Comparative Genomics of the First Resistant Candida auris Strain Isolated in Mexico: Phylogenomic and Pan-Genomic Analysis and Mutations Associated with Antifungal Resistance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Data and Isolates
2.2. Fungal Growth Conditions
2.3. DNA Extraction and Genome Sequencing
2.4. Thermotolerance and Halotolerance
2.5. Genome Assembly and Annotation
2.6. Phylogenomic Tree and Pan-Genome of Candida auris
2.7. Phylogenetic and Comparative Analysis of the Erg11 and Fks1 Proteins
2.8. Modeling the Erg11 Protein from Candida auris 20–1498
2.9. Molecular-Docking Study of Some Azoles on the Erg11 Protein
3. Results
3.1. C. auris 20–1498 Genome Assembly and Annotation
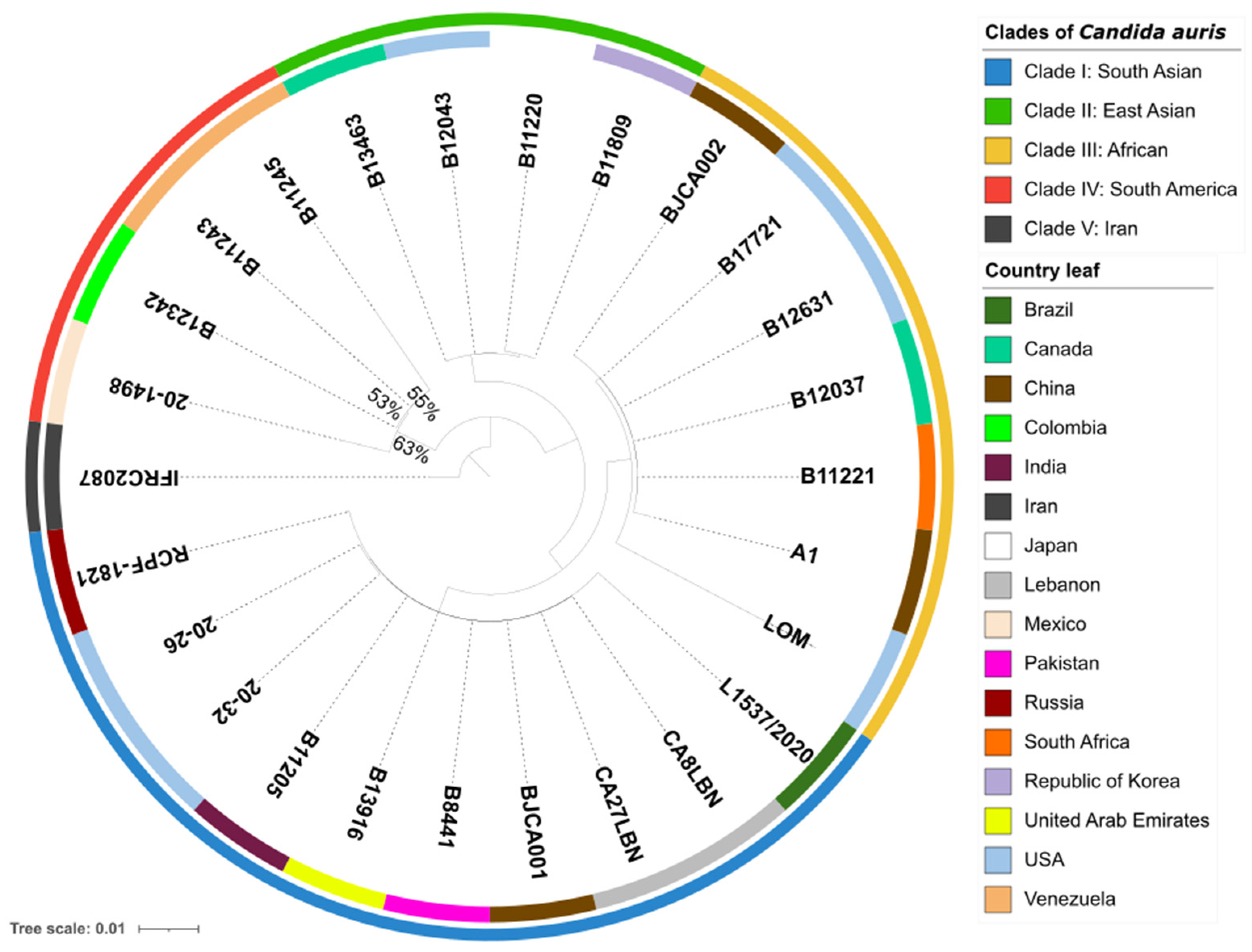
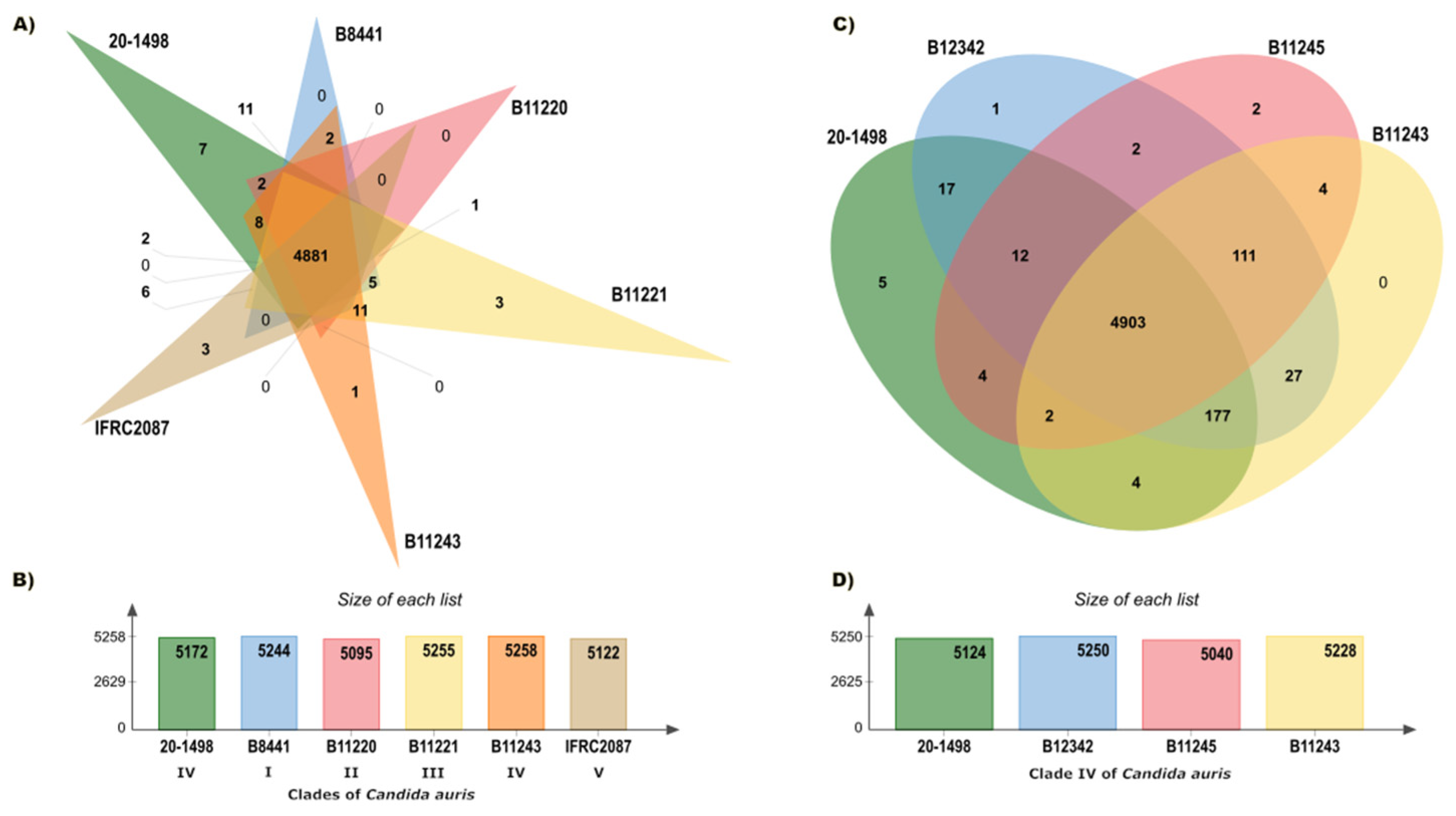
3.2. Phylogenetic Tree and Pan-Genome of Candida auris
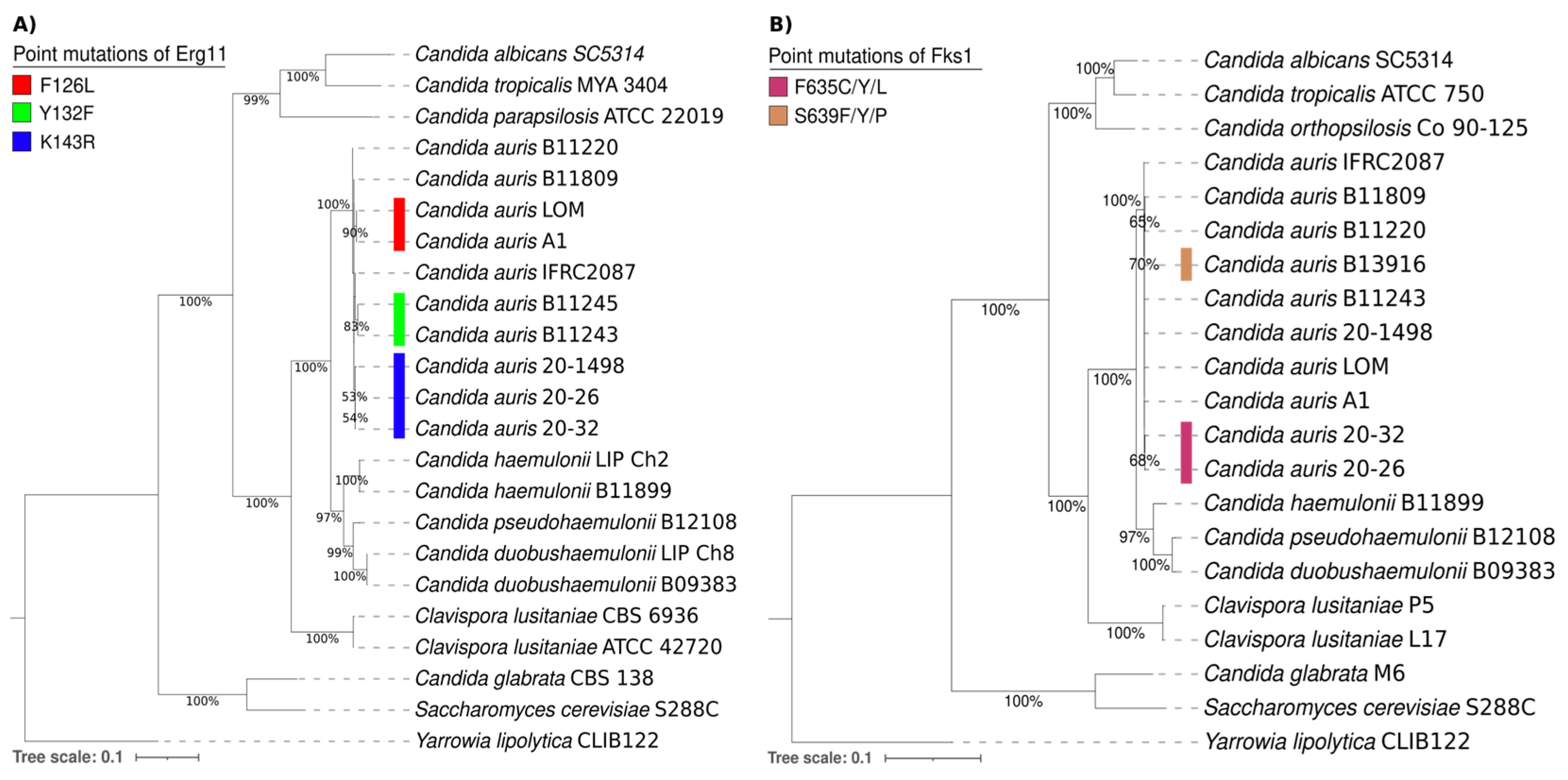
3.3. Phylogenetic and Comparative Analysis of Erg11 and Fks1 Proteins
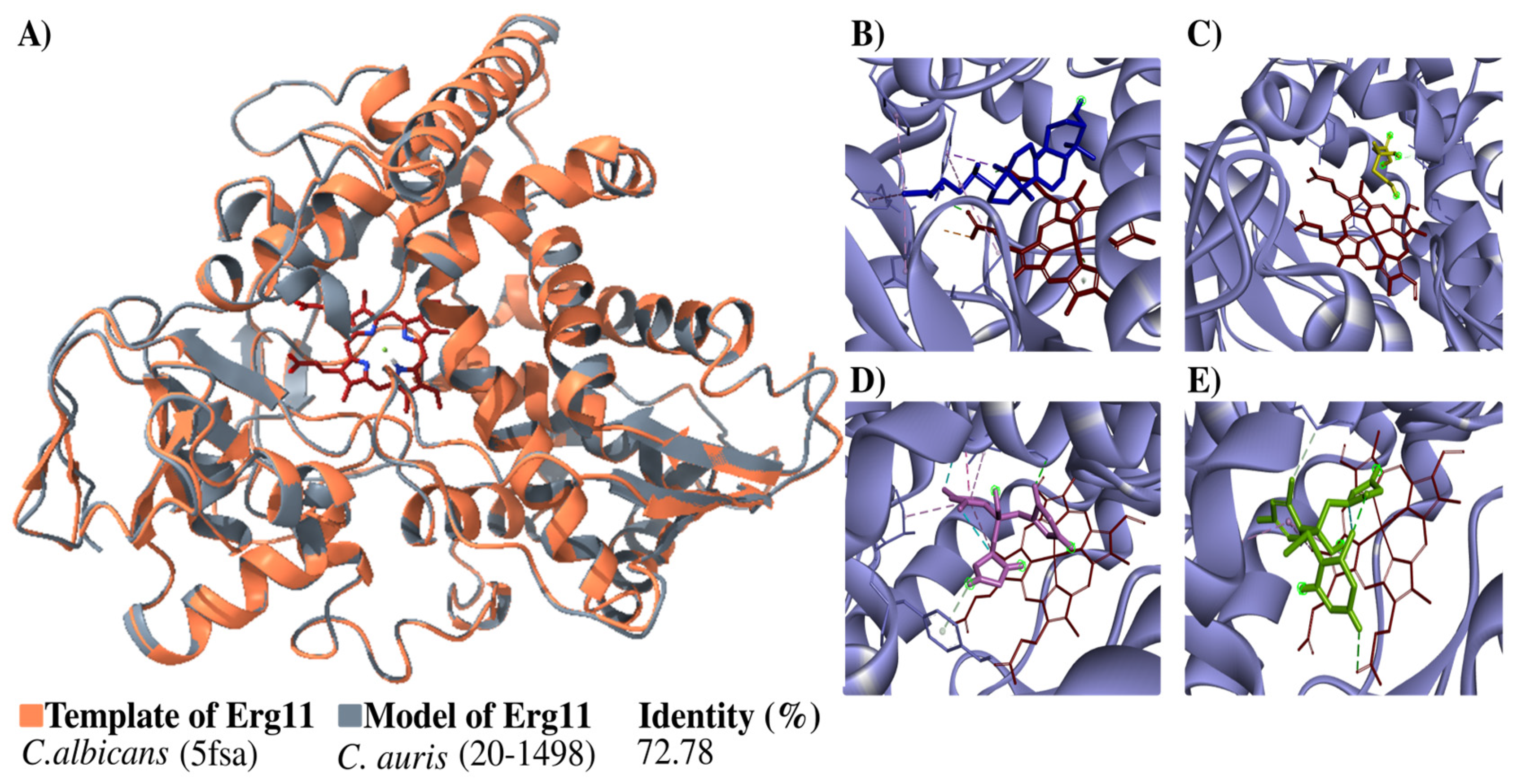
3.4. Modeling the Candida auris 20–1498 Erg11 Protein, and Its Use for the Molecular Docking of Some Azoles
3.5. Phenotypic Characteristics: Thermotolerance and Halotolerance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, K.; Woodworth, K.; Walters, M.; Berkow, E.L.; Jackson, B.; Chiller, T.; Vallabhaneni, S. Candida auris: The recent emergence of a multidrug-resistant fungal pathogen. Med. Mycol. 2019, 57, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chow, N.A.; Muñoz, J.F.; Gade, L.; Berkow, E.L.; Li, X.; Welsh, R.M.; Forsberg, K.; Lockhart, S.R.; Adam, R.; Alanio, A.; et al. Tracing the Evolutionary History and Global Expansion of Candida auris Using Population Genomic Analyses. mBio 2020, 11, e03364-19. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Chow, N.A.; de Groot, T.; Badali, H.; Abastabar, M.; Chiller, T.M.; Meis, J.F. Potential Fifth Clade of Candida auris, Iran, 2018. Emerg. Infect. Dis. 2019, 25, 1780–1781. [Google Scholar] [CrossRef] [PubMed]
- Spruijtenburg, B.; Badali, H.; Abastabar, M.; Mirhendi, H.; Khodavaisy, S.; Sharifisooraki, J.; Taghizadeh Armaki, M.; de Groot, T.; Meis, J.F. Confirmation of fifth Candida auris clade by whole genome sequencing. Emerg. Microbes Infect. 2022, 11, 2405–2411. [Google Scholar] [CrossRef] [PubMed]
- Suphavilai, C.; Ko, K.K.K.; Lim, K.M.; Tan, M.G.; Boonsimma, P.; Keat Chu, J.J.; Goh, S.S.; Rajandran, P.; Nagarajan, N. Discovery of the Sixth Candida auris Clade in Singapore.medRxiv. medRxiv. 2023. [Google Scholar] [CrossRef]
- Muñoz, J.F.; Welsh, R.M.; Shea, T.; Batra, D.; Gade, L.; Howard, D.; Rowe, L.A.; Meis, J.F.; Litvintseva, A.P.; Cuomo, C.A. Clade-specific chromosomal rearrangements and loss of subtelomeric adhesins in Candida auris. Genetics 2021, 218, iyab029. [Google Scholar] [CrossRef]
- Chowdhary, A.; Prakash, A.; Sharma, C.; Kordalewska, M.; Kumar, A.; Sarma, S.; Tarai, B.; Singh, A.; Upadhyaya, G.; Upadhyay, S.; et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: Role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 2018, 73, 891–899. [Google Scholar] [CrossRef]
- Ayala-Gaytán, J.J.; Montoya, A.M.; Martínez-Resendez, M.F.; Guajardo-Lara, C.E.; de JTreviño-Rangel, R.; Salazar-Cavazos, L.; Llaca-Díaz, J.M.; González, G.M. First case of Candida auris isolated from the bloodstream of a Mexican patient with serious gastrointestinal complications from severe endometriosis. Infection 2021, 49, 523–525. [Google Scholar] [CrossRef]
