Abstract
Penicillium species are ubiquitous in the environment and are of substantial importance, especially in industrial and medical aspects. During our investigation of the biodiversity of Penicillium, three new species were discovered in soil samples collected from East and Northeast China. They were determined as new to science based on morphological comparisons and phylogenetic analyses, and were found to belong to the subgenus Penicillium section Robsamsonia and subgenus Aspergilloides sections Aspergilloides and Citrina. Descriptions and illustrations of these species are provided, and their geographic distributions are also discussed.
1. Introduction
Species of Penicillium Link are ubiquitous. Some of them are of industrial and medical importance. A range of efficient plant-polysaccharide-degrading enzymes are secreted by Penicillium species, such as P. oxalicum Currie & Thom, and are very useful for sustainable bioproduction [1]. Penicillium sumatraense Svilv. has the potential to be adopted in algal bio-refinery processes for biofuel production because of its arsenal of degrading enzymes [2]. Enzymes from Penicillium species are also crucial for the enhanced saccharification of agro-industrial wastes, which can be used to build a bio-based economy [3]. In the medicinal field, the best-known antibiotic penicillin was produced by P. chrysogenum Thom [4]. And more than 280 compounds have been reported from this genus, exhibiting lots of bioactive effects, e.g., antimicrobial, anticancer, antiviral, and antioxidant effects [5]. On the other hand, P. digitatum (Pers.) Sacc. is not only a major source of postharvest decay in citrus fruits worldwide [6] but is also a rare human pathogen that can cause fatal pneumonia in immunocompromised hosts [7,8].
The genus Penicillium was established in 1809, and P. expansum Link was designated as the type species. It is the most speciose in the order Eurotiales. In a monography published in 2014, 354 species were accepted in this genus [9], and 483 species were recognized in one that was published in 2020 [10]. By the end of 2022, 64 species had been further added to this group [11]. In the last year, 54 new species were described, and 43 of them were discovered in Southwestern China [12]. This leads to the species number of the genus over 600 at this moment. In China, more than 170 Penicillium species have been recorded, of which 91 were originally described from this country [12].
During an investigation into the biodiversity of Penicillium, three new species were discovered from the soil samples collected in East and Northeast China. Their descriptions and illustrations are provided here.
2. Materials and Methods
2.1. Fungal Materials
Cultures were isolated from soil samples collected from Heilongjiang, Jiangsu and Shanghai provinces or province-level municipality during 2021 to 2023. Dried cultures were deposited in the Herbarium Mycologicum Academiae Sinicae (HMAS, Beijing, China), and the living ex-type strains were preserved at the China General Microbiological Culture Collection Center (CGMCC, Beijing, China).
2.2. Morphological Observations
Morphological characterization was conducted following standardized methods [9]. Four standard growth media were used: Czapek yeast autolysate agar (CYA, yeast extract Oxoid, Hampshire, UK), malt extract agar (MEA, Amresco, Solon, OH, USA), yeast extract agar (YES), and potato dextrose agar (PDA). The methods of inoculation, incubation, microscopic examination, and digital recording matched those described in our previous studies [12,13,14,15,16,17,18].
2.3. DNA Extraction, PCR Amplification, and Sequencing
DNA was extracted from the cultures grown on PDA for 7 days using the Plant Genomic DNA Kit (DP305, TIANGEN Biotech, Beijing, China). Polymerase chain reaction (PCR) amplifications of the internal transcribed spacer (ITS), beta-tubulin (BenA), calmodulin (CaM), and RNA polymerase II second largest subunit (RPB2) gene regions were conducted using the routine methods [9]. The products were purified and subjected to sequencing on an ABI 3730 DNA Sequencer (Applied Biosystems, Foster, CA, USA). Although the ITS region, the proposed universal DNA barcode for fungi, is helpful to classify a Penicillium species at the section or series level, it is not sufficient to distinguish them at species level. However, ITS sequences are still provided here, as they might be beneficial to other researchers.
2.4. Phylogenetic Analyses
The forward and reverse sequences newly generated in this study were assembled using Seqman v. 7.1.0 (DNASTAR Inc., Madison, WI, USA). The assembled sequences were deposited in GenBank. The sequences used for phylogenetic analyses are listed in Table 1, Table 2 and Table 3. Sequences of each of the three single gene datasets (BenA, CaM and RPB2) and the concatenated ones were aligned using MAFFT v. 7.221 [19], then manually edited and concatenated in BioEdit v. 7.1.10 [20] and MEGA v. 11.0.13 [21]. Maximum likelihood (ML) analyses were conducted using RAxML-HPC2 [22] on XSEDE 8.2.12 on CIPRES Science Gateway v. 3.3 [23] with the default GTRCAT model and bootstrap (BP) iteration setting. Bayesian inference (BI) analyses were performed with MrBayes v. 3.2.7 [24]. Appropriate nucleotide substitution models and parameters were determined using Modeltest v. 3.7 [25]. Four MCMC chains were run for at least 1 million generations, and posterior probability (PP) values were estimated with the remaining 75% of trees after a burn-in phase. The consensus trees were viewed in FigTree v. 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 28 December 2023)).

Table 1.
Species and sequences of Penicillium subgen. Penicillium sect. Robsamsonia ser. Robsamsonia used in phylogenetic analyses.

Table 2.
Species and sequences of Penicillium subgen. Aspergilloides sect. Aspergilloides ser. Glabra used in phylogenetic analyses.

Table 3.
Species and sequences of Penicillium subgen. Aspergilloides sect. Citrina ser. Sumatraensia used in phylogenetic analyses.
3. Results
To determine the phylogenetic positions of the new species, single-gene datasets (BenA, CaM and RPB2) and concatenated ones were compiled and analyzed for Penicillium subgen. Penicillium sect. Robsamsonia ser. Robsamsonia, subgen. Aspergilloides sect. Aspergilloides ser. Glabra, and sect. Citrina ser. Sumatraensia. Detailed characteristics of the datasets are listed in Table 4.

Table 4.
Detailed characteristics of the datasets.
