A.P. Wood as Cultural Heritage Material and its Deterioration by Biotic and Abiotic Agents. In Secondary Xylem Biology; Elsevier: Amsterdam, The Netherlands, 2016; pp. 233–257. [Google Scholar]
- Pedersen, N.B.; Łucejko, J.J.; Modugno, F.; Björdal, C. Correlation between bacterial decay and chemical changes in waterlogged archaeological wood analyzed by light microscopy and Py-GC/MS. Holzforschung 2020, 75, 635–645. [Google Scholar] [CrossRef]
- Tiano, P. Biodegradation of Cultural Heritage: Decay Mechanisms and Control Methods; CNR—Centro di Studio Sulle “Cause Deperimento e Metodi Conservazione Opere d’Arte”: Firenze, Italy, 2009; Volume 9. [Google Scholar]
- Sarkar, N.; Ghosh, S.K.; Bannerjee, S.; Aikat, K. Bioethanol production from agricultural wastes: An overview. Renew. Energy 2012, 37, 19–27. [Google Scholar] [CrossRef]
- Traoré, M.; Kaal, J.; Martínez Cortizas, A. Potential of pyrolysis-GC–MS molecular fingerprint as a proxy of Modern Age Iberian shipwreck wood preservation. J. Anal. Appl. Pyrolysis 2017, 126, 1–13. [Google Scholar] [CrossRef]
- Cornelissen, J.H.C.; Sass-Klaassen, U.; Poorter, L.; Van Geffen, K.; Van Logtestijn, R.S.P.; Van Hal, J.; Goudzwaard, L.; Sterck, F.J.; Klaassen, R.K.W.M.; Freschet, G.T.; et al. Controls on Coarse Wood Decay in Temperate Tree Species: Birth of the LOGLIFE Experiment. Ambio 2012, 41, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Björdal, C.G. Evaluation of microbial degradation of shipwrecks in the Baltic Sea. Int. Biodeterior. Biodegrad. 2012, 70, 126–140. [Google Scholar] [CrossRef]
- Sterflinger, K. Fungi: Their role in deterioration of cultural heritage. Fungal Biol. Rev. 2010, 24, 47–55. [Google Scholar] [CrossRef]
- Pyzik, A.; Ciuchcinski, K.; Dziurzynski, M.; Dziewit, L. The bad and the good—Microorganisms in cultural heritage environments—An update on biodeterioration and biotreatment approaches. Materials 2021, 14, 177. [Google Scholar] [CrossRef]