- Reséndiz-Sánchez, J.; Ortiz-Álvarez, J.; Casimiro-Ramos, A.; Hernández-Rodríguez, C.; Villa-Tanaca, L. First report of a catheter-related bloodstream infection by Candida haemulonii in a children’s hospital in Mexico City. Int. J. Infect. Dis. 2020, 92, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Steinbiss, S.; Silva-Franco, F.; Brunk, B.; Foth, B.; Hertz-Fowler, C.; Berriman, M.; Otto, T.D. Companion: A web server for annotation and analysis of parasite genomes. Nucleic Acids Res. 2016, 44, W29–W34. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Sun, J.; Lu, F.; Luo, Y.; Bie, L.; Xu, L.; Wang, Y. OrthoVenn3: An integrated platform for exploring and visualizing orthologous data across genomes. Nucleic Acids Res. 2023, 51, W397–W403. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Hargrove, T.Y.; Friggeri, L.; Wawrzak, Z.; Qi, A.; Hoekstra, W.J.; Schotzinger, R.J.; York, J.D.; Guengerich, F.P.; Lepesheva, G.I. Structural analyses of Candida albicans sterol 14α-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis. J. Biol. Chem. 2017, 292, 6728–6743. [Google Scholar] [CrossRef] [PubMed]
- Eswar, N.; Webb, B.; Marti-Renom, M.A.; Madhusudhan, M.S.; Eramian, D.; Shen, M.Y.; Pieper, U.; Sali, A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinform. 2006, 54, 5–6. [Google Scholar] [CrossRef]
- Gómez-García, O.; Andrade-Pavón, D.; Campos-Aldrete, E.; Ballinas-Indilí, R.; Méndez-Tenorio, A.; Villa-Tanaca, L.; Álvarez-Toledano, C. Synthesis, Molecular Docking, and Antimycotic Evaluation of Some 3-Acyl Imidazo [1,2-a] pyrimidines. Molecules 2018, 23, 599. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Rodrigues, L.S.; Gazara, R.K.; Passarelli-Araujo, H.; Valengo, A.E.; Pontes PV, M.; Nunes-da-Fonseca, R.; de Souza, R.F.; Venancio, T.M.; Dalla-Costa, L.M. First Genome Sequences of Two Multidrug-Resistant Candida haemulonii var. vulnera Isolates From Pediatric Patients With Candidemia. Front. Microbiol. 2020, 11, 1535. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, Y.; Hu, W.; Chen, F.; Zhao, J.; Chen, X.; Han, L. Application of Machine Learning Classifier to Candida auris Drug Resistance Analysis. Front. Cell. Infect. Microbiol. 2021, 11, 742062. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, S.E.; Jacobs, J.L.; Dennis, E.K.; Taimur, S.; Rana, M.; Patel, D.; Gitman, M.; Patel, G.; Schaefer, S.; Iyer, K.; et al. Candida auris Pan-Drug-Resistant to Four Classes of Antifungal Agents. Antimicrob. Agents Chemother. 2022, 66, e0005322. [Google Scholar] [CrossRef] [PubMed]
- Kiyohara, M.; Miyazaki, T.; Okamoto, M.; Hirayama, T.; Makimura, K.; Chibana, H.; Nakada, N.; Ito, Y.; Sumiyoshi, M.; Ashizawa, N.; et al. Evaluation of a Novel FKS1 R1354H Mutation Associated with Caspofungin Resistance in Candida auris Using the CRISPR-Cas9 System. J. Fungi 2023, 9, 529. [Google Scholar] [CrossRef]
- Reslan, L.; Araj, G.F.; Finianos, M.; El Asmar, R.; Hrabak, J.; Dbaibo, G.; Bitar, I. Molecular Characterization of Candida auris Isolates at a Major Tertiary Care Center in Lebanon. Front. Microbiol. 2022, 12, 770635. [Google Scholar] [CrossRef]
- Reyes-Montes MD, R.; Duarte-Escalante, E.; Martínez-Herrera, E.; Acosta-Altamirano, G.; Frías-De León, M.G. Current status of the etiology of candidiasis in Mexico. Rev. Iberoam. Micol. 2017, 34, 203–210. [Google Scholar] [CrossRef]