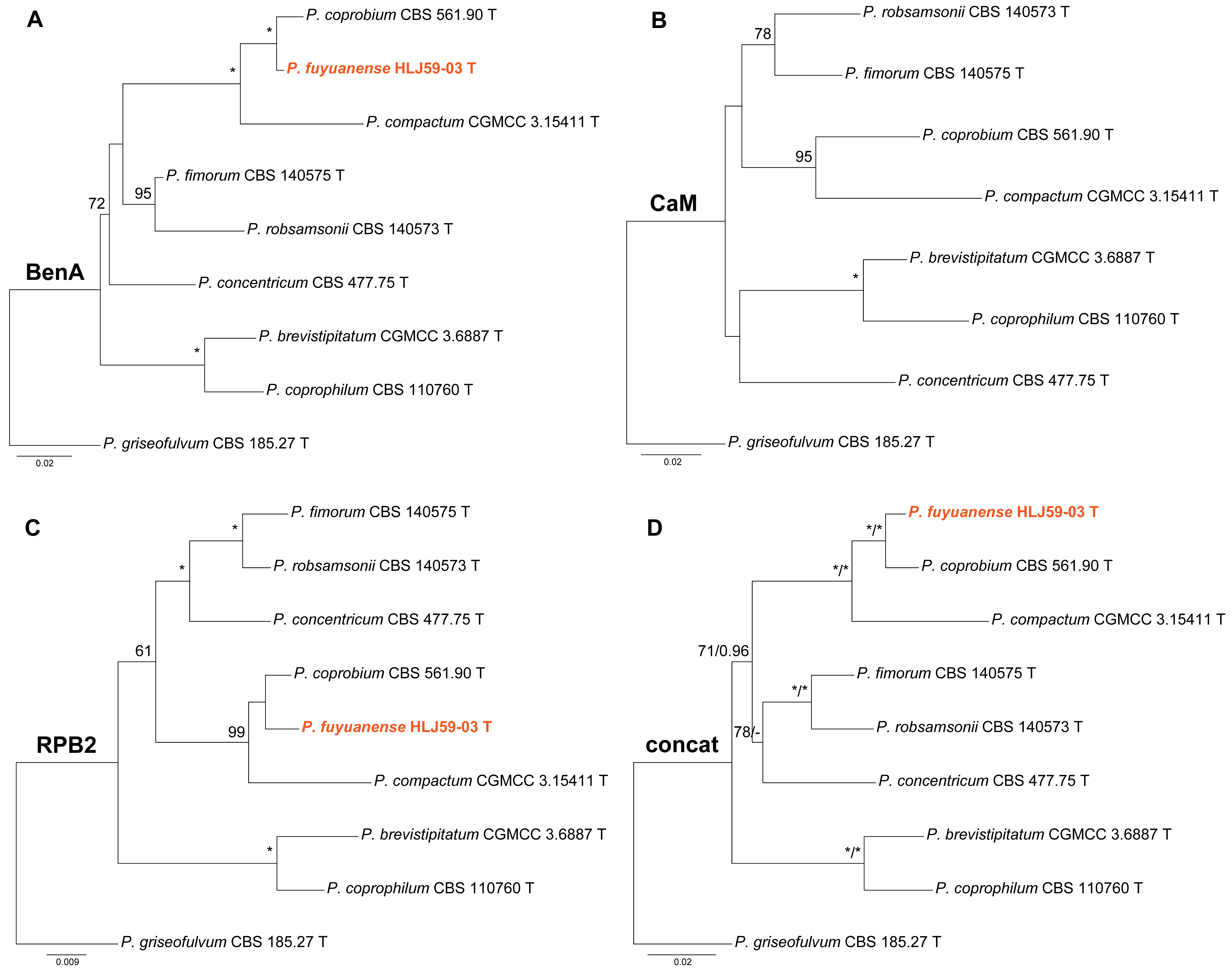
Phylogenies of Penicillium subgen. Penicillium sect. Robsamsonia ser. Robsamsonia are given in Figure 1. In the BenA and concatenated phylogenies (Figure 1A,D), the strain HLJ59-03 was closely related to P. coprobium with strong support (MLBP = 100% or BIPP = 1.00). Although HLJ59-03 was also a sister taxon of P. coprobium in the ML tree based on RPB2 sequences (Figure 1C), the statistical support between them was very weak. HLJ59-03 was absent in the CaM phylogeny (Figure 1B) because of the failure of PCR amplification.
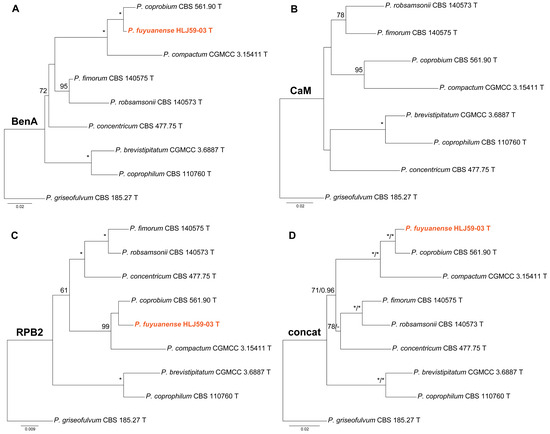
Figure 1.
Maximum likelihood phylogeny of Penicillium subgen. Penicillium section Robsamsonia series Robsamsonia inferred from (A) BenA, (B) CaM, (C) RPB2 and (D) concatenated datasets. Bootstrap values ≥ 50% are indicated at nodes of (A–C); bootstrap values ≥ 50% (left) or posterior probability values ≥ 0.90 (right) are indicated at nodes of (D). Asterisk denotes 100% bootstrap or 1.00 posterior probability. The new species is in color.
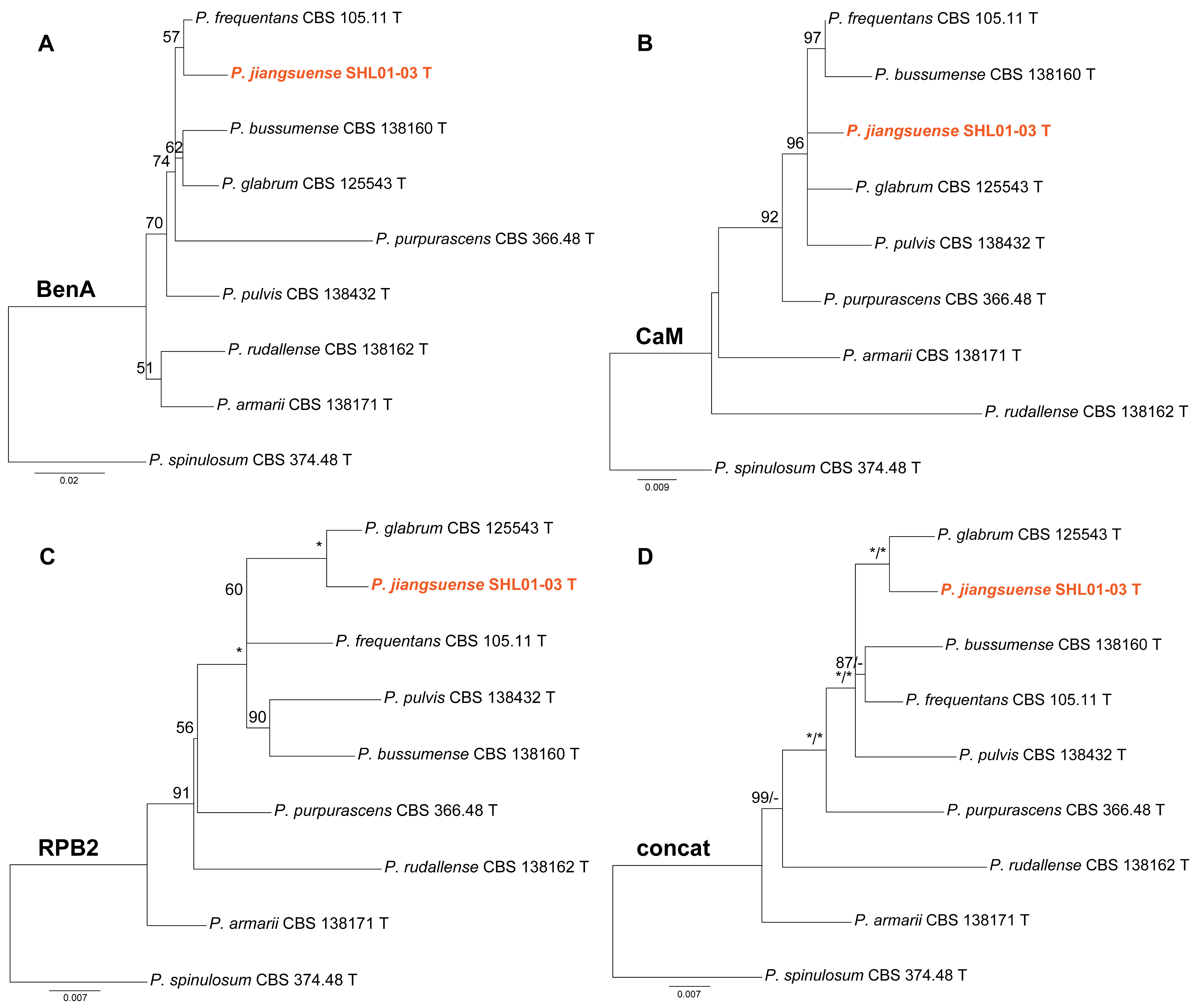
Phylogenies of Penicillium subgen. Aspergilloides sect. Aspergilloides ser. Glabra are shown in Figure 2. In the RPB2 and concatenated phylogenies (Figure 2C,D), strain SHL01-03 clustered with P. glabrum receiving strong support (MLBP = 100% or BIPP = 1.00). In contrast, SHL01-03 was a sister taxon of P. frequentans with weak support in the ML tree of BenA sequences (Figure 2A) and as an independent lineage in the CaM phylogeny (Figure 2B).