- High, K.E.; Penkman, K.E.H. A review of analytical methods for assessing preservation in waterlogged archaeological wood and their application in practice. Herit. Sci. 2020, 8, 83. [Google Scholar] [CrossRef]
- Singh, A.P.; Kim, Y.S.; Chavan, R.R. Advances in understanding microbial deterioration of buried and waterlogged archaeological woods: A review. Forests 2022, 13, 394. [Google Scholar] [CrossRef]
- Milovanović, B.; Mrđić, N.; Danković, I. Arheološka istraživanja na lokalitetu Rit (Viminacijum) u 2020. godini. In Arheologija u Srbiji. Projekti Arheološkog Instituta u 2020; Vitezović, S., Radišić, M., Petrović, Đ., Eds.; Arheološkog Instituta: Beograd, Serbia, 2023; pp. 107–117. [Google Scholar]
- Huisman, D.J.; Manders, M.R.; Kretschmar, E.I.; Klaassen, R.K.W.M.; Lamersdorf, N. Burial conditions and wood degradation at archaeological sites in The Netherlands. Int. Biodeter. Biodegr. 2008, 61, 33–44. [Google Scholar] [CrossRef]
- Nikolić, E.V. Creation of the Mammoth Park at Viminacium, Serbia. Geoheritage 2019, 11, 935–947. [Google Scholar] [CrossRef]
- Ellis, M.B.; Ellis, J.P. Microfungi on Land Plants: An Identification Handbook, New Enlarged ed.; The Richmond Publishing Co., Ltd.: Slough, UK, 1997. [Google Scholar]
- Watanabe, T. Pictorial Atlas of Soil and Seed Fungi: Morphologies of Cultured Fungi and Key to Species, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2002. [Google Scholar]
- Samson, R.A.; Houbraken, J.; Thrane, U.; Frisvad, J.C.; Andersen, B. Food and Indoor Fungi, CBS Laboratory Manual Series; CBS Fungal Biodiversity Centre: Utrecht, The Netherlands, 2010; p. 2. [Google Scholar]
- Woudenberg, J.H.C.; Groenewald, J.Z.; Binder, M.; Crous, P.W. Alternaria redefined. Stud. Mycol. 2013, 75, 171–212. [Google Scholar] [CrossRef] [PubMed]
- Savković, Ž.; Stupar, M.; Unković, N.; Ivanović, Ž.; Blagojević, J.; Vukojević, J.; Ljaljević Grbić, M. In vitro biodegradation potential of airborne Aspergilli and Penicillia. Sci. Nat. 2019, 106, 8. [Google Scholar] [CrossRef] [PubMed]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Borrego, S.; Guiamet, P.; de Saravia, S.G.; Batistini, P.; Garcia, M.; Lavin, P.; Perdomo, I. The quality of air at archives and the biodeterioration of photographs. Int. Biodeterior. Biodegrad. 2010, 64, 139–145. [Google Scholar] [CrossRef]