- Muñoz, J.F.; Gade, L.; Chow, N.A.; Loparev, V.N.; Juieng, P.; Berkow, E.L.; Farrer, R.A.; Litvintseva, A.P.; Cuomo, C.A. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat. Commun. 2018, 9, 5346. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Antifungal Susceptibility Testing and Interpretation|Candida auris|Fungal Diseases|CDC. 2023. Available online: https://www.cdc.gov/candida-auris/hcp/laboratories/antifungal-susceptibility-testing.html (accessed on 28 May 2024).
- Healey, K.R.; Kordalewska, M.; Jiménez Ortigosa, C.; Singh, A.; Berrío, I.; Chowdhary, A.; Perlin, D.S. Limited ERG11 Mutations Identified in Isolates of Candida auris Directly Contribute to Reduced Azole Susceptibility. Antimicrob. Agents Chemother. 2018, 62, e01427-18. [Google Scholar] [CrossRef] [PubMed]
- Carolus, H.; Pierson, S.; Muñoz, J.F.; Subotić, A.; Cruz, R.B.; Cuomo, C.A.; Van Dijck, P. Genome-Wide Analysis of Experimentally Evolved Candida auris Reveals Multiple Novel Mechanisms of Multidrug Resistance. mBio 2021, 12, e03333-20. [Google Scholar] [CrossRef]
- Li, J.; Aubry, L.; Brandalise, D.; Coste, A.T.; Sanglard, D.; Lamoth, F. Upc2-mediated mechanisms of azole resistance in Candida auris. Microbiol. Spectr. 2024, 12, e0352623. [Google Scholar] [CrossRef] [PubMed]
- Akinbobola, A.B.; Kean, R.; Hanifi, S.M.A.; Quilliam, R.S. Environmental reservoirs of the drug-resistant pathogenic yeast Candida auris. PLoS Pathog. 2023, 19, e1011268. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.R.; Chow, N.; Forsberg, K.; Litvintseva, A.P.; Lockhart, S.R.; Welsh, R.; Vallabhaneni, S.; Chiller, T. On the Origins of a Species: What Might Explain the Rise of Candida auris? J. Fungi 2019, 5, 58. [Google Scholar] [CrossRef]