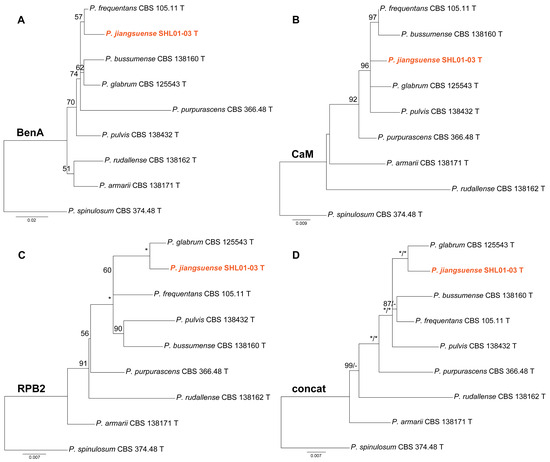
Figure 2.
Maximum likelihood phylogeny of Penicillium subgen. Aspergilloides section Aspergilloides series Glabra inferred from (A) BenA, (B) CaM, (C) RPB2 and (D) concatenated datasets. Bootstrap values ≥ 50% are indicated at nodes of (A–C); bootstrap values ≥ 50% (left) or posterior probability values ≥ 0.90 (right) are indicated at nodes of (D). Asterisk denotes 100% bootstrap or 1.00 posterior probability. The new species is in color.
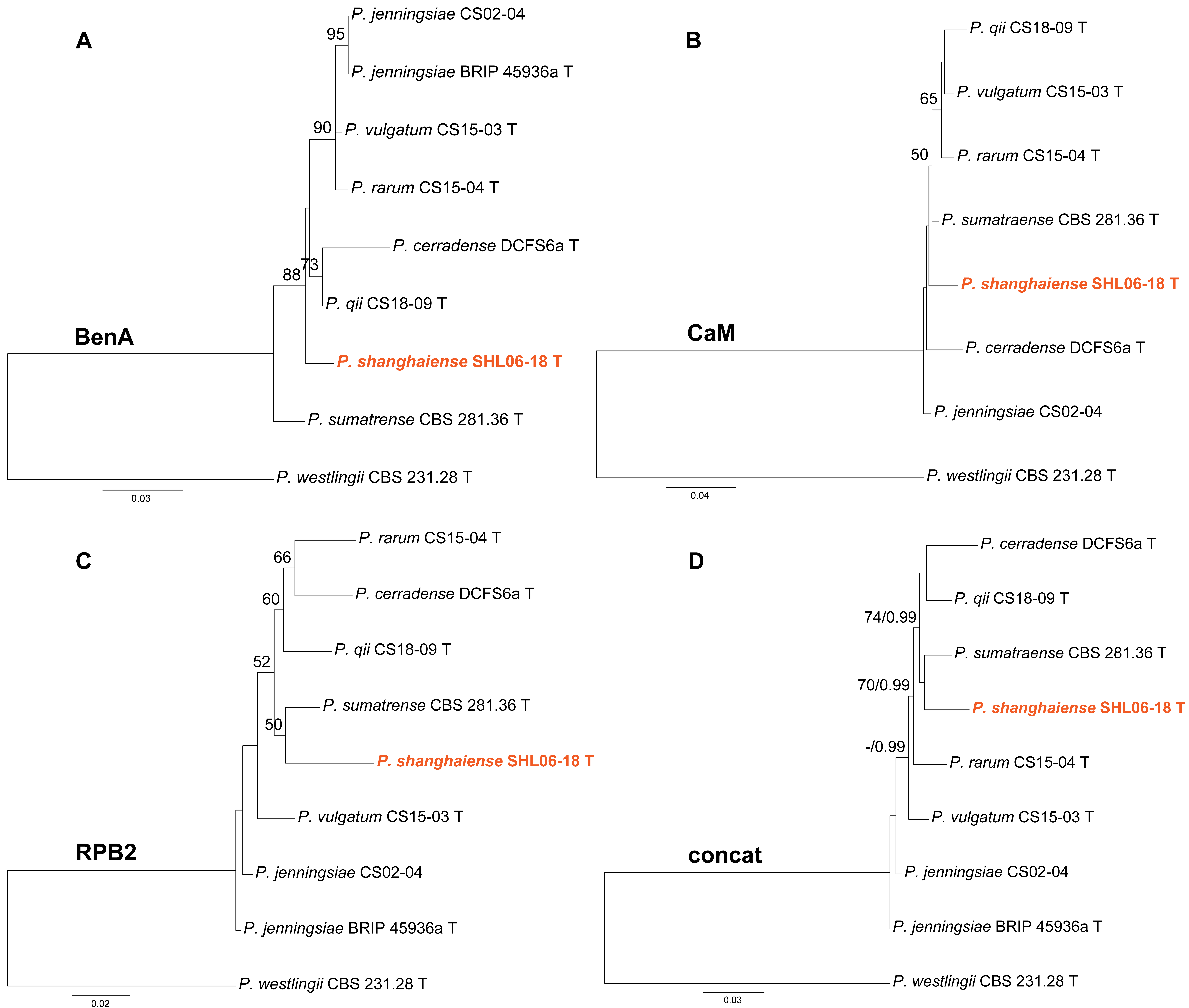
Phylogenies of Penicillium subgen. Aspergilloides sect. Citrina ser. Sumatraensia were depicted in Figure 3. Strain SHL06-18 was shown as an independent lineage in the BenA and CaM phylogenies (Figure 3A,B), while it was clustered with P. sumatraense in the RPB2 analysis and the concatenated ML tree (Figure 3C,D).
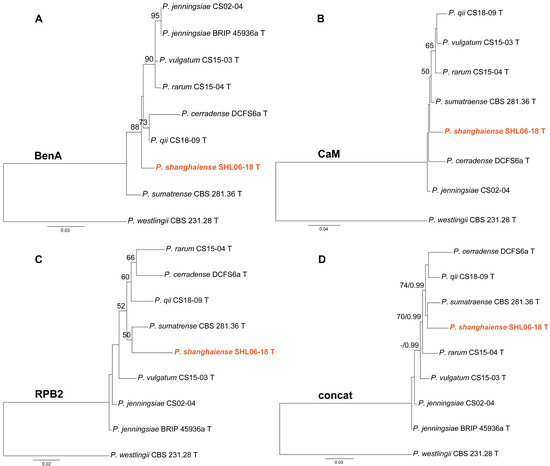
Figure 3.