- Liang, Y.L.; Zhang, Z.; Wu, M.; Wu, Y.; Feng, J.X. Isolation, screening, and identification of cellulolytic bacteria from natural reserves in the subtropical region of China and optimization of cellulase production by Paenibacillus terrae ME27-1. Biomed. Res. Int. 2014, 13, 512497. [Google Scholar] [CrossRef] [PubMed]
- Senthivelan, T.; Kanagaraj, J.; Panda, R.C.; Narayani, T. Screening and production of a potential extracellular fungal laccase from Penicillium chrysogenum: Media optimization by response surface methodology (RSM) and central composite rotatable design (CCRD). Biotechnol. Rep. 2019, 23, e00344. [Google Scholar] [CrossRef]
- Pangallo, D.; Šimonovičová, A.; Chovanová, K.; Ferianc, P. Wooden art objects and the museum environment: Identification and biodegradative characteristics of isolated microflora. Lett. Appl. Microbiol. 2007, 45, 87–94. [Google Scholar] [CrossRef]
- Sharma, A.; Aggarwal, N.K.; Yadav, A. Isolation and Screening of Lignolytic fungi from Various Ecological Niches. UMYU J. Microbiol. Res. 2017, 5, 25–34. [Google Scholar] [CrossRef]
- Pedersen, N.B.; Matthiesen, H.; Blanchette, R.A.; Alfredsen, G.; Held, B.W.; Westergaard-Nielsen, A.; Hollesen, J. Fungal Attack on Archaeological Wooden Artefacts in the Arctic—Implications in a Changing Climate. Sci. Rep. 2020, 10, 14577. [Google Scholar] [CrossRef]
- Pournou, A.; Bogomolova, E. Fungal Colonization on Excavated Prehistoric Wood: Implications for in-Situ Display. Int. Biodeterior. Biodegrad. 2009, 63, 371–378. [Google Scholar] [CrossRef]
- Abdel-Azeem, A.M.; Held, B.W.; Richards, J.E.; Davis, S.L.; Blanchette, R.A. Assessment of Biodegradation in Ancient Archaeological Wood from the Middle Cemetery at Abydos, Egypt. PLoS ONE 2019, 14, e0213753. [Google Scholar] [CrossRef]