- Villanueva-Lozano, H.; Treviño-Rangel, R.J.; González, G.M.; Ramírez-Elizondo, M.T.; Lara-Medrano, R.; AlemanBocanegra, M.C.; Guajardo-Lara, C.E.; Gaona-Chávez, N.; Castilleja-Leal, F.; Torre-Amione, G.; et al. Outbreak of Candida auris infection in a COVID-19 hospital in Mexico. Clin. Microbiol. Infect. 2021, 27, 813–816. [Google Scholar] [CrossRef]

| Features | Values |
|---|---|
| Quality of reads | QC 1 > 30 |
| Genome assembly size (bp) | 12,869,713 |
| Number of contigs | 70 |
| N50 (bp) | 1,604,526 |
| Genome coverage | 99.68 |
| GC content (%) | 45.5 |
| Number of predicted genes | 5432 |
| Number of coding genes | 5304 |
| Number of genes with multiple CDSs | 565 |
| Number of hypothetical proteins | 5515 |
| Molecules | Binding Energy (kcal/mol) | Interacting Residues | Interactions |
|---|---|---|---|
| Lanosterol | K143; R143 WT; M −12; −10.4 | Val A: 304, HEM A: 525, Gly A: 307, Leu A: 376, Met A: 504, Pro A: 230, Leu A: 121, Phe A: 380, His A: 377, Phe A: 233, Ser A: 378, Tyr A: 118, Thr A: 122, Ile A: 131, Gly A: 303, Phe A: 126, Leu A: 300 | Van Der Waals Pi-sigma Alkyl Pi-alkyl |
| Mevalonate | −5–5; −4.4 | Gly A: 308, Phe A: 126, Tyr A: 132, Ile A: 131, Val A: 304, HEM A: 525, Leu A: 300, Gly A: 303, Gly A: 307 | Van Der Waals Carbon hydrogen bond |
| Fluconazole | −8.8; −8.6 | Leu A: 300, Val A: 304, Gly A: 303, Ile A: 131, Tyr A: 132, HEM A: 525, Phe A: 228, Tyr A: 118, Leu A: 376, Pro A: 375, Thr A: 311, His A: 310, Gly A: 307, Phe A: 126 | Van Der Waals Conventional hydrogen bond Halogen (fluorine) Pi-donor hydrogen bond Amide-pi stacked Pi-alkyl |