Maximum likelihood phylogeny of Penicillium subgen. Aspergilloides section Citrina series Sumatraensia inferred from (A) BenA, (B) CaM, (C) RPB2 and (D) concatenated datasets. Bootstrap values ≥ 50% are indicated at nodes of (A–C); bootstrap values ≥ 50% (left) or posterior probability values ≥ 0.90 (right) are indicated at nodes of (D). The new species is in color.
4. Taxonomy
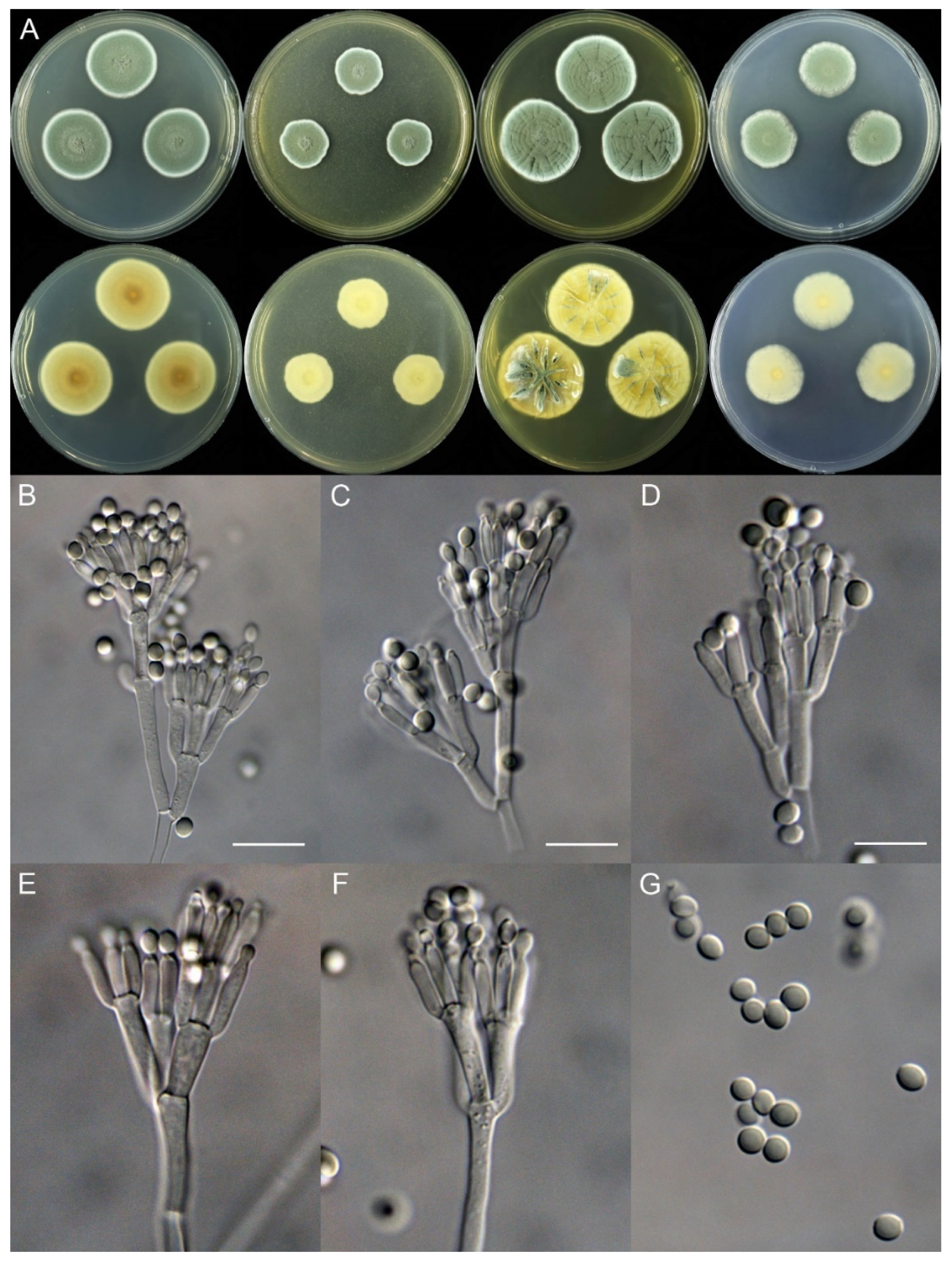
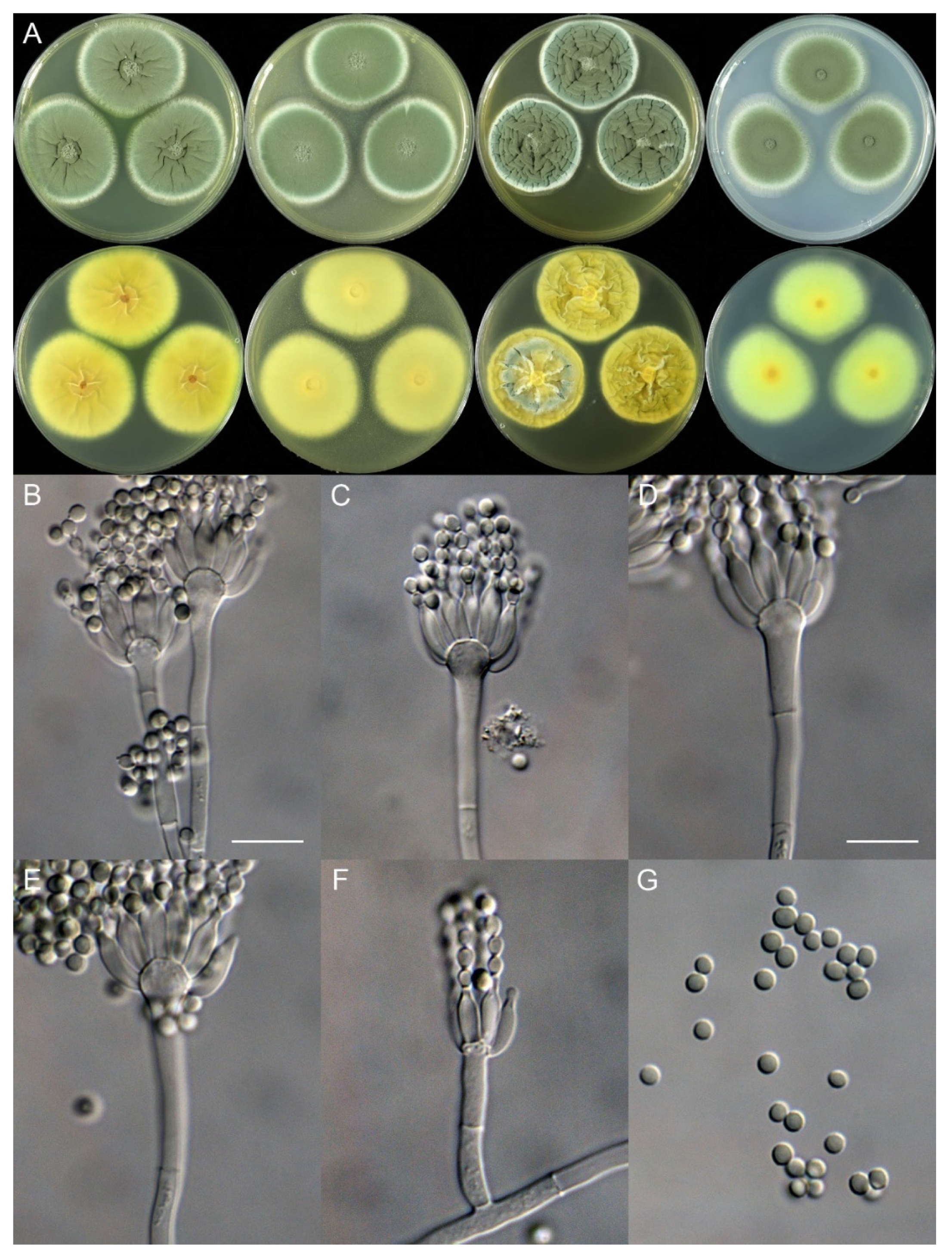
Penicillium fuyuanense X.C. Wang & W.Y. Zhuang, sp. nov. Figure 4.