- Van Der Wal, A.; De Boer, W.; Smant, W.; Van Veen, J.A. Initial Decay of Woody Fragments in Soil Is Influenced by Size, Vertical Position, Nitrogen Availability and Soil Origin. Plant Soil 2007, 301, 189–201. [Google Scholar] [CrossRef]
- Hamed, S.A.M. In-vitro studies on wood degradation in soil by soft-rot fungi: Aspergillus niger and Penicillium chrysogenum. Int. Biodeterior. Biodegrad. 2013, 78, 98–102. [Google Scholar] [CrossRef]
- Blanchette, R.A.; Held, B.W.; Jurgens, J.; Stear, A.; Dupont, C. Fungi Attacking Historic Wood of Fort Conger and the Peary Huts in the High Arctic. PLoS ONE 2021, 16, e0246049. [Google Scholar] [CrossRef]
- Kim, J.S.; Kim, M.; Lim, J.W.; Cha, M.Y.; Lee, K.H.; Yoon, Y.H.; Kim, Y.S. Characterization of Microbial Decay and Microbial Communities in Waterlogged Archaeological Rosewood (Dalbergia Species). Forests 2023, 14, 1992. [Google Scholar] [CrossRef]
- Rojas, T.I.; Aira, M.J.; Batista, A.; Cruz, I.L.; González, S. Fungal Biodeterioration in Historic Buildings of Havana (Cuba). Grana 2012, 51, 44–51. [Google Scholar] [CrossRef]
- Robinson, S.C.; Tudor, D.; MacDonald, G.; Mansourian, Y.; Cooper, P.A. Repurposing Mountain Pine Beetle Blue Wood for Art through Additional Fungal Colonization. Int. Biodeterior. Biodegrad. 2013, 85, 372–374. [Google Scholar] [CrossRef]
- Mansour, M.M.; Hamed, S.A.E.K.M.; Salem, M.Z.; Ali, H.M. Illustration of the effects of five fungi on Acacia saligna wood organic acids and ultrastructure alterations in wood cell walls by HPLC and TEM examinations. Appl. Sci. 2020, 10, 8–2886. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Smedsgaard, J.; Larsen, T.O.; Samson, R.A. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud. Mycol. 2004, 49, 201–241. [Google Scholar]
- Mustapha, K.; Rabah, S.; Anouar, A.; Ahmed, L.; Abdelrhafour, T.E.; Mohammed, E.; Maria Emilia, C.; Ela, C.; Alain, B. Antibacterial Effects and Toxigenesis of Penicillium Aurantiogriseum and P. Viridicatum. Afr. J. Biotechnol. 2007, 6, 2314–2318. [Google Scholar] [CrossRef]