| Voriconazole | −9.5; −7.3 | Leu A: 376, Ile A: 379, Arg A: 381, Thr A: 311, Gly A: 308, Ile A: 131, Leu A: 300, Val A: 304, Gly A: 303, Phe A: 126, Thr A:122, Gly A: 307, Tyr A: 132, HEM A: 525 | Van Der Waals Conventional hydrogen bond Carbon hydrogen bond Alkyl Pi-alkyl |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casimiro-Ramos, A.; Bautista-Crescencio, C.; Vidal-Montiel, A.; González, G.M.; Hernández-García, J.A.; Hernández-Rodríguez, C.; Villa-Tanaca, L. Comparative Genomics of the First Resistant Candida auris Strain Isolated in Mexico: Phylogenomic and Pan-Genomic Analysis and Mutations Associated with Antifungal Resistance. J. Fungi 2024, 10, 392. https://doi.org/10.3390/jof10060392
Casimiro-Ramos A, Bautista-Crescencio C, Vidal-Montiel A, González GM, Hernández-García JA, Hernández-Rodríguez C, Villa-Tanaca L. Comparative Genomics of the First Resistant Candida auris Strain Isolated in Mexico: Phylogenomic and Pan-Genomic Analysis and Mutations Associated with Antifungal Resistance. Journal of Fungi. 2024; 10(6):392. https://doi.org/10.3390/jof10060392
Chicago/Turabian StyleCasimiro-Ramos, Arturo, Celia Bautista-Crescencio, Alvaro Vidal-Montiel, Gloria M. González, Juan Alfredo Hernández-García, César Hernández-Rodríguez, and Lourdes Villa-Tanaca. 2024. "Comparative Genomics of the First Resistant Candida auris Strain Isolated in Mexico: Phylogenomic and Pan-Genomic Analysis and Mutations Associated with Antifungal Resistance" Journal of Fungi 10, no. 6: 392. https://doi.org/10.3390/jof10060392
APA StyleCasimiro-Ramos, A., Bautista-Crescencio, C., Vidal-Montiel, A., González, G. M., Hernández-García, J. A., Hernández-Rodríguez, C., & Villa-Tanaca, L. (2024). Comparative Genomics of the First Resistant Candida auris Strain Isolated in Mexico: Phylogenomic and Pan-Genomic Analysis and Mutations Associated with Antifungal Resistance. Journal of Fungi, 10(6), 392. https://doi.org/10.3390/jof10060392