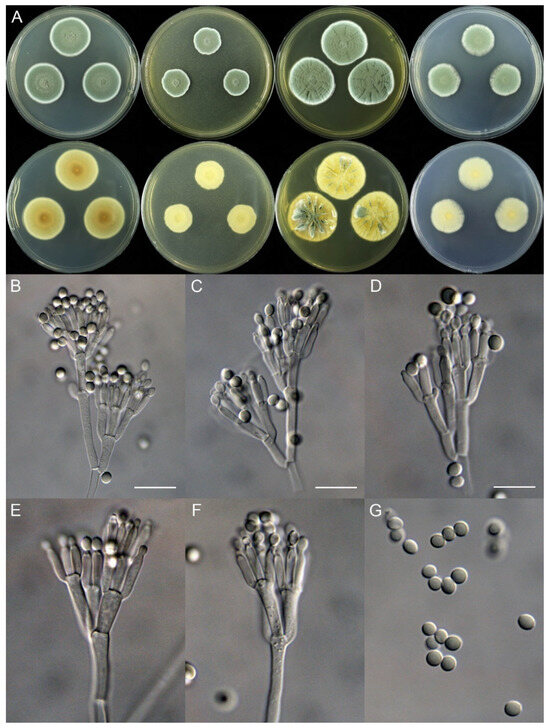
Figure 4.
Penicillium fuyuanense (HLJ59-03). (A) Colonies: top row left to right, obverse CYA, MEA, YES, and PDA; bottom row left to right, reverse CYA, MEA, YES, and PDA; (B–F) Conidiophores; (G) Conidia. Bars: (B) = 15 µm; (C) = 12.5 µm; (D) = 10 µm, also for (E–G).
Fungal Names: FN571813.
Etymology: The specific epithet refers to the type locality.
In Penicillium subgenus Penicillium section Robsamsonia series Robsamsonia.
Typification: China. Heilongjiang Province, Jiamusi City, Fuyuan City, Fuyuan Town, Dongjige, 48°21′27″ N 134°16′58″ E, in soil under Rhododendron dauricum L., 14 May 2023, Xin-Cun Wang and He Song, culture, He Song, HLJ59-03 (holotype HMAS 247927, ex-type strain CGMCC 3.27293).
DNA barcodes: ITS PP357618, BenA PP373069, RPB2 PP373080.
Colony diam., 7 days, 25 °C (unless stated otherwise): CYA 27–28 mm; CYA 37 °C no growth; CYA 5 °C germinated, 3–4 mm; MEA 19–20 mm; YES 32–34 mm; PDA 22–23 mm.
Colony characteristics: On CYA 25 °C, 7 days: Colonies nearly circular, plain, protuberant and funiculose at centers, slightly concentrically sulcate; margins narrow, entire; mycelia white; texture velutinous; sporulation dense; conidia en masse dull green; soluble pigments absent; exudates absent; reverse yellow brown, white at margins.
On MEA 25 °C, 7 days: Colonies irregular, plain, protuberant and funiculose at centers; margins narrow, entire; mycelia white; texture velutinous; sporulation dense; conidia en masse dull green; soluble pigments absent; exudates absent; reverse white to buff, darker at centers.
On YES 25 °C, 7 days: Colonies nearly circular, plain or protuberant, concentrically and radially sulcate, funiculose at centers; margins narrow, entire; mycelia white; texture velutinous; sporulation dense; conidia en masse dull green; soluble pigments absent; exudates absent; reverse yellow brown.
On PDA 25 °C, 7 days: Colonies nearly circular to irregular, slightly protuberant at centers; margins narrow, entire or irregular; mycelia white; texture velutinous; sporulation dense; conidia en masse dull green; soluble pigments absent; exudates absent; reverse white, yellow at centers.
Micromorphology: Conidiophores biverticillate, terverticillate, quaterverticillate or more branched; stipes smooth-walled, 20–375 × 2.0–3.5 μm; branches 2, 16.5–28.5 × 2.5–5 μm; rami 2–3, 10–14.5 × 2.5–5.5 μm; metulae 2–5, 8–14.5 × 2.5–5 μm; phialides acerose, tapering into very thin neck, 4–6 per metula, 7.5–11 × 2.0–3.0 μm; conidia subglobose to ellipsoidal, smooth-walled, 3.5–4.5 × 3.0–4.0 μm.
Notes: This species is closely related to P. coprobium and P. compactum (Figure 1) phylogenetically. It differs from P. coprobium in 10 bp for BenA and 13 bp for RPB2, and from P. compactum in 25 bp for BenA and 29 bp for RPB2. Morphologically, it differs from P. coprobium in lacking of white sclerotia on MEA and producing quaterverticillate or even more branched conidiophores, shorter rami (10–14.5 vs. 12–20 μm), and larger conidia (3.5–4.5 × 3.0–4.0 vs. 3.2–4.0 × 2.5–3.0 μm) [26]. It differs from P. compactum in yellow brown other than dark brown or blackish brown colony on reverse view of CYA and YES at 25 °C, quaterverticillate or rich-branched conidiophores, longer stipes (20–375 vs. 40–100 μm) and shorter phialides (7.5–11 vs. 9–13 μm) [27]. Their morphological distinctions are summarized in Table 5.

Table 5.
Morphological comparisons of new species and their closely related species in series Robsamsonia.
Penicillium jiangsuense X.C. Wang & W.Y. Zhuang, sp. nov. Figure 5.
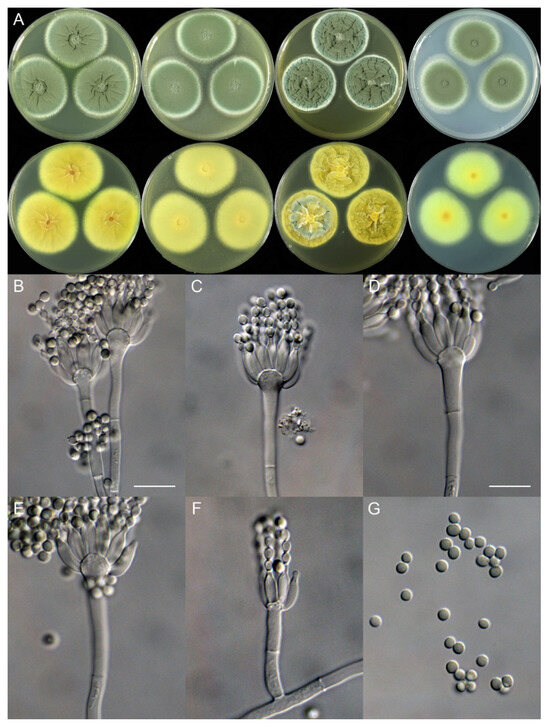
Figure 5.