- Bouhoudan, A.; Chidi, F.; Khaddor, M. Effect of sucrose on the physiology and terrestric acid production of Penicillium aurantiogriseum. Agric. For. 2020, 66, 179–189. [Google Scholar] [CrossRef]
- Wang, B.; Qi, M.; Ma, Y.; Zhang, B.; Hu, Y. Microbiome Diversity and Cellulose Decomposition Processes by Microorganisms on the Ancient Wooden Seawall of Qiantang River of Hangzhou, China. Microb. Ecol. 2023, 86, 2109–2119. [Google Scholar] [CrossRef]
- Zyani, M.; Mortabit, D.; Mostakim, M.; Iraqui, M.; Haggoud, A.; Ettayebi, M.; Koraichi, S.I. Cellulolytic Potential of Fungi in Wood Degradation from an Old House at the Medina of Fez. Ann. Microbiol. 2009, 59, 699–704. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Hong, S.-B.; Ko, S.-J.; Kim, S.-H. Detection of Extracellular Enzyme Activity in Penicillium Using Chromogenic Media. Mycobiology 2007, 35, 166–169. [Google Scholar] [CrossRef]
- Hirose, D.; Shirouzu, T.; Hirota, M.; Ohtsuka, T.; Senga, Y.; Du, M.; Shimono, A.; Zhang, X. Species richness and species composition of fungal communities associated with cellulose decomposition at different altitudes on the Tibetan Plateau. J. Plant Ecol. 2009, 2, 217–224. [Google Scholar] [CrossRef]
- Tarayre, C.; Bauwens, J.; Brasseur, C.; Mattéotti, C.; Millet, C.; Guiot, P.A.; Destain, J.; Vandenbol, M.; Portetelle, D.; De Pauw, E.; et al. Isolation and Cultivation of Xylanolytic and Cellulolytic Sarocladium kiliense and Trichoderma virens from the Gut of the Termite Reticulitermes Santonensis. Environ. Sci. Pollut. Res. 2015, 22, 4369–4382. [Google Scholar] [CrossRef]
- Woudenberg, J.H.C.; Sandoval-Denis, M.; Houbraken, J.; Seifert, K.A.; Samson, R.A. Cephalotrichum and Related Synnematous Fungi with Notes on Species from the Built Environment. Stud. Mycol. 2017, 88, 137–159. [Google Scholar] [CrossRef]
- Petlamul, W.; Sripornngam, T.; Buakwan, N.; Buakaew, S.; Mahamad, K. The Capability of Beauveria bassiana for Cellulase Enzyme Production. In Proceedings of the 7th International Conference on Bioscience, Biochemistry and Bioinformatics, Bangkok, Thailand, 21–23 January 2017; ACM: Bangkok, Thailand, 2017; pp. 62–66. [Google Scholar]
- Korniłłowicz-Kowalska, T.; Rybczyńska, K. Screening of Microscopic Fungi and Their Enzyme Activities for Decolorization and Biotransformation of Some Aromatic Compounds. Int. J. Environ. Sci. Technol. 2015, 12, 2673–2686. [Google Scholar] [CrossRef]
- Sista Kameshwar, A.K.; Qin, W. Systematic Review of Publicly Available Non-Dikarya Fungal Proteomes for Understanding Their Plant Biomass-Degrading and Bioremediation Potentials. Bioresour. Bioprocess. 2019, 6, 30. [Google Scholar] [CrossRef]
- Lisov, A.; Belova, O.; Zavarzina, A.; Konstantinov, A.; Leontievsky, A. The Role of Laccase from Zygomycetous Fungus Mortierella Elasson in Humic Acids Degradation. Agronomy 2021, 11, 2169. [Google Scholar] [CrossRef]
- Ben Younes, S.; Karray, F.; Sayadi, S. Isolation of Thermophilic Fungal Strains Producing Oxido-Reductase and Hydrolase Enzymes from Various Tunisian Biotopes. Int. Biodeterior. Biodegrad. 2011, 65, 1104–1109. [Google Scholar] [CrossRef]
- Asemoloye, M.D.; Tosi, S.; Daccò, C.; Wang, X.; Xu, S.; Marchisio, M.A.; Gao, W.; Jonathan, S.G.; Pecoraro, L. Hydrocarbon Degradation and Enzyme Activities of Aspergillus Oryzae and Mucor Irregularis Isolated from Nigerian Crude Oil-Polluted Sites. Microorganisms 2020, 8, 1912. [Google Scholar] [CrossRef]
- Sijinamanoj, V.; Muthukumar, T.; Muthuraja, R.; Rayappan, K.; Karmegam, N.; Saminathan, K.; Govarthanan, M.; Kethireswari, P. Ligninolytic valorization of agricultural residues by Aspergillus nomius and Trichoderma harzianum isolated from gut and comb of Odontotermes obesus (Termitidae). Chemosphere 2021, 284, 131384. [Google Scholar] [CrossRef]
- Anastasi, A.; Tigini, V.; Varese, G.C. The bioremediation potential of different ecophysiological groups of fungi. In Fungi as Bioremediators; Goltapeh, E.M., Ed.; Springer: Berlin, Heidelberg, 2013; pp. 29–49. [Google Scholar]