Penicillium jiangsuense (SHL01-03). (A) Colonies: top row left to right, obverse CYA, MEA, YES, and PDA; bottom row left to right, reverse CYA, MEA, YES, and PDA; (B–F) Conidiophores; (G) Conidia. Bars: (B) = 12.5 µm, also for (C); (D) = 10 µm, also for (E–G).
Fungal Names: FN571814.
Etymology: The specific epithet refers to the type locality.
In Penicillium subgenus Aspergilloides section Aspergilloides series Glabra.
Typification: China. Jiangsu Province, Lianyungang City, Haizhou District, Huaguoshan Mountain, 34°38′22″ N 119°16′55″ E, in soil, 20 April 2021, Xin-Cun Wang, culture, Yi-Jing Ding, SHL01-03 (holotype HMAS 247928, ex-type strain CGMCC 3.27294).
DNA barcodes: ITS PP357619, BenA PP373070, CaM PP373075, RPB2 PP373081.
Colony diam., 7 days, 25 °C (unless stated otherwise): CYA 44–45 mm; CYA 37 °C 8–9 mm; CYA 5 °C germinated, 2–3 mm; MEA 42–43 mm; YES 40–41 mm; PDA 40–41 mm.
Colony characteristics: On CYA 25 °C, 7 days: Colonies nearly circular, protuberant at centers, radially sulcate; margins moderately wide, entire; mycelia white; texture velutinous; sporulation dense; conidia en masse dull green; soluble pigments absent; exudates absent; reverse yellow brown, red brown at centers.
On CYA 37 °C, 7 days: Colonies irregular, protuberant, cerebroid, strongly sulcate; margins narrow, irregular; mycelia white; texture tight; sporulation absent; conidia en masse not seen; soluble pigments absent; exudates absent; reverse carneous or flesh-colored, lighter at centers.
On MEA 25 °C, 7 days: Colonies nearly circular, plain, funiculose at centers; margins moderately wide, entire; mycelia white; texture velutinous; sporulation dense; conidia en masse dull green; soluble pigments absent; exudates absent; reverse light yellow brown.
On YES 25 °C, 7 days: Colonies nearly circular, strongly sulcate, concave at centers; margins narrow to moderately wide, entire; mycelia white; texture velutinous; sporulation dense; conidia en masse dull green; soluble pigments absent; exudates absent; reverse yellow brown.
On PDA 25 °C, 7 days: Colonies nearly circular, plain, slightly funiculose at centers; margins wide, fimbriate; mycelia white; texture velutinous; sporulation dense; conidia en masse dull green; soluble pigments absent; exudates absent; reverse white, orange brown at centers.
Micromorphology: Conidiophores monoverticillate; stipes smooth-walled, rarely rough-walled, 35–85 × 2.5–4.0 μm; phialides ampulliform, tapering into a very thin neck, 8–12 per metula, 8.5–11 (–13.5) × 2.5–4.5 μm; conidia subglobose to ellipsoidal, smooth-walled, 3.0–3.5 × 2.5–3.0 μm.
Notes: This species is closely related to P. frequentans (Figure 2A) and P. glabrum phylogenetically (Figure 2C,D). It differs from P. frequentans in 7 bp for BenA, 5 bp for CaM and 22 bp for RPB2; and from P. glabrum in 10 bp for BenA, 8 bp for CaM and 10 bp for RPB2. Morphologically, it differs from the above two species in being able to grow on CYA at 37 °C and having smooth-walled and slightly larger conidia (3.0–3.5 vs. 2.5–3.0 μm) [28,29]. Their morphological distinctions are listed in Table 6.

Table 6.
Morphological comparisons of new species and their closely related species in the series Glabra.
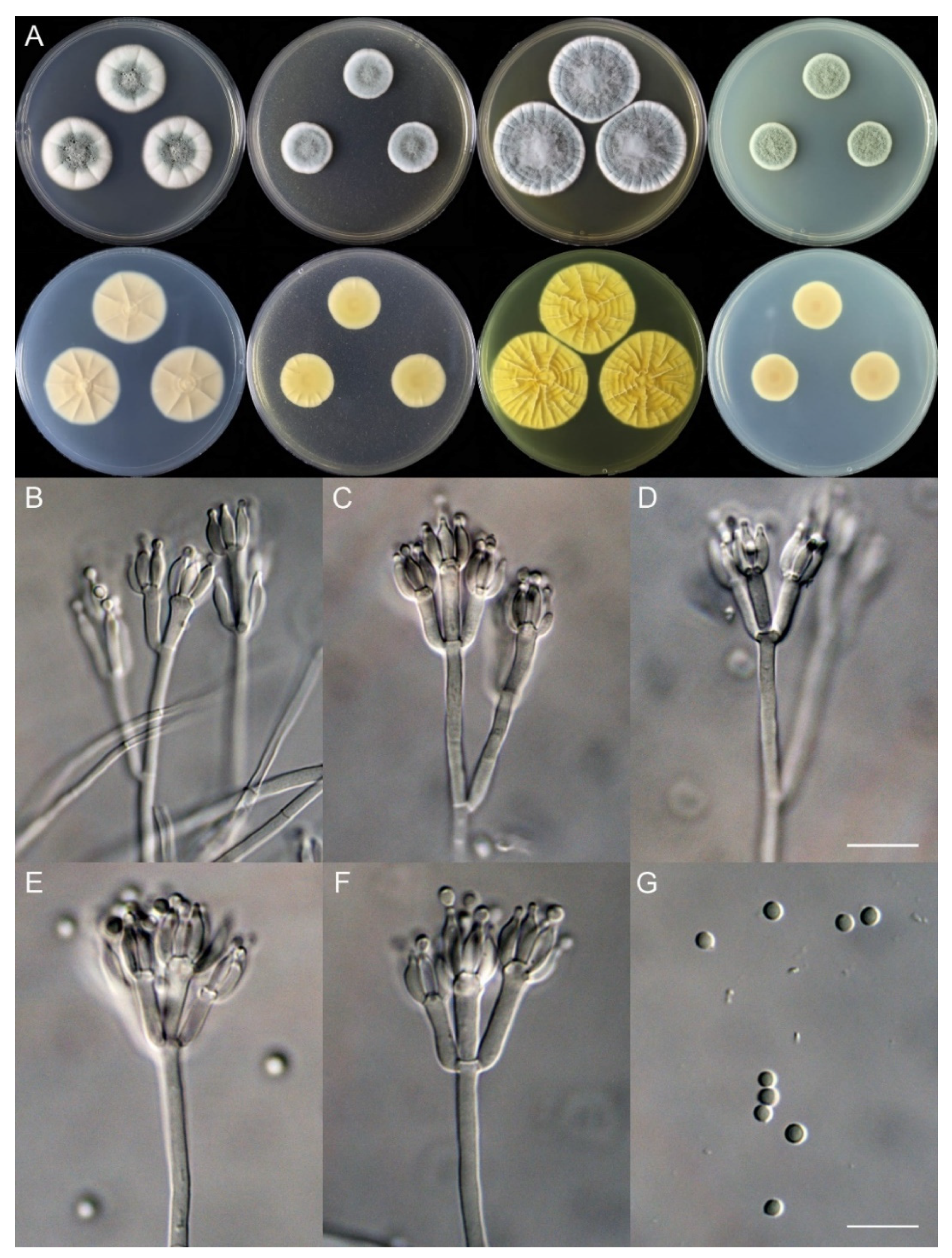
Penicillium shanghaiense X.C. Wang & W.Y. Zhuang, sp. nov. Figure 6.