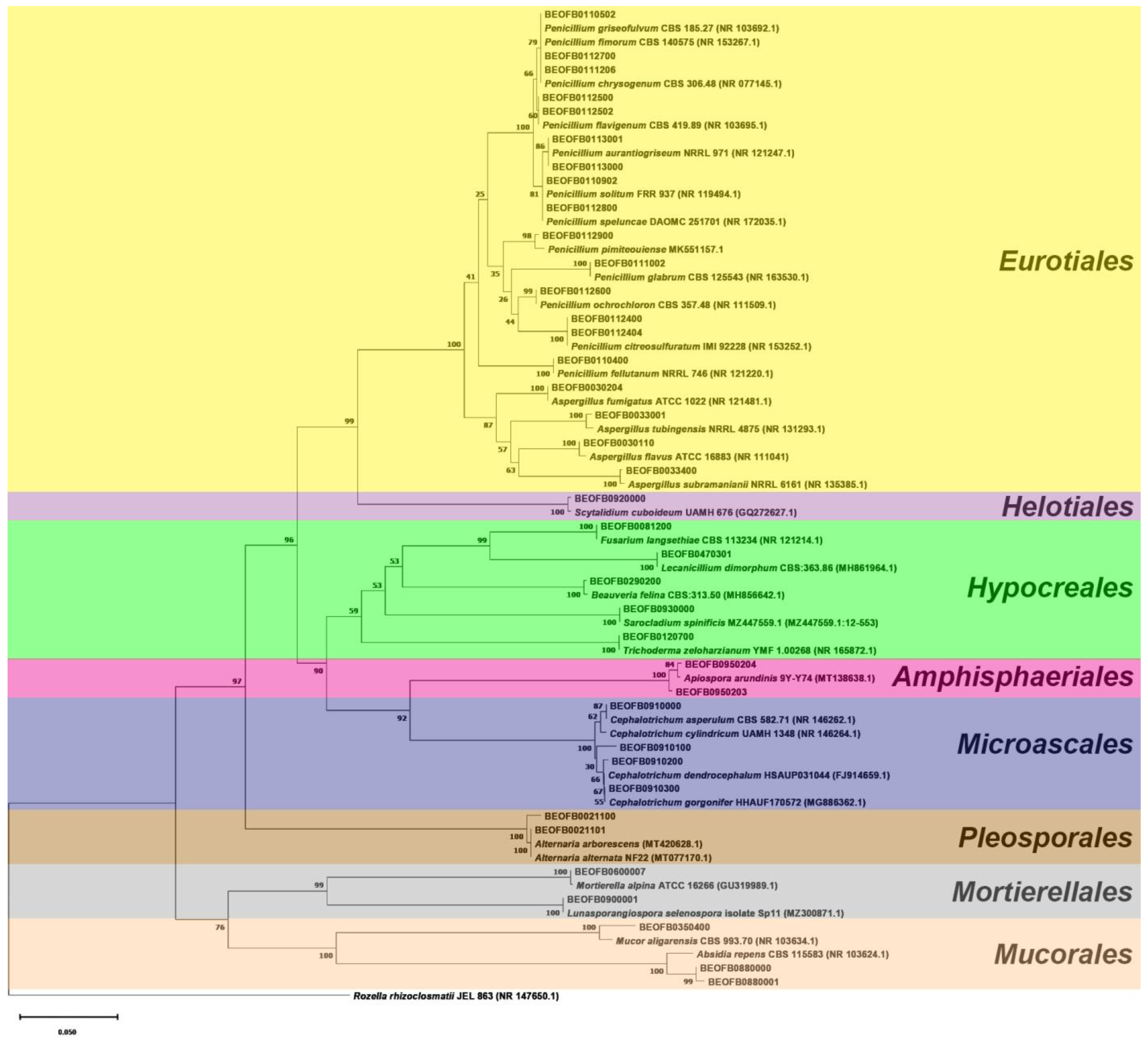
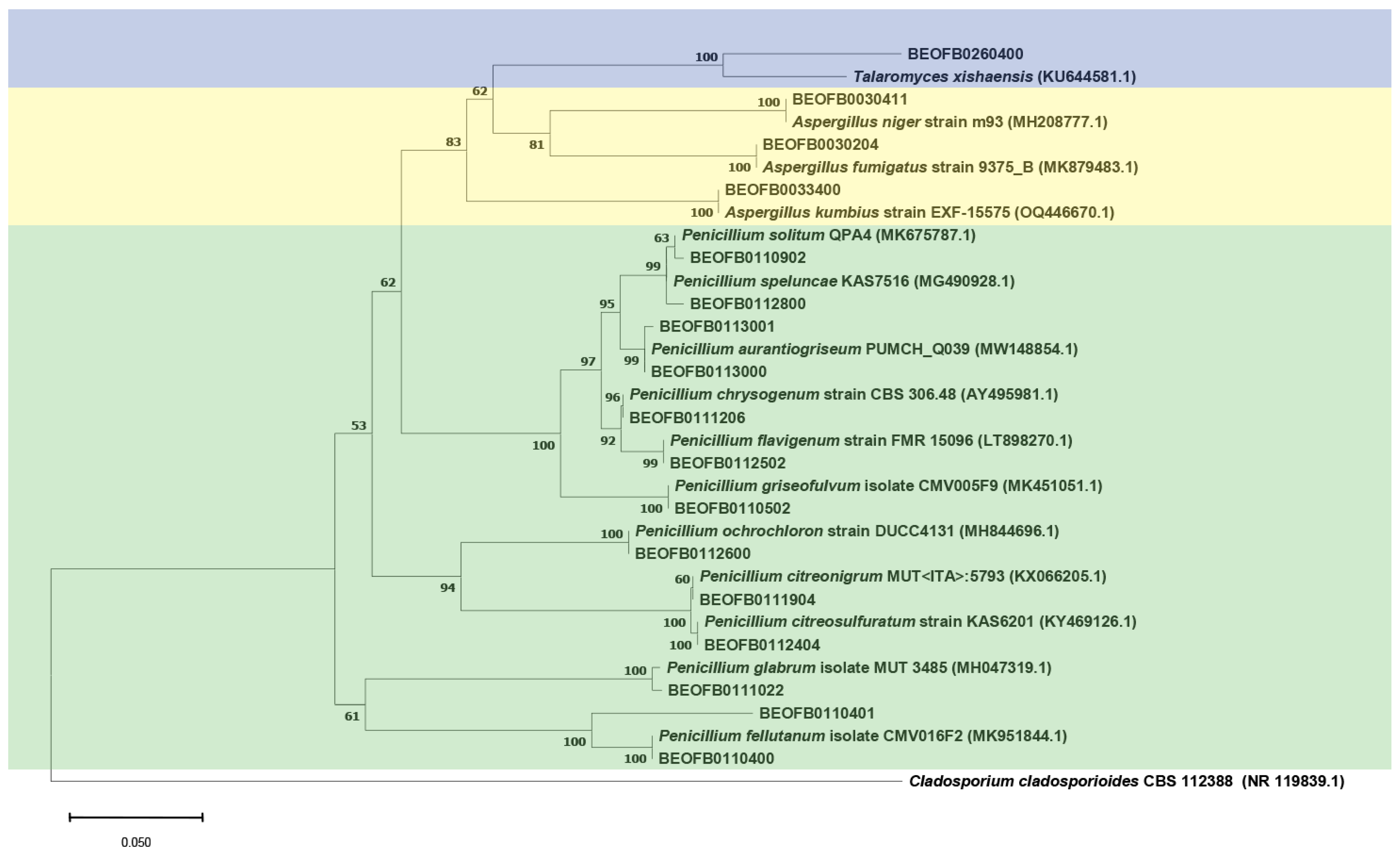
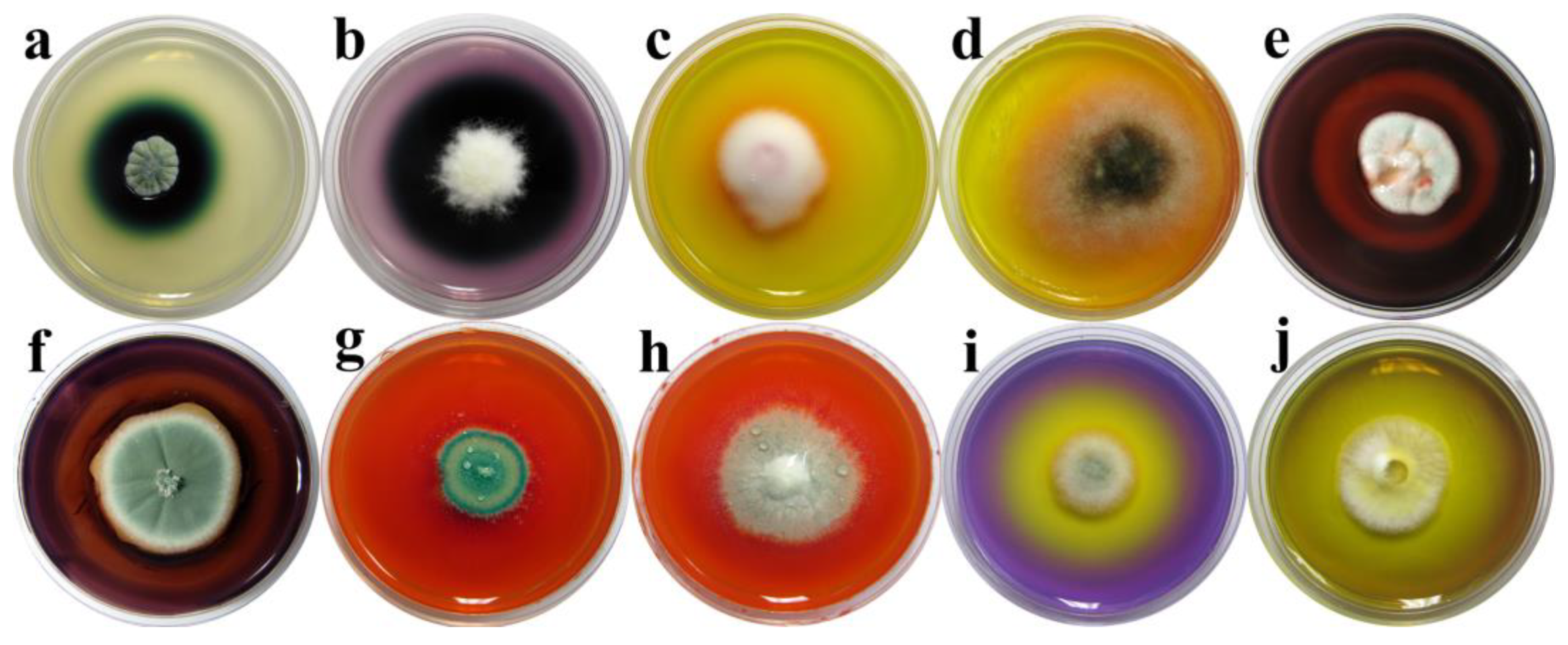
| Number | Strain | Sample Site | Isolate | Biodegradative Profile |
|---|---|---|---|---|
| 1 | BEOFB0880000 | M1 | Absidia repens | HA |
| 2 | BEOFB0880001 | B1 | Absidia repens | MnP, Lac, AP |
| 3 | BEOFB0021100 | M10 | Alternaria sp. | MnP, Lac |
| 4 | BEOFB0021101 | B4 | Alternaria sp. | HA, AP, PP |
| 5 | BEOFB0950204 | M7 | Apiospora arundinis | Lac |
| 6 | BEOFB0950203 | B3 | Apiospora arundinis | Lac |
| 7 | BEOFB0030110 | M11 | Aspergillus flavus var. flavus | Lac, HA |
| 8 | BEOFB0030204 | B4 | Aspergillus fumigatus | - |
| 9 | BEOFB0033400 | M9 | Aspergillus kumbius | - |
| 10 | BEOFB0030411 | M11 | Aspergillus niger | AP |
| 11 | BEOFB0290200 | M10 | Beauveria felina | CA |
| 12 | BEOFB0910000 | M3 | Cephalotrichum asperulum | MnP, Lac, HA, CA |
| 13 | BEOFB0910100 | M6 | Cephalotrichum cylindricum | Lac, HA, CA |
| 14 | BEOFB0910200 | B1 | Cephalotrichum dendrocephalum | MnP, HA, AP, PP |
| 15 | BEOFB0910300 | M9 | Cephalotrichum gorgonifer | HA |
| 16 | BEOFB0081200 | B4 | Fusarium langsethiae | Lac |
| 17 | BEOFB0470301 | M2 | Lecanicillium dimorphum | HA, CA, MnP |
| 18 | BEOFB0900001 | M3 | Lunasporangiospora selenospora | HA, CA |
| 19 | BEOFB0600007 | M2 | Mortierella alpina | MnP, Lac, CA |
| 20 | BEOFB0350400 | B2 | Mucor aligarensis | - |
| 21 | BEOFB0113000 | M10 | Penicillium aurantiogriseum | Lac, HA, AP |
| 22 | BEOFB0113001 | B3 | Penicillium aurantiogriseum | Lac, HA, AP, PP |
| 23 | BEOFB0111206 | B1 | Penicillium chrysogenum | HA, AP |
| 24 | BEOFB0111904 | M2 | Penicillium chrysogenum | Lac, HA |
| 25 | BEOFB0112404 | B1 | Penicillium citreosulfuratum | Lac, HA |
| 26 | BEOFB0110400 | M1 | Penicillium fellutanum | Lac |
| 27 | BEOFB0110401 | M6 | Penicillium fellutanum | Lac, HA |
| 28 | BEOFB0112500 | M3 | Penicillium flavigenum | MnP, Lac, HA, AP |