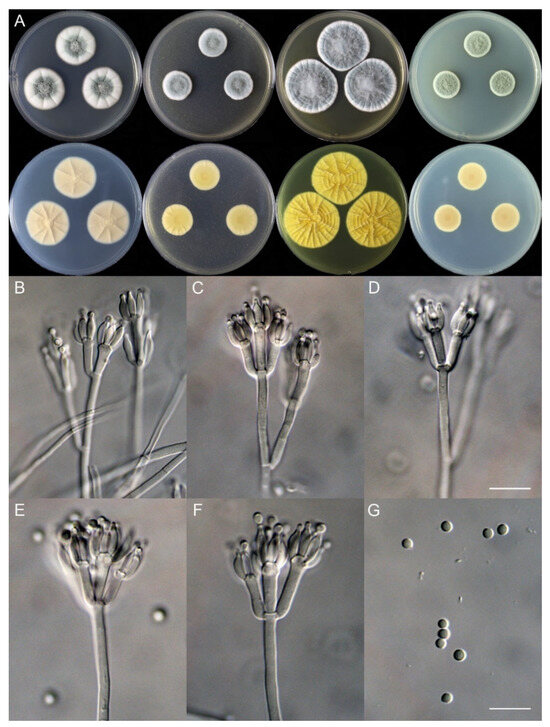
Figure 6.
Penicillium shanghaiense (SHL06-18). (A) Colonies: top row left to right, obverse CYA, MEA, YES, and PDA; bottom row left to right, reverse CYA, MEA, YES, and PDA; (B–F) Conidiophores; (G) Conidia. Bars: (D) = 12.5 µm, also for (B,C); (G) = 10 µm, also for (E,F).
Fungal Names: FN571815.
Etymology: The specific epithet refers to the type locality.
In Penicillium subgenus Aspergilloides section Citrina series Sumatraensia.
Typification: China. Shanghai City, Chongming District, Dongtan National Nature Reserve, 31°31′6″ N 121°56′58″ E, in soil under Camphora officinarum Nees ex Wall., 23 April 2021, Xin-Cun Wang, culture, Yi-Jing Ding, SHL06-18 (holotype HMAS 247929, ex-type strain CGMCC 3.27295).
DNA barcodes: ITS PP357620, BenA PP373071, CaM PP373076, RPB2 PP373082.
Colony diam., 7 days, 25 °C (unless stated otherwise): CYA 29–31 mm; CYA 37 °C no growth; CYA 5 °C no growth; MEA 20–21 mm; YES 37–39 mm; PDA 18–19 mm.
Colony characteristics: On CYA 25 °C, 7 days: Colonies nearly circular, slightly protuberant at centers, radially sulcate; margins wide, entire; mycelia white; texture velutinous; sporulation moderately dense; conidia en masse bluish green to dull green; soluble pigments absent; exudates abundant, clear; reverse carneous or flesh-colored.
On MEA 25 °C, 7 days: Colonies nearly circular, protuberant; margins narrow to moderately wide, entire; mycelia white; texture floccose; sporulation dense; conidia en masse greyish green; soluble pigments absent; exudates absent; reverse yellow brown, buff at margins.
On YES 25 °C, 7 days: Colonies nearly circular, strongly sulcate; margins narrow to moderately wide, undulated; mycelia white; texture floccose, velutinous at margin areas; sporulation dense; conidia en masse greyish green; soluble pigments absent; exudates absent; reverse buff to yellow brown.
On PDA 25 °C, 7 days: Colonies nearly circular; margins narrow, entire; mycelia white; texture floccose; sporulation dense; conidia en masse greyish green; soluble pigments absent; exudates absent; reverse light brown.
Micromorphology: Conidiophores biverticillate or terverticillate; stipes smooth-walled, 85–415 × 2.0–3.0 μm; rami 2, 21.5–32.5 × 2.0–3.0 μm; metulae 2–5, 9.5–17.5 × 2.0–5.0 μm; phialides ampulliform, tapering into a very thin neck, 3–8 per metula, 6.0–9.0 × 2.0–3.0 μm; conidia subglobose, smooth-walled, 2.0–3.0 μm.
Notes: This species is a member of the series Sumatraensia and a sister of the other six species in the group (Figure 3). Phylogenetically, it differs from P. cerradense in 97 bp for the three gene fragments (the details can be found in Table 7), from P. jenningsiae in 71 bp, from P. qii in 66 bp, from P. rarum in 76 bp, from P. sumatraense in 72 bp, and from P. vulgatum in 69 bp. Morphologically, the new species differs from P. cerradense in terms of its faster growth rate on MEA (20–21 vs. 15 mm), slower growth rate on PDA (18–19 vs. 30 mm), terverticillate instead of predominantly monoverticillate conidiophores, and absence of sclerotia [30]; from P. jenningsiae in slower growth rates on the four media, terverticillate conidiophores, longer stipes (85–415 vs. 100–250 μm), and shorter phialides (6–9 vs. 8–12 μm) [12,31]; from P. qii in terverticillate conidiophores [12]; from P. rarum in without monoverticillate conidiophores and longer rami (21.5–32.5 vs. 14–20 μm) [12]; from P. sumatraense in its slower growth rate on MEA (20–21 vs. 30–45 mm) [29]; and from P. vulgatum in its slower growth rates on the four media and terverticillate conidiophores [12]. Table 7 provides the detailed distinctions among taxa in the series.

Table 7.
Morphological and molecular comparisons of new species and their closely related species in the series Sumatraensia.
5. Discussion
The three new species were isolated from soil samples collected from East China (Jiangsu Province and Shanghai City) and Northeast China (Heilongjiang Province). They are morphologically and phylogenetically distinguishable from any existing species of the genus and belong to the Penicillium subgenus Penicillium section Robsamsonia series Robsamsonia, subgenus Aspergilloides section Aspergilloides series Glabra and section Citrina series Sumatraensia.
The three new species are from different climate zones. Penicillium fuyuanense, along with its closest sisters P. coprobium and P. compactum, are all from boreal (cold–temperate) zone and high-latitude areas. Penicillium jiangsuense, P. shanghaiense and their relatives are from a temperate zone. Most species of Penicillium are usually found in the same climate zone. Penicillium choerospondiatis X.C. Wang & W.Y. Zhuang occurred in subtropical China [15], and was later reported to be found in Kolkata, India [32]. For the time being, it seems that samples of individual new species are expected to be seen in similar climates.
Northeast China and East China encompass 10 provinces and Shanghai Municipal City, accounting for 17% area of this country, in which nine Penicillium species were originally described, e.g., P. compactum from Heilongjiang, P. jianxiense H.Z. Kong & Z.Q. Liang from Jiangxi, and P. formosanum H.M. Hsieh, H.J. Su & Tzean, and P. ulaiense H.M. Hsieh, H.J. Su & Tzean from Taiwan. Compared with the fact that 37 new species of the genus have recently been recorded in Chongqing Municipal City [12], covering only 0.86% of China, more intensive investigations are desperately needed to extend our knowledge on the biodiversity of Penicillium in our country.
Author Contributions
Conceptualization, W.-Y.Z., G.-Z.D. and X.-C.W.; Methodology, X.-C.W.; Software, X.-C.W.; Validation, X.-C.W.; Formal analysis, H.S., Y.-J.D. and X.-C.W.; Investigation, H.S. and X.-C.W.; Resources, W.-Y.Z., G.-Z.D. and X.-C.W.; Data curation, X.-C.W.; Writing—original draft, X.-C.W.; Writing—review & editing, W.-Y.Z. and X.-C.W.; Visualization, X.-C.W.; Supervision, W.-Y.Z., G.-Z.D. and X.-C.W.; Project administration, W.-Y.Z., G.-Z.D. and X.-C.W.; Funding acquisition, W.-Y.Z., G.-Z.D. and X.-C.W. All authors have read and agreed to the published version of the manuscript.
Funding
This project was supported by the National Key Research and Development Program of China (2022YFC2303000), the National Natural Science Foundation of China (32270008) and Heilongjiang province economic crop industry technology collaborative innovation system construction project (2018711).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ning, Y.N.; Tian, D.; Zhao, S.; Feng, J.X. Regulation of genes encoding polysaccharide-degrading enzymes in Penicillium. Appl. Microbiol. Biotechnol. 2024, 108, 16. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, M.; Larini, I.; Scafati, V.; Scortica, A.; Compri, M.; Pontiggia, D.; Zapparoli, G.; Vitulo, N.; Benedetti, M.; Mattei, B. A novel Penicillium sumatraense isolate reveals an arsenal of degrading enzymes exploitable in algal bio-refinery processes. Biotechnol. Biofuels 2021, 14, 180. [Google Scholar] [CrossRef] [PubMed]
- Karp, S.G.; Rozhkova, A.M.; Semenova, M.V.; Osipov, D.O.; de Pauli, S.T.Z.; Sinitsyna, O.A.; Zorov, I.N.; de Souza Vandenberghe, L.P.; Soccol, C.R.; Sinitsyn, A.P. Designing enzyme cocktails from Penicillium and Aspergillus species for the enhanced saccharification of agro-industrial wastes. Bioresour. Technol. 2021, 330, 124888. [Google Scholar] [CrossRef] [PubMed]
- Houbraken, J.; Frisvad, J.C.; Seifert, K.A.; Overy, D.P.; Tuthill, D.M.; Valdez, J.G.; Samson, R.A. New penicillin-producing Penicillium species and an overview of section Chrysogena. Persoonia 2012, 29, 78–100. [Google Scholar] [CrossRef] [PubMed]
- Toghueo, R.M.K.; Boyom, F.F. Endophytic Penicillium species and their agricultural, biotechnological, and pharmaceutical applications. 3 Biotech 2020, 10, 107. [Google Scholar] [CrossRef]
- Costa, J.H.; Bazioli, J.M.; Pontes, J.G.D.; Fill, T.P. Penicillium digitatum infection mechanisms in citrus: What do we know so far? Fungal Biol. 2019, 123, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Oshikata, C.; Tsurikisawa, N.; Saito, A.; Watanabe, M.; Kamata, Y.; Tanaka, M.; Tsuburai, T.; Mitomi, H.; Takatori, K.; Yasueda, H.; et al. Fatal pneumonia caused by Penicillium digitatum: A case report. BMC Pulm. Med. 2013, 13, 16. [Google Scholar] [CrossRef] [PubMed]
- Iturrieta-Gonzalez, I.; Giacaman, A.; Godoy-Martinez, P.; Vega, F.; Sepulveda, M.; Santos, C.; Toledo, V.; Rivera, G.; Ortega, L.; San Martin, A.; et al. Penicillium digitatum, first clinical report in Chile: Fungal co-infection in COVID-19 patient. J. Fungi 2022, 8, 961. [Google Scholar] [CrossRef]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.B.; Klaassen, C.H.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samson, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef]
- Houbraken, J.; Kocsube, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef]
- Visagie, C.M.; Yilmaz, N.; Kocsubé, S.; Frisvad, J.C.; Hubka, V.; Samson, R.A.; Houbraken, J. A review of recently introduced Aspergillus, Penicillium, Talaromyces and other Eurotiales species. Stud. Mycol. 2024, 107, 1–66. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Zhang, Z.K.; Zhuang, W.Y. Species diversity of Penicillium in Southwest China with discovery of forty-three new species. J. Fungi 2023, 9, 1150. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Chen, K.; Xia, Y.W.; Wang, L.; Li, T.H.; Zhuang, W.Y. A new species of Talaromyces (Trichocomaceae) from the Xisha Islands, Hainan, China. Phytotaxa 2016, 267, 187–200. [Google Scholar] [CrossRef]
- Wang, X.C.; Chen, K.; Qin, W.T.; Zhuang, W.Y. Talaromyces heiheensis and T. mangshanicus, two new species from China. Mycol. Prog. 2017, 16, 73–81. [Google Scholar] [CrossRef]
- Wang, X.C.; Chen, K.; Zeng, Z.Q.; Zhuang, W.Y. Phylogeny and morphological analyses of Penicillium section Sclerotiora (Fungi) lead to the discovery of five new species. Sci. Rep. 2017, 7, 8233. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, Z.K.; Wang, X.C.; Zhuang, W.Y.; Cheng, X.H.; Zhao, P. New species of Talaromyces (Fungi) isolated from soil in southwestern China. Biology 2021, 10, 745. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Zhuang, W.Y. New Species of Aspergillus (Aspergillaceae) from tropical islands of China. J. Fungi 2022, 8, 225. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Zhuang, W.Y. New Species of Talaromyces (Trichocomaceae, Eurotiales) from Southwestern China. J. Fungi 2022, 8, 647. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of Penicillium subgenus Penicillium—A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Stud. Mycol. 2004, 49, 1–173. [Google Scholar]
- Houbraken, J.; Wang, L.; Lee, H.B.; Frisvad, J.C. New sections in Penicillium containing novel species producing patulin, pyripyropens or other bioactive compounds. Persoonia 2016, 36, 299–314. [Google Scholar] [CrossRef]
- Houbraken, J.; Visagie, C.M.; Meijer, M.; Frisvad, J.C.; Busby, P.E.; Pitt, J.I.; Seifert, K.A.; Louis-Seize, G.; Demirel, R.; Yilmaz, N.; et al. A taxonomic and phylogenetic revision of Penicillium section Aspergilloides. Stud. Mycol. 2014, 78, 373–451. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.I. The Genus Penicillium and Its Teleomorphic States Eupenicillium and Talaromyces; Academic Press Inc.: London, UK, 1979; p. 634. [Google Scholar]
- Andrade, K.C.R.; Fernandes, R.A.; Pinho, D.B.; de Freitas, M.M.; Filho, E.X.F.; Pessoa, A.; Silva, J.I.; Magalhaes, P.O. Sequencing and characterization of an L-asparaginase gene from a new species of Penicillium section Citrina isolated from Cerrado. Sci. Rep. 2021, 11, 17861. [Google Scholar] [CrossRef]
- Tan, Y.P.; Shivas, R.G. Index of Australian Fungi No. 3; Zenodo: Geneva, Switzerland, 2022; p. 21. [Google Scholar]
- Dutta, M.; Hazra, A.; Bhattacharya, E.; Bose, R.; Mandal Biswas, S. Characterization and metabolomic profiling of two pigment producing fungi from infected fruits of Indian Gooseberry. Arch. Microbiol. 2023, 205, 141. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).