| 29 | BEOFB0112502 | B3 | Penicillium flavigenum | AP, HA, CA |
| 30 | BEOFB0111002 | B4 | Penicillium glabrum | AP, HA, PP |
| 31 | BEOFB0110502 | M7 | Penicillium griseofulvum | AP, CA, MnP, Lac |
| 32 | BEOFB0112600 | M3 | Penicillium ochrochloron | - |
| 33 | BEOFB0112900 | M10 | Penicillium pimiteouiense | Lac, AP |
| 34 | BEOFB0110902 | B3 | Penicillium solitum | MnP, Lac, HA, AP, CA |
| 35 | BEOFB0112800 | M6 | Penicillium speluncae | MnP, Lac, AP, CA |
| 36 | BEOFB0930000 | M3 | Sarocladium spinificis | Lac, HA, CA |
| 37 | BEOFB0260400 | M3 | Talaromyces xishaensis | - |
| 38 | BEOFB0120700 | M5 | Trichoderma zeloharzianum | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Djokić, I.; Knežević, A.; Savković, Ž.; Ljaljević Grbić, M.; Dimkić, I.; Bukvički, D.; Gavrilović, D.; Unković, N. Characterization of Culturable Mycobiome of Newly Excavated Ancient Wooden Vessels from the Archeological Site of Viminacium, Serbia. J. Fungi 2024, 10, 343. https://doi.org/10.3390/jof10050343
Djokić I, Knežević A, Savković Ž, Ljaljević Grbić M, Dimkić I, Bukvički D, Gavrilović D, Unković N. Characterization of Culturable Mycobiome of Newly Excavated Ancient Wooden Vessels from the Archeological Site of Viminacium, Serbia. Journal of Fungi. 2024; 10(5):343. https://doi.org/10.3390/jof10050343
Chicago/Turabian StyleDjokić, Ivana, Aleksandar Knežević, Željko Savković, Milica Ljaljević Grbić, Ivica Dimkić, Danka Bukvički, Dragana Gavrilović, and Nikola Unković. 2024. "Characterization of Culturable Mycobiome of Newly Excavated Ancient Wooden Vessels from the Archeological Site of Viminacium, Serbia" Journal of Fungi 10, no. 5: 343. https://doi.org/10.3390/jof10050343
APA StyleDjokić, I., Knežević, A., Savković, Ž., Ljaljević Grbić, M., Dimkić, I., Bukvički, D., Gavrilović, D., & Unković, N. (2024). Characterization of Culturable Mycobiome of Newly Excavated Ancient Wooden Vessels from the Archeological Site of Viminacium, Serbia. Journal of Fungi, 10(5), 343. https://doi.org/10.3390/jof10050343








